Abstract
The aim of this study was to detect the epidermal growth factor receptor (EGFR) gene type at pre- and postchemotherapy to evaluate the impact of platinum-based chemotherapy on EGFR gene mutations and provide a theoretical foundation for clinical treatment.
Around 40 serum DNA samples were collected from advanced nonsmall cell lung cancer patients who received platinum-based chemotherapy as first-line treatment in our hospital from August 1, 2014 to June 1, 2015. The EGFR gene exons 19 and 21 were amplified by polymerase chain reaction (PCR) and detected by direct sequencing. The outcomes were analyzed with SPSS 17.0.
Of 40 patients, 38 were included in the analysis. An EGFR gene mutation was detected in 17 cases (44.7%) at prechemotherapy compared with 19 cases (50.0%) at postchemotherapy. The EGFR gene mutation differences were not statistically significantly (P = .165) during pre- and postchemotherapy. The EGFR gene type was consistent in 26 cases (68.4%). Among the 12 discordant cases, 5 cases changed from mutant type to wild type, while 7 cases changed from wild type to mutant type. EGFR mutation positive patients had a disease control rate (DCR) of 88.2% (15/17), whereas it was only 57.1% in EGFR mutation negative patients, which was statistically significant (P = 0.01) indicating a better curative effect in EGFR mutation positive patients.
Platinum-based chemotherapy may change the serum EGFR gene type in advanced lung adenocarcinoma.
Keywords: chemotherapy, EGFR, lung adenocarcinoma, platinum
1. Introduction
Lung cancer is one of the most common malignant tumors in the world. According to the World Health Organization (WHO) released data of 2012, 1.8 million new cases of lung cancer occurred in the world and 1.59 million people died of lung cancer.[1] Nonsmall cell lung cancer (NSCLC) accounts for about 85% of all lung cancers.[2] National Comprehensive Cancer Network (NCCN) guidelines recommend epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) in EGFR mutation positive patients with advanced metastasis as first-line treatment. The patients with negative mutations should receive normal chemotherapy. The best second-line treatment for EGFR-mutation negative patients is still controversial. In clinical studies, it was found that EGFR-TKIs have a significant curative effect for some EGFR mutation negative patients as second-line treatment. In addition, the EGFR-TKI clinical remission rate was not the same for EGFR mutation positive patients used as a first-line treatment (70%) or second-line treatment (30–40%).[3–6] Therefore, we speculated that EGFR gene mutations might be affected by chemotherapy. However, tissue samples were not easy to obtain before and after chemotherapy. Fortunately, studies[7] have shown that during the formation and development of solid tumors, some tumor cells enter the blood circulation frequently and these cancer cells reach an apotheosis in the blood circulation and release genetic material into the blood. Several large-scale studies[8,9] have found that compared with tissue, serum DNA test results of EGFR gene mutation status had a consistency of 79.7% to 94% in NSCLC patients. These studies suggested that serum DNA can be used in EGFR gene mutation detection. In our study, we monitored EGFR gene mutation status using serum DNA before and after chemotherapy to provide a theoretical basis for clinical chemotherapy and targeted therapy.
2. Materials and methods
2.1. Patient population
Around 40 advanced lung adenocarcinoma patients were identified who were treated in the First Affiliated Hospital of Shihezi University from August 2014 to June 2015. Patients who met the following inclusion criteria were included in the study: (1) diagnosed with lung adenocarcinoma stage IIIB to IV by histology or cytological assessment and EGFR gene mutations detected by histopathological examination (amplification refractory mutation system, ARMs); (2) age ≥20 years; (3) Eastern Cooperative Oncology Group (ECOG) score 0 to 2 with measurable lesions; (4) normal viscera function; and (5) accepted cisplatin-based chemotherapy voluntarily and was able to comply with the research protocol. The study protocol was approved by The First Affiliated Hospital of Shihezi University. Written informed consents were obtained from all the patients enrolled in this study.
