Abstract
The enzyme acetyl-CoA carboxylase (ACC) plays a crucial role in fatty acid metabolism. In recent years, ACC has been recognized as a promising drug target for treating different diseases. However, the role of ACC in vascular endothelial cells (ECs) has been neglected so far. To characterize the role of ACC, we used the ACC inhibitor, soraphen A, as a chemical tool, and also a gene silencing approach. We found that ACC1 was the predominant isoform in human umbilical vein ECs as well as in human microvascular ECs and that soraphen A reduced the levels of malonyl-CoA. We revealed that ACC inhibition shifted the lipid composition of EC membranes. Accordingly, membrane fluidity, filopodia formation, and migratory capacity were reduced. The antimigratory action of soraphen A depended on an increase in the cellular proportion of PUFAs and, most importantly, on a decreased level of phosphatidylglycerol. Our study provides a causal link between ACC, membrane lipid composition, and cell migration in ECs. Soraphen A represents a useful chemical tool to investigate the role of fatty acid metabolism in ECs and ACC inhibition offers a new and valuable therapeutic perspective for the treatment of EC migration-related diseases.
Keywords: membranes/fluidity, lipidomics, fatty acid/biosynthesis, phospholipids/phosphatidylglycerol, vascular biology/endothelial cells, soraphen, filopodia
The multi-domain enzyme acetyl-CoA carboxylase (ACC) is crucially involved in the fatty acid metabolism of eukaryotes. In the recent years, ACC has been increasingly recognized as a promising drug target for the treatment of different diseases (1, 2): i) The persistent increase in the incidence of the metabolic syndrome has aroused renewed interest in ACC after its discovery about 60 years ago (1, 3). ACC inhibition or knockdown promises beneficial effects to control lipid synthesis in an effort to treat metabolic diseases like obesity and diabetes (4–9). ii) Several studies reported that ACC is upregulated in different human cancers. Because this phenomenon is associated with increased lipogenesis and rapid cancer cell growth, the inhibition of ACC has been suggested as a promising therapeutic strategy (10–13). iii) Furthermore, both the pharmacological inhibition and the deletion of ACC1 impaired TH17 cell-mediated autoimmune disease in mice (14).
ACC catalyzes the carboxylation of acetyl-CoA into malonyl-CoA. This reaction is the first and rate-limiting step in the biosynthesis of fatty acids in all organisms (7, 15, 16). Malonyl-CoA is an essential substrate for fatty acid synthesis and chain elongation and functions as a strong inhibitor of mitochondrial fatty acid β-oxidation. There are two tissue-specific isoforms of ACC in mammalian cells: ACC1, present in lipogenic tissue (liver, adipose), and ACC2, present in oxidative tissues (liver, heart, skeletal muscle) (1). Cytosolic ACC1 generates malonyl-CoA for de novo lipogenesis, while mitochondrial ACC2 generates malonyl-CoA that acts as an inhibitor of carnitine palmitoyltransferase. This enzyme transfers fatty acids into the mitochondria for β-oxidation to acetyl-CoA (16, 17).
Despite the great interest in ACC as pharmacological drug target, no attention has as yet been given to the role of ACC in vascular endothelial cells (ECs). A monolayer of ECs forms the inner lining of each blood vessel and provides a barrier that tightly controls the passage of fluid, solutes, macromolecules, and cells from the blood to the underlying tissue (18, 19). Besides its function as a dynamic barrier, the vascular endothelium can also be regarded as a multifunctional paracrine and endocrine organ within the cardiovascular system (19).
A very important function of ECs is their ability to migrate, for example, during developmental and growth processes as well as during wound healing and tissue regeneration. Besides its importance as a fundamental physiological process, altered EC migration is part of the pathogenesis and progression of many severe disorders, such as ischemia reperfusion injury, diabetic angiopathy, macular degeneration, rheumatoid arthritis, and cancer (20, 21). Thus, interfering with EC migration offers an interesting therapeutic approach. Surprisingly, the requirement of ACC for the migration of ECs has been neglected so far. Therefore, this study aimed to investigate the role of ACC in primary human ECs with a focus on plasma membrane properties and the migratory capacity of ECs.
To inhibit the function of ACC, we used the well-established ACC inhibitor, soraphen A, a macrocyclic polyketidic natural compound originally isolated from the myxobacterium Sorangium cellulosum, as well as a gene silencing-based approach. Soraphen A is known to inhibit eukaryotic ACC by binding to the biotin carboxylase domain of ACC1 and ACC2. The binding of soraphen A interferes with the oligomerization of this domain, which is required for ACC activity (22, 23).
Our findings show for the first time that ACC1 regulates endothelial filopodia formation and, most importantly, EC migration by rearranging the cells’ phospholipid composition. The present study fills a gap in the literature by showing the relationship between the fatty acid metabolism and the migration of ECs.
MATERIALS AND METHODS
Materials
Soraphen A was isolated by Gerth et al. (24), as described previously, and dissolved in DMSO (Sigma-Aldrich, St. Louis, MO). Oleic acid and linolenic acid (Sigma-Aldrich) were dissolved in ethanol; linoleic acid sodium salt (Sigma-Aldrich) was dissolved in ethanol supplemented with 0.1% Tween 20 (Carl Roth, Karlsruhe, Germany). The 5-tetradecyloxy-2-furonic acid (TOFA; Santa Cruz Biotechnology, Dallas, TX), stearic acid (Sigma-Aldrich), and PD98059 (Cell Signaling Technology/New England Biolabs, Frankfurt am Main, Germany) were dissolved in DMSO. For their further use, compounds were diluted in growth medium without exceeding a final concentration of 0.1% (v/v) DMSO or ethanol in cell culture experiments.
Preparation of multilaminar vesicles
Multilaminar vesicles were prepared by the thin-film hydration method. The 1,2-dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt [dioleoylphosphatidylglycerol (DOPG)] {PG[18:1(9Z)/18:1(9Z)]; Sigma-Aldrich} was diluted in dichloromethane (Acros Organics, Morris Plains, NJ). The rotary evaporator, Laborota 4003 (Heidolph Instruments, Schwabach, Germany), was used to generate a lipid film. Starting at a 500 mbar vacuum, the pressure was reduced by 100 mbar every 10 min at room temperature to obtain a thin film of lipids. After 1 h at constant 50 mbar, the lipid film was rehydrated with PBS by vortexing for 10 min. Subsequently, the liposome solution was sterilized by filtration.