2.2. Methods
All patients were administered platinum-based chemotherapy, cisplatin 75 mg/m2, intravenous glucose tolerance test (IVGTT) for 21 days/cycle; nedaplatin 80 mg/m2, IVGTT for 21 days/cycle; and carboplatin (accounted by the area under the curve, AUC), IVGTT for 21 days/cycle. All patients were given hydration therapy to reduce adverse events the day before chemotherapy and antinausea therapy was given before injections. A chest computed tomography scan was checked after every 2 cycles. Curative effects were evaluated using response evaluation criteria in solid tumors (RECIST, version 1.1).
2.3. Experimental procedures
About 5 ml each of venous blood samples were collected from each patient during 2 days prior to chemotherapy, after 2-cycles of chemotherapy, and after 4-cycles of chemotherapy. The samples were centrifuged for 10 minutes to separate the serum. DNA samples were extracted with a QIAampBloodMinKit according to the manufacturer's instructions. The DNA quantification assessment at A260/A280 was 1.6 to 2.2. The DNA concentration was 2.4 to 27.6 ng/μL. We obtained the EGFR gene sequence from the GenBank database, and designed EGFR exon 19 and 21 primers after a NCBIBLAST search. Primers were synthesized by Shanghai Sangon Biological Company, China. Primer sequences are shown in Table 1. Gene sequencing was carried out with 5 μL of the PCR product, which was detected by 2% agarose gel electrophoresis. Finally, sequencing was performed by the Beijing BGI Company, China. The detected EGFR gene mutations are shown in Figures 1–5.
Table 1.
Primer sequences used for PCR of exons 19 and 21 for the EGFR gene.
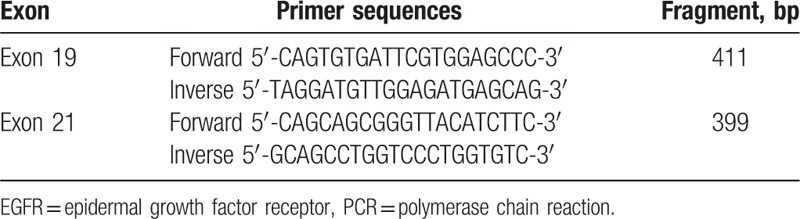
Figure 1.
EGFR exon 19 wild type. EGFR = epidermal growth factor receptor.
Figure 5.
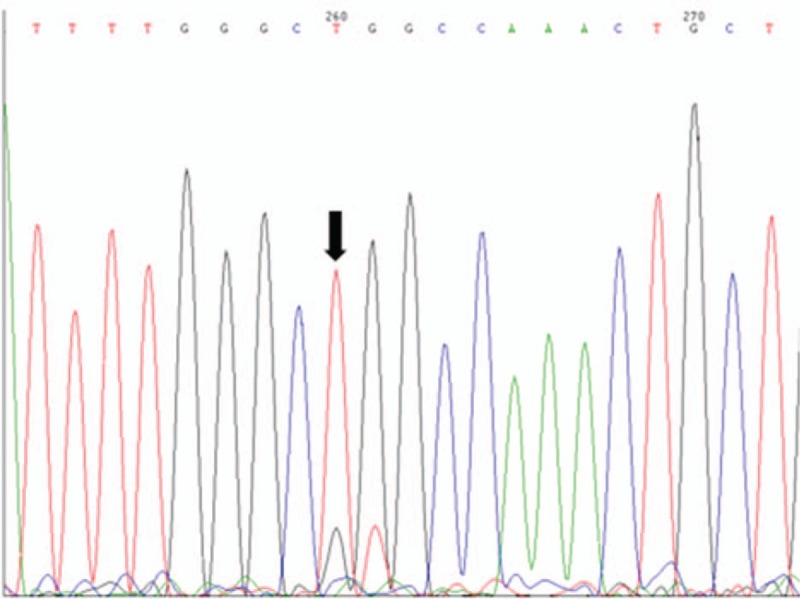
EGFR exon 21 mutant type. EGFR = epidermal growth factor receptor.