Cell culture
Human umbilical vein ECs (HUVECs) were obtained from PELOBiotech (Martinsried, Germany) and the human dermal microvascular EC line, HMEC-1 (25), was purchased from ATCC (Manassas, VA). Both cell types were cultured in EC growth medium (PELOBiotech) containing 10% heat-inactivated FBS (Biochrom, Berlin, Germany), 100 U/ml penicillin (PAN-Biotech, Aidenbach, Germany), 100 μg/ml streptomycin (PAN-Biotech), and 0.25 μg/ml amphotericin B (PAN-Biotech) at 37°C in an atmosphere containing 5% CO2. HepG2 cells were purchased from the Leibniz Institute DSMZ/German Collection of Microorganisms and Cell Cultures (Leipzig, Germany) and cultivated in DMEM (low glucose) supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Cell viability assays
The CellTiter-Blue cell viability assay (Promega, Mannheim, Germany) was used to determine the metabolic activity according to the manufacturer’s instructions. This assay uses the nonfluorescent dye, resazurin, which is reduced to the fluorescent dye, resorufin, by viable cells, as an indicator for metabolic activity. Fluorescence intensity (ex: 560 nm; em: 590 nm) was detected by a Tecan Infinite F200 plate reader (Tecan, Männedorf, Switzerland).
The quantification of the apoptosis rate was performed according to Nicoletti et al. (26). Cells undergoing apoptosis were assessed by staining with propidium iodide (50 μg/ml; Sigma-Aldrich). Staurosporine (1 μM; Sigma-Aldrich) was used as positive control. The number of cells with subdiploidic DNA content was analyzed with a FACSVerse flow cytometer (BD Biosciences, Heidelberg, Germany).
Cell death was measured using the CytoTox 96 nonradioactive cytotoxicity assay kit (Promega). The amount of released lactate dehydrogenase (LDH) from dying cells was determined according to the manufacturer’s protocol by a Tecan Infinite F200 plate reader. Lysis solution was used for maximum LDH release as positive control.
Quantification of cellular malonyl-CoA levels
HUVECs (1 × 107) were suspended in methanol/water (70/30) supplemented with the internal standard, 13C-malonyl-CoA (1 nmol; Sigma-Aldrich), and proteins precipitated at −20°C for 1 h. After centrifugation (20,000 g, 5 min, 4°C), the supernatant was evaporated to dryness. The residue was extracted with 50 μl water. Chromatography was carried out on an Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 100 mm) using an Acquity ultra-performance LC (UPLC) system from Waters (Milford, MA). Acyl-CoA esters were separated at 0.75 ml/min and 50°C by isocratic elution with 100% eluent A (10 mM aqueous ammonium acetate) for 1 min followed by a linear gradient to 100% eluent B (acetonitrile/10 mM aqueous ammonium acetate, 95/5) within 3 min. The chromatography system was coupled to a QTRAP 5500 mass spectrometer (Sciex, Darmstadt, Germany) equipped with an electrospray ionization source. Malonyl-CoA ([M+H]+) was quantified by multiple reaction monitoring in the positive ion mode after neutral loss of 2′-phospho-ADP ([M+H-507]+). The ion spray voltage was set to 3,000 V, the heated capillary temperature to 600°C, the curtain gas pressure to 30 psi, the sheath gas pressure to 45 psi, the auxiliary gas pressure to 55 psi, the declustering potential to 60 V, the entrance potential to 10 V, and the collision energy to 45 V. Analyst software 1.6 (Sciex) was used for the processing of analytical data.
Analysis of phospholipid profiles
Extraction and analysis of phospholipids were performed as described previously (27). In brief, extracted phospholipids were separated on an Acquity UPLC BEH C8 column (1.7 μm, 1 × 100 mm) using an Acquity UPLC system (Waters), which was coupled to a QTRAP 5500 mass spectrometer (Sciex) equipped with an electrospray ionization source. The fatty acid anion fragments of glycerophospholipids were detected by multiple reaction monitoring in the negative ion mode. The most intensive transition was used for quantification. Mass spectra were processed using Analyst 1.6 software (Sciex).
Cholesterol assay
The Amplex Red cholesterol assay kit (catalog number A12216; Invitrogen, Carlsbad, CA) was used to determine the amount of cholesterol in HUVECs according to the manufacturer’s protocol. The assay is based on an enzyme-coupled reaction that detects free cholesterol and cholesterol esters. H2O2, which is generated during the reaction, is detected using 10-acetyl-3,7-dihydroxyphenoxazine (Amplex Red reagent). In brief, HUVECs were treated with soraphen A (30 μM, 24 h) or left untreated. Cell pellets were homogenized in 200 μl of the organic solvent mixture (chloroform, isopropanol, IGEPAL CA-630; 7:11:0.1). After sonication and centrifugation (13,000 g, 10 min) the supernatants were evaporated to dryness. The residues were dissolved in 200 μl 1× reaction buffer. Fifty microliters of the lipids in 1× reaction buffer were mixed with 50 μl of the Amplex Red reagent (2 U/ml HRP, 2 U/ml cholesterol oxidase, 2 U/ml cholesterol esterase). The fluorescence intensity (ex: 560 nm; em: 590 nm) was detected by a Tecan Infinite F200 plate reader.
Membrane fluidity determination
The LS 55 fluorescence spectrometer (Perkin-Elmer) was used to perform steady-state fluorescence anisotropy measurements (L-configuration). A rotated sample holder allowed adjustment of the cell monolayer within the cuvette to an angle of 30° relative to the incident light beam. The height of the cuvette was adjusted allowing the light beam to pass through the monolayer.
The excitation and emission wavelengths were set to 360 nm (slit 5 nm) and 430 nm (slit 10 nm), respectively. The emission polarizer was set vertically and the absorption polarizer was set vertically or horizontally allowing intensity measurements of polarized light signals parallel (II) by vertical and vertical (v/v) and perpendicular (⊥) by vertical and horizontal (v/h) settings. HUVECs were cultivated on glass cover slips with a size of 13.5 × 13.5 mm (Paul Marienfeld, Lauda-Königshofen, Germany), which perfectly fit into a quartz glass cuvette. After treatment with soraphen A, the cells were rinsed with HBSS (without phenol red; Thermo Fisher Scientific, Schwerte, Germany) and placed vertically into the cuvette. The cuvette was filled with 2 ml of HBSS for individual background (Bkg) determination parallel and perpendicular to the initial polarized light beam. After labeling the cells with 30 μM trimethylammonium diphenylhexatriene (TMA-DPH; Sigma-Aldrich) for 30 min at 37°C, cells were rinsed again. In the following 30 min, up to seven individual measurements were performed at constant 37°C. The measured intensities were corrected by subtracting the Bkg signals v/v and v/h from the measured intensity v/v and v/h. Fluorescence anisotropy (r) was calculated using the following equations:
where I is intensity, I v/v is measured I v/v – Bkg v/v and I v/h is measured Iv/h – Bkg v/h.
The grating factor (Gf) was determined prior to the anisotropy measurement. It represents the correcting factor to compensate for the polarization bias of the detection system. Results are normalized to the untreated control and presented as the relative change in anisotropy values indicating alterations in membrane fluidity.
siRNA transfection
To downregulate ACC gene expression, HUVECs were transfected with the Amaxa HUVEC Nucleofector kit (Lonza, Cologne, Germany) using ON-TARGET plus human ACACA (ACC1) siRNA smart pool, ON-TARGET human ACACB (ACC2) siRNA smart pool, or siGENOME control pool (Thermo Scientific Dharmacon, Lafayette, CO). According to the manufacturer’s protocol, 1 × 106 HUVECs were resuspended in 100 μl Nucleofector solution and mixed with 300 nmol siRNA. This cell solution was transfected using the electroporation device, Nucleofector 2b (Lonza).
F-actin staining and measurement of filopodia formation
Cells were fixed with Roti-Histofix (4%, Carl Roth) and subsequently permeabilized with 0.2% Triton X-100 (Sigma-Aldrich) in PBS. Unspecific binding sites were blocked with 0.2% BSA in PBS. The actin cytoskeleton was stained by incubating cells with rhodamine-phalloidin (1:800, 1 h, room temperature, catalog number R415; Life Technologies, Darmstadt, Germany). To count the number of filopodia, HUVECs were pretreated with soraphen A (30 μM, 22 h). Subsequently, cells were detached and seeded out in low density (15,000 cells/well, μ-Slide 8 Well; Ibidi, Martinsried, Germany) in cell culture medium containing 30 μM soraphen A for 2 h. HUVECs transfected with siRNA were seeded out again after 46 h post transfection and plated for 2 h. Images were taken using a Leica DMI6000 B fluorescence microscope (Leica Microsystems, Wetzlar, Germany). The number of filopodia was counted manually. Twenty-five cells per experiment were analyzed.
Microtubule staining
HUVECs were incubated in cell extraction buffer [80 mM PIPES (pH 6.8); 1 mM MgCl2, 5 mM EGTA, 0.5% Triton X-100] to remove monomeric and dimeric tubulin subunits. After fixation with 0.5% glutaraldehyde, probes were quenched with 0.1% NaBH4 in PBS for 7 min. Cells were stained with an α-tubulin antibody (Abcam, Cambridge, UK) and with Hoechst 33342 (Sigma-Aldrich).
Migration assays
The undirected migration of HUVECs or HMECs was determined using a wound healing/scratch assay. A confluent monolayer was treated for 12 h, as indicated, or siRNA-transfected cells were used. A wound of approximately 1 mm was inflicted into a monolayer of 12 h-pretreated ECs by using a pipette tip. The cells were incubated for an additional 12 h either in starvation medium (Medium 199 with 1% FBS, representing 0% migration), culture medium (EC growth medium with 10% FBS, representing 100% migration), or culture medium containing a compound. After 12 h, cells were washed with PBS, fixed with Roti-Histofix (4%; Carl Roth) for 10 min at room temperature, and analyzed microscopically using a Leica DMIL LED microscope (Leica Microsystems). Images were analyzed using ImageJ (version 1.49k) by determining the covered and uncovered area. Relative migration was calculated related to the starvation control.
A Boyden chamber assay was used to determine the migration of ECs triggered by a chemotactic gradient. HUVECs or HMECs were seeded onto collagen G-coated (Biochrom) Millicell cell culture inserts (polyethylene terephthalate) with a pore size of 8 μm (Merck, Darmstadt, Germany). After reaching confluency, cells were stimulated with 30 μM soraphen A for 18 h and were allowed to migrate toward a chemoattractant gradient (20% FBS) for 6 h. The migration process of HUVECs was started 42 h after cells were transfected with siRNA. Cells were fixed with Accustain (Sigma-Aldrich) and visualized with crystal violet staining solution. Nonmigratory cells from the upper compartment of the cell culture inserts were removed using a cotton swab. Crystal violet was solubilized by addition of dissolving buffer [50% ethanol (v/v), 0.1 M sodium citrate in water] under constant agitation for 10 min. The optical density of each solution was measured at 540 nm using a Tecan Infinite F200 Pro plate reader.
To further investigate the chemotactic behavior of migrating ECs, μ-Slide Chemotaxis chambers coated with collagen IV (Ibidi) were used according to the manufacturer’s instructions. HUVECs were treated as indicated or transfected with siRNA and used 28 h after transfection. Cells were observed for 20 h in an FBS gradient (0–20%) in a climate chamber at 37°C and 5% CO2. Images were taken every 10 min with a Leica DMI6000 B fluorescence microscope (Leica Microsystems). The ImageJ plugins, “Manual Tracking” and “Chemotaxis Analysis,” were used to track cells and calculate the parameters forward migration index toward the Y-axis direction (FMI:Y), directness, accumulated distance, Euclidean distance, and the velocity of cells.
Quantitative PCR
HUVECs were treated as indicated or transfected with siRNA. According to the manufacturer’s instructions, RNA isolation was performed with the RNeasy mini kit (Qiagen, Hilden, Germany), including on-column DNase digestion (RNase-Free DNase set; Qiagen). SuperScript II reverse transcriptase (Life Technologies) was used to reverse-transcribe 1 μg of RNA. For quantitative PCR (qPCR), the StepOnePlus system (Applied Biosystems, Foster City, CA) with SYBR Green PCR Master Mix (Life Technologies) was used. Based on the 2−ΔΔCT method, the mRNA values were normalized to the GAPDH housekeeping gene. The following primers were used: ACC1 (forward, 5′-TCC ACT TGG CTG AGC GAT TG-3′ reverse, 5′-CAA GTC AGC AAA CTG CAC GG-3′), ACC2 (forward, 5′-TCT CGT GAG TCT ACC CGG AA-3′ reverse, 5′-AGC TTC TTG TGT TCC CGT CC-3′), and GAPDH (forward, 5′-CCA CAT CGC TCA GAC ACC AT-3′ reverse, 5′-TGA AGG GGT CAT TGA TGG CAA-3′).