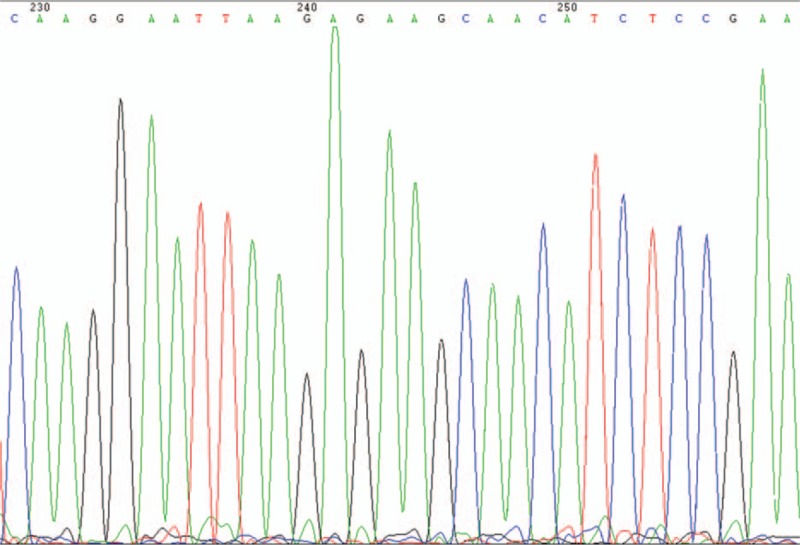
Figure 2.
EGFR exon 19 mutant type. EGFR = epidermal growth factor receptor.
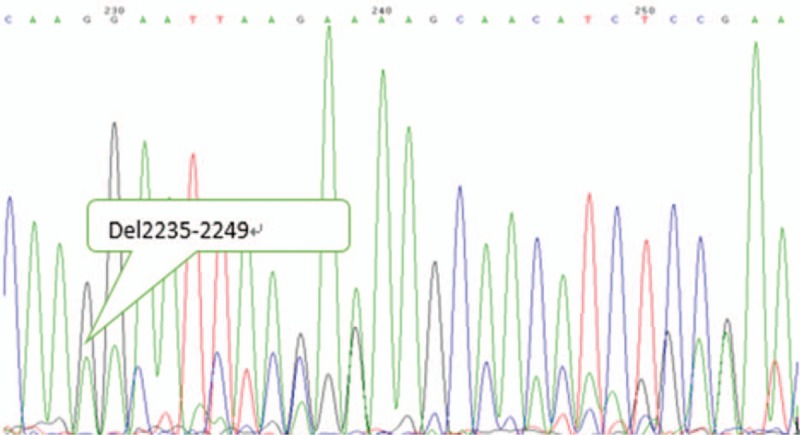
Figure 3.
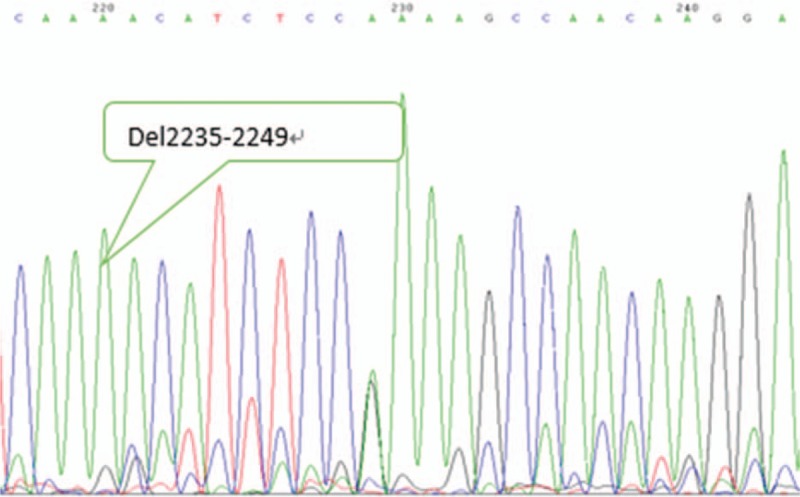
EGFR exon 19 mutant type. EGFR = epidermal growth factor receptor.