Transcriptome analysis
Transcriptome analysis was performed by library preparation via massive analysis of cDNA ends (GenXPro, Frankfurt, Germany) followed by sequencing via MiSeq (Illumina, San Diego, CA). This method allows sequencing only from the 3′-ends of cDNAs. Massive analysis of cDNA ends was performed according to the manufacture’s protocol. In brief, 5 μg mRNA per sample were converted into cDNA using biotinylated primers to tag cDNAs with barcoded adapters. After a purification step with silica columns for DNA purification (QIAquick PCR purification kit; Qiagen), samples were pooled and sheared into ≤200 bp fragments. Biotinylated cDNAs were captured with streptavidin-coated paramagnetic beads to wash off unbound fragments. After end repair and adaptor ligation, PCR amplification followed. For Illumina sequencing, 16 pM cDNA was used according to the manufacturer’s protocol. The results are expressed as normalized mRNA amount, i.e., the quotient of the average raw count of each gene within the library and the geometric mean of all counts in all samples. The median of the quotient is calculated per library. According to Anders and Huber (28), each single raw count is divided by the library-specific median value. The average of the normalized values of all replicates is represented by the displayed number.
Western blot analysis
HUVECs were treated as indicated in the respective figure legends. Cells were washed with ice-cold PBS and lysed with RIPA buffer. Laemmli sample buffer was added to the supernatants and heated at 95°C for 5 min. Equal amounts of proteins were separated by SDS polyacrylamide gel electrophoresis (Bio-Rad Laboratories, Munich, Germany) and transferred to polyvinylidene fluoride membranes by tank blotting (Bio-Rad Laboratories). The following primary antibodies were used: anti-ACC1 (catalog number 4190, dilution: 1:1,000, Cell Signaling Technology/New England Biolabs), anti-ACCβ (F-9) (catalog number sc-3773313, dilution: 1:500, Santa Cruz Biotechnology). The following HRP-conjugated secondary antibodies were used: goat anti-rabbit (catalog number sc-2004, dilution: 1:10,000, Santa Cruz Biotechnology), goat anti-mouse (catalog number sc-2005, dilution: 1:10,000, Santa Cruz Biotechnology), mouse monoclonal anti-β-actin antibody (catalog number A3854, dilution: 1:100,000, Sigma-Aldrich). Densitometric analysis was performed with ImageJ software version 1.49k.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 5.0 software (San Diego, CA). One-way ANOVA with Tukey’s post hoc test or unpaired t-test was used for significance analysis. Statistically significant differences were assumed at P ≤ 0.05. Bar graph data represent mean values ± SEM. The number of independently performed experiments is mentioned in the respective figure legends. For HUVECs, each number represents a different HUVEC donor. For HMECs and HepG2, each number represents a new cell passage.
RESULTS
ACC1 is the predominant isoform in ECs
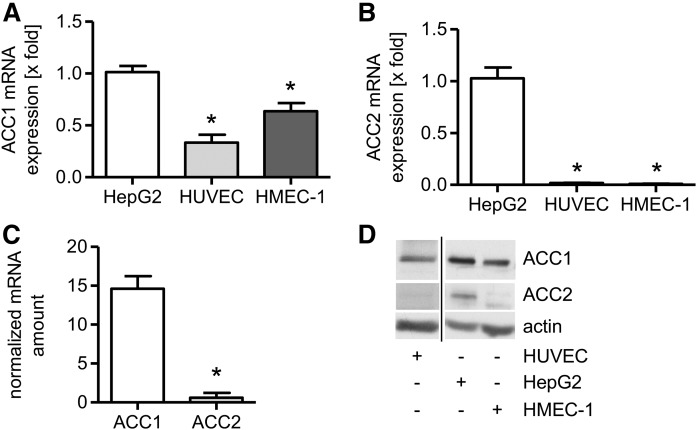
To characterize the role of ACC in ECs, we first investigated which isoform of ACC dominates in untreated quiescent ECs. The literature reports divergent results regarding the functional redundancy between the isoform ACC1 and ACC2 in liver cells (29–31). Hence, mRNA as well as protein levels of ACC1 and ACC2 were determined by qPCR, mRNA counting (transcriptome analysis), and immunoblotting, respectively. ACC1 and ACC2 levels in HUVECs and HMECs were compared with the human liver cancer cell line, HepG2 (positive control), by qPCR and Western blot analysis. Figure 1A shows that the ACC1 mRNA expression in HUVECs was about one-third of the expression in HepG2 cells and the expression in HMECs was about two-thirds of the expression in HepG2 cells. In contrast, the expression of ACC2 mRNA in both HUVECs and HMECs was less than 2% of the levels found in HepG2 cells (Fig. 1B). Accordingly, mRNA counting by transcriptome analysis showed that the normalized ACC1 mRNA amount in HUVECs was about 24 times higher than the expression of ACC2 mRNA (Fig. 1C). The results of the immunoblotting experiments (Fig. 1D) were in accordance with these data. Taken together, these results clearly show that ACC1 is the predominant isoform in ECs.
Fig. 1.
ACC1 is the predominant isoform in ECs. The mRNA expression of ACC1 (A) and ACC2 (B) was determined in untreated ECs (HUVECs, HMECs) and compared with the liver cancer cell line HepG2 (positive control) by qPCR analysis. Data are expressed as mean ± SEM (n = 3) (*P ≤ 0.05 vs. HepG2). C: The numbers of ACC1 and ACC2 transcripts were determined by transcriptome analysis. Data are expressed as mean ± SEM (n = 3) (*P ≤ 0.05 vs. ACC1). D: ACC1, ACC2, and β-actin protein levels were measured in HUVECs, HMECs, and the liver cancer cell line HepG2 (positive control). One representative Western blot out of three independently performed experiments is shown.