Figure 4.
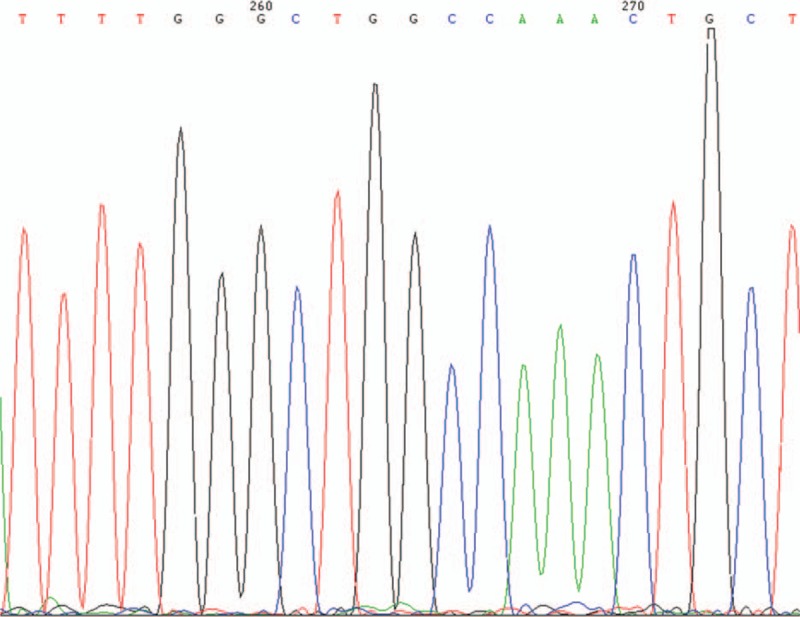
EGFR exon 21 wild type. EGFR = epidermal growth factor receptor.
2.4. Statistical analysis
SPSS 17.0 software was used for statistical analysis. Using Kappa score analysis, the EGFR gene consistency between tissue and serum was assessed. Using Fisher's analysis, the differences in EGFR mutations before and after chemotherapy were determined and the relationship between EGFR gene mutations and clinical curative effects were assessed with single factor logistic regression analysis (significance level α=0.05).
3. Results
3.1. Clinical features and evaluation of effects
Among 40 patients, 38 patients were successfully analyzed, 1 was lost to follow-up, and 1 had failed DNA extraction. Of 38 patients, 25 were males and 13 were females. Ages were between 47 and 74 years, and the median age was 60.2 years. There were patients with a history of smoking and 18 cases of nonsmokers. All patients accepted platinum-based chemotherapy. Evaluation of effects was carried out during every second cycle. A partial response (PR) was found in 6 cases, stable disease (SD) was found in 21 cases, and progressive disease (PD) was observed in 11 cases.
3.2. EGFR gene state before chemotherapy in the tissues
Before chemotherapy, the tissue EGFR gene mutation rate was 44.7% (17/38). There were 6 cases of mutations in exon 19 and the mutation rate was 15.8% (6/38). There were 11 cases of mutations in exon 21 and the mutation rate was 29.0% (11/38). For males, the mutation rate was 36.0% (9/25) and in females it was 61.5 (8/13). The mutation rate in female was higher than in males, but the difference was not statistically significant (F = 2.189, P = .139). The EGFR gene mutation rate in smoking patients was 20.0% (4/20) and, in contrast, the rate for nonsmoking patients was 72.2% (13/18), which was significantly different (F = 7.01, P = .008).