Soraphen A shifts the membrane lipid composition of HUVECs
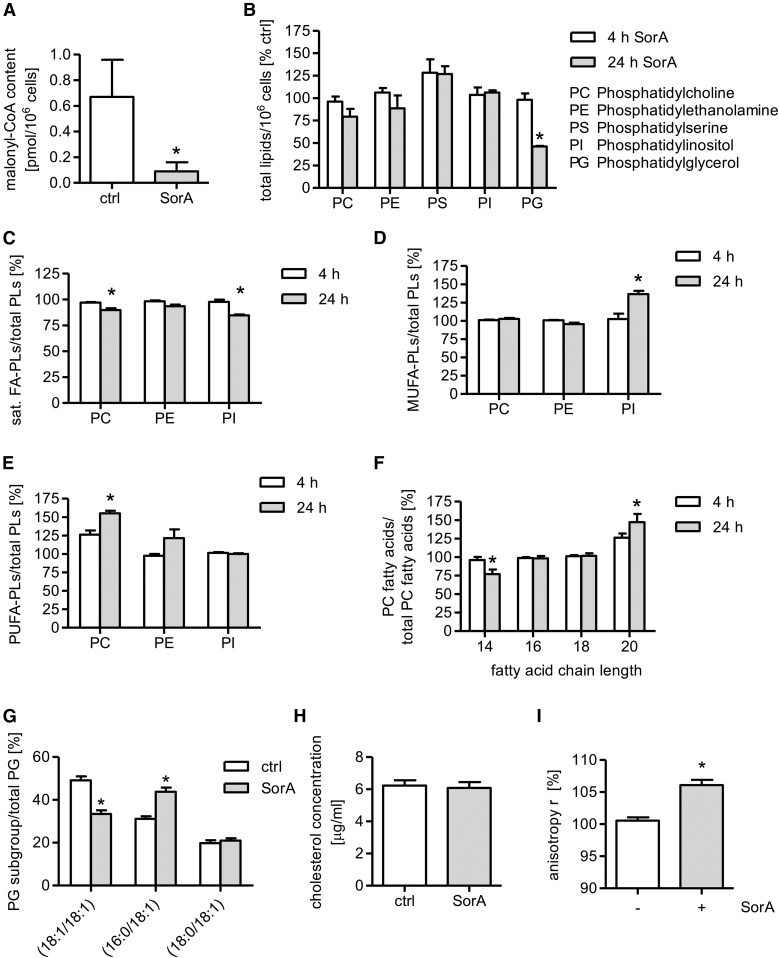
Several studies have reported that soraphen A is a potent inhibitor of ACC (32, 33). To prove the functionality of soraphen A in ECs, we measured the amount of malonyl-CoA after soraphen A treatment (30 μM, 24 h) by UPLC-MS/MS. Malonyl-CoA levels were strongly reduced upon soraphen A treatment by almost 90% (Fig. 2A). Soraphen A at the applied concentration did not show any cytotoxic effects in HUVECs after 24 or 48 h of incubation (supplemental Fig. S1A–C), as shown by analyzing the metabolic activity, the apoptosis rate (cells with subdiploid DNA content), and the percentage of cells with compromised membrane integrity (release of LDH). These findings demonstrate that soraphen A inhibits ACC and does not impair the viability of ECs.
Fig. 2.
Soraphen A (SorA) shifts the membrane lipid composition of HUVECs. A: HUVECs were either left untreated [control (ctrl)] or were treated with soraphen A (30 μM, 24 h). The content of malonyl-CoA was measured by UPLC-MS/MS. Data are expressed as mean ± SEM (n = 3) (*P ≤ 0.05 vs. ctrl). B–G: HUVECs were either left untreated or were treated with soraphen A (30 μM) for 4 h (B–F) or 24 h (B–G). The membrane lipid composition, grade of membrane lipid desaturation, and length of fatty acid chains were measured by UPLC-MS/MS (sat. FA-PLs, saturated fatty acid-containing phospholipids; MUFA-PLs, MUFA-containing phospholipids; PUFA-PLs, phospholipids containing PUFAs with more than two double bonds). Untreated cells are defined as 100% (B–F). Data are expressed as mean ± SEM (n = 3, n = 2 for PI) (*P ≤ 0.05 vs. control). H: HUVECs were either left untreated [control (ctrl)] or were treated with soraphen A (30 μM, 24 h). The amount of cholesterol was measured by the Amplex Red cholesterol assay. Data are expressed as mean ± SEM (n = 3) (*P ≤ 0.05 vs. ctrl). I: HUVECs were either left untreated or were treated with soraphen A (30 μM, 24 h). The fluorescence anisotropy (r) was determined upon loading of the cells with the dye, TMA-DPH. Data are expressed as mean ± SEM (n = 3) (*P ≤ 0.05 vs. ctrl).
Because malonyl-CoA is crucial for the synthesis of fatty acids and because the majority of de novo generated fatty acids are incorporated into membrane phospholipids (34–36), we hypothesized that soraphen A treatment might have an impact on the membrane lipid composition of ECs. Therefore, we analyzed the phospholipid profiles of HUVECs treated with soraphen A (30 μM, 4 or 24 h) by UPLC-MS/MS, whereas untreated cells are defined as 100%. Short-time treatment with soraphen A (30 μM, 4 h) neither significantly affected the cellular phospholipid composition nor the grade of fatty acid saturation (Fig. 2B–E). Long-term inhibition of endothelial ACC, however, resulted in a marked shift of the phospholipid composition: Soraphen A (30 μM, 24 h) significantly lowered the levels of phosphatidylglycerol (PG) by 54% (Fig. 2B). Moreover, soraphen A significantly lowered the cellular ratio of phosphatidylinositol (PI)- and phosphatidylcholine (PC)-containing saturated fatty acids (Fig. 2C) relative to PI with MUFAs (Fig. 2D) and PC with PUFAs (Fig. 2E). Besides the grade of saturation, ACC inhibition also affects the fatty acid chain length. As shown in Fig. 2F, soraphen A significantly increased the portion of PC phospholipids with longer fatty acid chains (C-20), whereas the portion of PC with C-14 fatty acids was significantly reduced. Within the PG species, the distribution of the fatty acid chain lengths was altered: Soraphen A decreased the relative amount of PG(18:1/18:1) and increased the amount of PG(16:0/18:1) (Fig. 2G). Besides phospholipids, we also investigated cholesterol levels, because this steroid is a crucial component of mammalian cell membranes (37). ACC inhibition by soraphen A did not alter cellular cholesterol concentrations (Fig. 2H). Taken together, soraphen A treatment increased the grade of fatty acid unsaturation as well as the portion of phospholipids with longer fatty acid chains for PC and led to reduced fatty acid chain lengths for PG.