3.3. Serum and tissue EGFR gene status before chemotherapy
Before chemotherapy, tissue EGFR gene mutations were detected in 17 patients (44.74%) and serum mutations were found in 13 patients (34.3%). The results from a Kappa test showed a false positive rate of 0%, a false negative rate of 16%, and the specific degree of 100%. The sensitivity detected was 84% and the Kappa value was 0.782 (Kappa >0.75). In conclusion, serum and tissue EGFR gene testing were showing consistent results.
3.4. Serum EGFR gene status after chemotherapy
After chemotherapy, the serum EGFR gene mutation rate was 50% (19/38). Twelve cases were all positive before and after chemotherapy and 14 cases were negative. Five patients changed from positive to negative and 7 patients changed from negative to positive. However, before and after chemotherapy, the EGFR mutation differences were not statistically significantly in general (F = 3.60, P = .165; Table 2).
Table 2.
The EGFR gene type of 38 patients prechemotherapy and postchemotherapy in serum.

3.5. EGFR mutations state the relationship with the effect of chemotherapy
All 38 patients accepted platinum-based chemotherapy. Complete response (CR) was observed in 0 cases, PR in 6 (15.8%), SD in 21 (55.3%), PD in 11 (28.9%), and disease control rate (DCR) in 27 (71.1%). EGFR mutation positive patients had a DCR of 88.2% (15/17). In contrast, the DCR of EGFR mutation negative patients was 57.1% (12/21), which was significantly different (P = .01). The platinum-based chemotherapy in EGFR mutation positive patients had a curative effect that was better than that of patients with a negative status.
3.6. Relationship between the curative effect and EGFR gene changes
Chemotherapy effects and EGFR gene status changes were assessed with single factor logistic regression analysis. The regression coefficient was 0.465 (P = .227, OR = 1.592 [0.748–3.387]). The chemotherapy effect showed no significant difference from the EGFR gene changes (Table 3).
Table 3.
The relationship between chemotherapy and the changes in EGFR gene status.

4. Discussion
This study analyzed the effect of platinum-based chemotherapy on EGFR mutations in patients with advanced NSCLC. The results showed that chemotherapy may increase the EGFR mutation probability (44.7% vs 50%) in patients. The causes of EGFR gene mutations were quite high. Chin et al,[10] have found that NSCLC cells with EGFR exon 19 mutations had shown relatively negative sensitivity to EGFR-TKIs during platinum resistance. This study showed that the EGFR gene of patients during platinum chemotherapy was not highly stable. Additionally, Gow et al[11] have reported that the EGFR gene status of some NSCLC patients was not the same between the primary tumor and metastatic tumors. These results suggested that the EGFR gene status of tumors could change during the process of tumor development. This conclusion was strongly supported by Jiang's study,[12] which had reported one positive case of a serum EGFR gene mutation that was negative with tissue gene mutation, out of 16 cases. Clinically, most patients on advanced metastasis only have EGFR gene detection before first-line treatment and just for a single part. This may be the reason why some EGFR-TKI treatment efficacy is not consistent for different EGFR gene types.
Currently, clinical EGFR gene testing mainly uses tissue for pathology inspection to determine a diagnosis. However, using tissue has many disadvantages clinically, such as discomfort for the patient, high risk, and patient unwillingness to accept. This study suggests that serum and tissue EGFR gene mutation assessment has good consistency. Moreover, the serum sampling method has many advantages,[13] such as less trauma, low risk, and ease in obtaining the specimen. These advantages make the detection of EGFR gene mutations more acceptable and this will have a large effect on the evaluation and outcome of treatment.
In this study, we also showed that the experimental results inevitably had false negatives. The main reason was the serum DNA content is low and is affected by patient individual differences. The process of extraction requires sophisticated technology. Therefore, this method may not replace the use of tissue samples.[14,15] If an easier experimental method with a higher sensitivity can be developed, serum EGFR gene testing can be implemented in clinical pathological diagnosis.