Because the membrane fluidity depends, among other factors, on the fatty acid composition (chain length, degree of saturation), we hypothesized that ACC inhibition might alter this parameter. Therefore, we analyzed the membrane fluidity of HUVECs by fluorescence anisotropy measurements. The fluorescent dye, TMA-DPH, anchors itself at the membrane/water interface and, thus, allows the determination of rotational changes close to the phospholipid head groups (38). Soraphen A treatment (30 μM, 24 h) led to increased anisotropy values (Fig. 2I), indicating that the EC membrane becomes more rigid upon inhibition of ACC.
In summary, soraphen A reduces the amount of malonyl-CoA, which leads to a shift in the phospholipid composition and to decreased membrane fluidity. The amount of PG and saturated fatty acids is reduced, whereas the amount of MUFAs and PUFAs and the fatty acid chain length within the PC subgroup is increased. Additionally, soraphen A led to a reduction of fatty acid chain lengths of the PG species.
ACC inhibition decreases the number of filopodia in migrating ECs
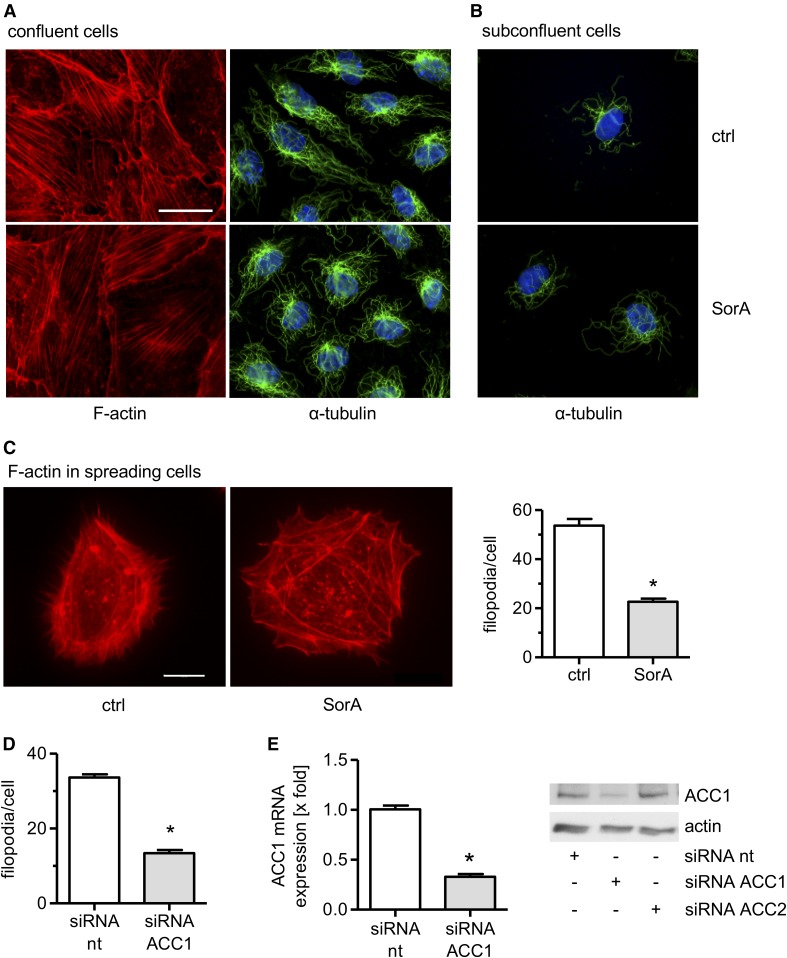
Because F-actin fibers are anchored to the cell membrane, changes of the membrane stiffness can lead to alterations of the actin cytoskeleton (39). Thus, we analyzed the action of soraphen A on the actin cytoskeleton. Moreover, we also investigated the tubulin cytoskeleton. Confluent HUVECs were treated with soraphen A (30 μM, 24 h) and stained with rhodamine-phalloidin or an anti-α-tubulin antibody. Soraphen A treatment did not affect the actin or the tubulin cytoskeleton in confluent quiescent cells (Fig. 3A). Also the tubulin cytoskeleton was not influenced in spreading/moving ECs (Fig. 3B). Most importantly, however, soraphen A treatment significantly altered the distribution of F-actin in spreading/moving ECs (Fig. 3C): The number of filopodia was strongly decreased by approximately 60%. This effect was confirmed in ACC1-silenced HUVECs (Fig. 3D). The successful knockdown of ACC1 was confirmed on the mRNA and the protein level (Fig. 3E). Filopodia are projections of bundled cross-linked actin filaments with a finger-like structure. They play an important role in several fundamental physiological processes, especially in cell motility (40, 41). These findings show that ACC inhibition does not affect the cytoskeleton of quiescent ECs, whereas it leads to a diminished formation of filopodia, a crucial F-actin structure in moving ECs.
Fig. 3.
ACC inhibition decreases the number of filopodia in migrating ECs. HUVECs were grown to confluency (A) or in low density (B) and were treated with soraphen A (SorA) (30 μM, 24 h). F-actin (red), α-tubulin (green), and nuclei (blue) were visualized by immunocytochemistry and fluorescence microscopy. Magnification: 64×. The scale bar represents 25 μm. One representative image out of three independently performed experiments is shown. C: HUVECs were left untreated or treated with soraphen A (30 μM, 24 h), detached, and allowed to spread at low density for 2 h. D: HUVECs were treated with nontargeting (nt) or ACC1-targeting siRNA. Forty-six hours after transfection, cells were allowed to spread at low density for 2 h. C: Magnification: 64×. The scale bar represents 10 μm. One representative image out of three independently performed experiments is shown. C, D: F-actin (red) was visualized by immunocytochemistry and fluorescence microscopy. The number of filopodia was quantified by counting. Data are expressed as mean ± SEM (n = 3) [*P ≤ 0.05 vs. control (ctrl) or siRNA nt]. E: Gene silencing controls. HUVECs were transfected with nontargeting (nt) siRNA, ACC1-targeting siRNA, or ACC2-targeting siRNA. Forty-eight hours after transfection, ACC1 mRNA levels were analyzed by qPCR. ACC1 and β-actin protein levels were determined by Western blot analysis. Data are expressed as mean ± SEM (n = 3) (*P ≤ 0.05 vs. siRNA nt). One representative image out of three independently performed experiments is shown.