The results of this study suggested that platinum-based chemotherapy may affect EGFR genes in patients with advanced NSCLC. There were few limitations in our study due to the limited research funding and methodology. First, small sample sizes inevitably led to statistical error; multicenter and large-scale studies are needed to confirm our findings. Second, due to the limited research funding and methodology, amplifying all the exons by PCR was too difficult and unrealistic. Moreover, we observed that the mutation sensitivity of exons 19 and 21 were the highest, and therefore, we chose these 2 exons for our study. Finally, the results would be more convincing if a tissue sample after chemotherapy could have been obtained, as comparative evaluation might have derived more convincing results. Although this study had varied results, improvements were still demonstrated. If further studies confirm our findings, then the negative EGFR gene patients are expected to benefit from the TKI treatment after chemotherapy and additional EGFR gene detection after chemotherapy will be necessary.
Footnotes
Abbreviations: ARMS = amplification refractory mutation system, AUC = area under the curve, CR = complete response, DCR = disease control rate, ECOG = Eastern Cooperative Oncology Group, EGFR = epidermal growth factor receptor, IVGTT = Intravenous Glucose Tolerance Test, NCCN = National Comprehensive Cancer Network, NSCLC = nonsmall cell lung cancer, NSFC = National Natural Science Foundation of China, PCR = polymerase chain reaction, PD = partial disease, PR = partial response, RECIST = response evaluation criteria in solid tumors, SD = stable disease, TKI = tyrosine kinase inhibitor, WHO = World Health Organization.
YW and XM contributed equally to this work.
Funding: National Natural Science Foundation of China (NSFC) (81560381)
Author contributions: YW and XM carried out the experimental studies, participated in data collection, and drafted the manuscript. YW helped in collection of data. DM performed the statistical analysis of the data and helped in study design. PG helped in drafting the manuscript. All authors read and approved the final manuscript.
The authors have no conflicts of interest to disclose.
References
- [1].Soerjomataram I, Lortet-Tieulent J, Parkin DM, et al. Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet 2012;380:1840–50. [DOI] [PubMed] [Google Scholar]
- [2].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [3].Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958–67. [DOI] [PubMed] [Google Scholar]
- [4].Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–57. [DOI] [PubMed] [Google Scholar]
- [5].Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med 2005;353:133–44. [DOI] [PubMed] [Google Scholar]
- [6].Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123–32. [DOI] [PubMed] [Google Scholar]
- [7].Nie K, Jia Y, Zhang X. Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol 2015;36:7–19. [DOI] [PubMed] [Google Scholar]
- [8].Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chin TM, Quinlan MP, Singh A, et al. Reduced Erlotinib sensitivity of epidermal growth factor receptor-mutant non-small cell lung cancer following cisplatin exposure: a cell culture model of second-line erlotinib treatment. Clin Cancer Res 2008;14:6867–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009;27:2653–9. [DOI] [PubMed] [Google Scholar]
- [11].Gow CH, Chang YL, Hsu YC, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol 2009;20:696–702. [DOI] [PubMed] [Google Scholar]
- [12].Jiang B, Li J, Gong P. EGFR gene mutation detection with circulating DNA and the targeting therapy response in advanced NSCLC patients. Chin J Cancer Prev Treat 2014;21:29–33. [Google Scholar]
- [13].Brevet M, Johnson ML, Azzoli CG, et al. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer 2011;73:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li Z, Zhang Y, Bao W, et al. Insufficiency of peripheral blood as a substitute tissue for detecting EGFR mutations in lung cancer: a meta-analysis. Target Oncol 2014;9:381–8. [DOI] [PubMed] [Google Scholar]
- [15].Mazurek A, Pierzyna M, Giglok M, et al. Quantification of concentration and assessment of EGFR mutation in circulating DNA. Cancer Biomark 2015;15:515–24. [DOI] [PubMed] [Google Scholar]