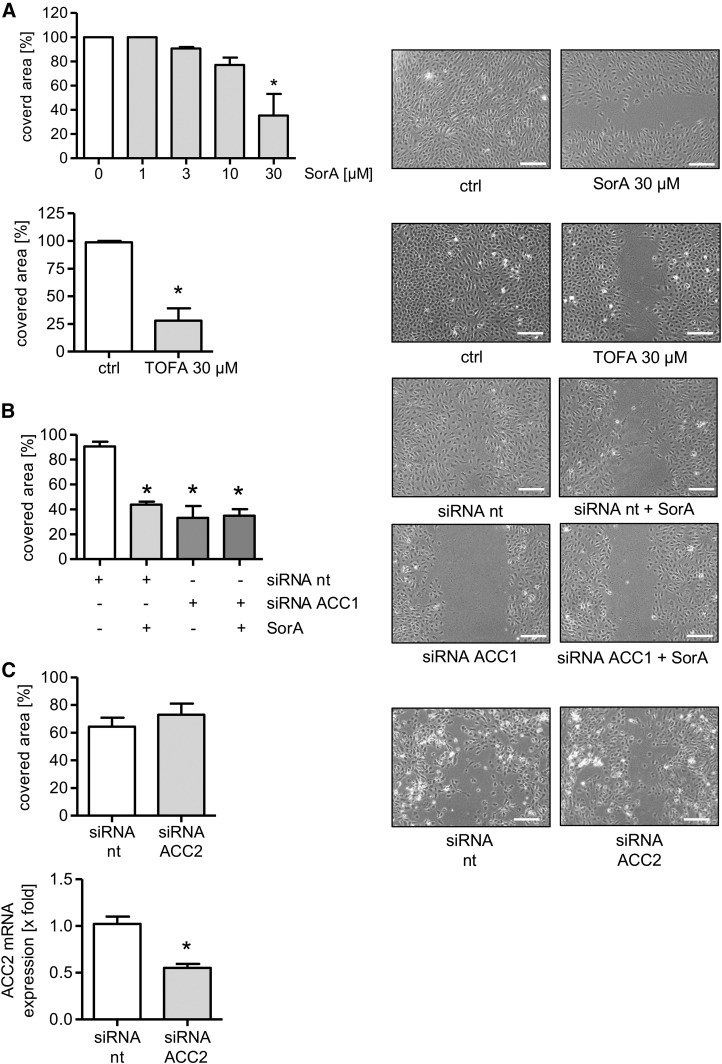
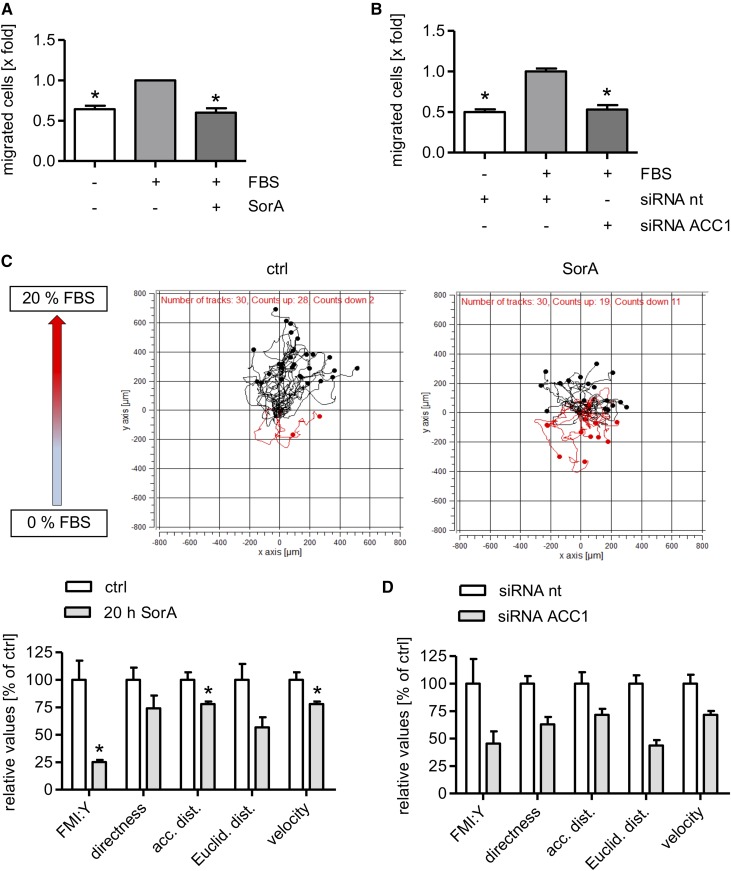
ACC1 inhibition attenuates undirected EC migration
Because of the role of filopodia in cell movement, we analyzed the action of soraphen A on the migratory capacity of ECs. Figure 4A shows that soraphen A inhibited the process of undirected EC migration (scratch assay) in a concentration-dependent manner. To exclude a compound-specific effect, we used TOFA, another well-established ACC inhibitor, and found that TOFA also blocked EC migration (Fig. 4A) without influencing endothelial viability (supplemental Fig. S1D).
Fig. 4.
ACC1 inhibition attenuates undirected EC migration. A: HUVECs were left untreated or were treated with different concentrations of soraphen A (SorA) or TOFA for 24 h and a scratch assay was performed. B: HUVECs were transfected with nontargeting (nt) and ACC1-targeting siRNA. Thirty-six hours after transfection, a scratch assay was performed. Moreover, cells were left untreated or were additionally treated with soraphen A for 24 h. C: HUVECs were transfected with nontargeting (nt) or ACC2-targeting siRNA. Forty-eight hours after transfection, ACC2 mRNA levels were analyzed by qPCR. A–C: Cells were allowed to migrate for 12 h. The covered area was determined. Data are expressed as mean ± SEM (n = 3) [*P ≤ 0.05 vs. control (ctrl) or siRNA nt]. One representative image out of three independently performed experiments is shown. Magnification: 10×. Scale bars represent 200 μm.
To verify the crucial involvement of ACC, we analyzed the impact of the knockdown of both ACC1 and ACC2 on the undirected migration of HUVECs. ACC1 silencing strongly decreased endothelial migration (Fig. 4B). Of note, Fig. 4B also shows that soraphen A exerted no additional effect in HUVECs treated with ACC1 siRNA on cell migration. The knockdown of ACC2, as expected due to its very low expression, had no influence (Fig. 4C). The successful knockdown of ACC2 was confirmed on the mRNA level (Fig. 4C). To exclude that ACC2 knockdown affects ACC1 protein levels, we analyzed ACC1 expression in ACC2-silenced HUVECs. No counterregulatory effects were detected (Fig. 3E). These data verify the predominant role of the ACC1 isoform and, most importantly, its relevance for the undirected migration of ECs.
ACC1 inhibition impairs chemotactic migration
Besides undirected movement, we were also interested in the role of ACC in the process of chemotactic migration. Boyden chamber assays revealed that soraphen A (30 μM, 24 h) (Fig. 5A) and ACC1 knockdown (Fig. 5B) completely blocked migration in the direction of an FBS gradient. In addition, we analyzed the chemotactic migration of ECs via live-cell imaging. Several parameters were calculated to quantify the influence of soraphen A: The FMI:Y indicates the efficiency of the migration of cells toward the chemoattractant. The parameter directness is calculated by dividing the Euclidian distance by the accumulated distance. Soraphen A treatment led to a significant reduction of the parameters FMI:Y, accumulated distance, and velocity (Fig. 5C). The parameter directness and Euclidean distance were also attenuated by ACC inhibition (Fig. 5C), albeit not statistically significant. ACC1 silencing showed similar results (Fig. 5D). Taken together, ACC1 inhibition leads to a reduced migratory capacity toward a chemotactic stimulus.
Fig. 5.
ACC inhibition attenuates chemotactic migration. A: HUVECs were seeded onto the porous filter membrane insert of a Boyden chamber. Cells were either left untreated or were pretreated with soraphen A (SorA) (30 μM, 18 h). B: HUVECs were transfected with nontargeting (nt) or ACC1-targeting siRNA and seeded onto the porous filter membrane insert of a Boyden chamber. Forty-two hours after transfection, the Boyden chamber assay was started. A, B: Cells were allowed to migrate toward a chemotactic gradient (20% FBS) for 6 h. Migrated cells were quantified by crystal violet staining. Data are expressed as mean ± SEM (n = 4 for A, n = 3 for B) [*P ≤ 0.05 vs. control (ctrl)]. C: HUVECs were either left untreated or were treated with 30 μM soraphen A. D: HUVECs were transfected with nontargeting (nt) or ACC1-targeting siRNA. Twenty-eight hours after transfection, cells were seeded into chemotaxis chambers. C, D: Chemotactic migration within a 20% FBS gradient was monitored for 20 h. The parameters FMI:Y, directness, accumulated distance, Euclidean distance, and the velocity of cells were determined. Data are expressed as mean ± SEM (n = 3) (*P ≤ 0.05 vs. ctrl or siRNA nt).
Increased PUFA levels mimic the antimigratory action of ACC inhibition
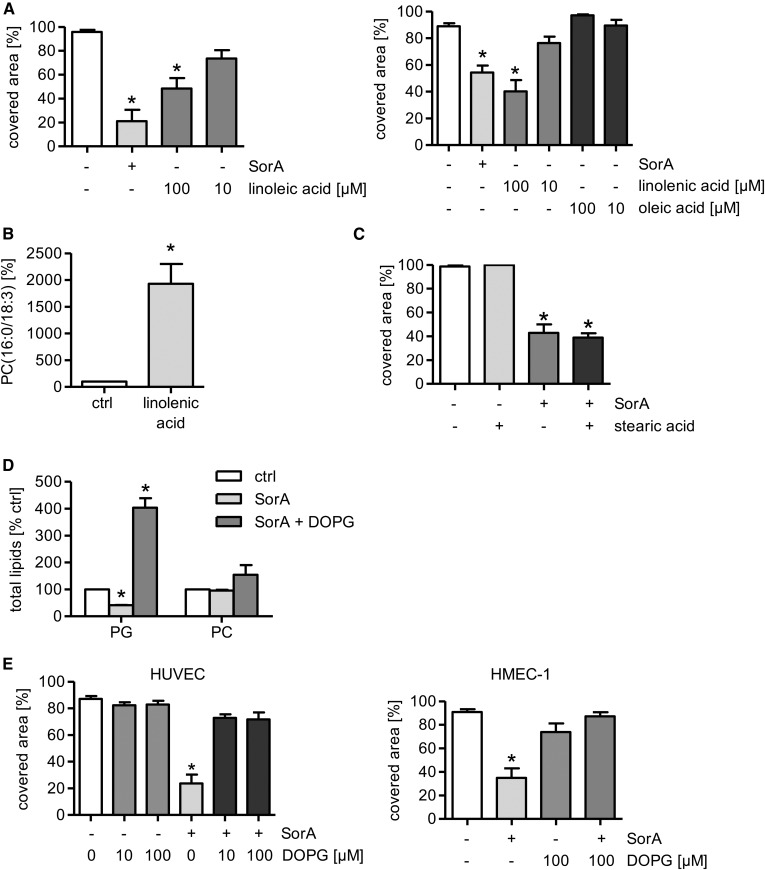
The lipidomics analysis showed that the cellular proportion of MUFA and PUFA species was upregulated in distinct phospholipid subgroups after soraphen A treatment (Fig. 2D, E). To verify their importance for EC migration, HUVECs were treated with representative MUFAs or PUFAs and were analyzed for their migratory capacity. The MUFA, oleic acid (10 and 100 μM, 24 h), had no effect on the migratory capacity (Fig. 6A), whereas the PUFAs, linoleic and linolenic acid (100 μM, 24 h), mimicked the antimigratory effect of soraphen A (Fig. 6A). The successful incorporation of PUFAs was exemplarily verified for linolenic acid (18:3) by measuring the amount of PC(16:0/18:3) (Fig. 6B). These results demonstrate the importance of PUFAs in the ACC-mediated reduction of EC migration.
Fig. 6.
The addition of PUFAs mimics and PG reverses the anti-migratory action of soraphen A (SorA). A–E: HUVECs or HMECs (E) were either left untreated or were treated with soraphen A (30 μM, 24 h), different concentrations of linoleic acid, linolenic acid [(B) 100 μM], oleic acid, stearic acid [(C) 1 μM], or DOPG [(D) 100 μM] for 24 h. A, C, E: Cells were allowed to migrate for 12 h after a scratch was inflicted into an endothelial monolayer. The covered area was determined. B, D: The membrane lipid composition was measured by UPLC-MS/MS. Data are expressed as mean ± SEM [n = 5 for A (left) and E (HUVEC); n = 3 for all others; *P ≤ 0.05 vs. control (ctrl)].
PG abolishes the antimigratory action of soraphen A
Because soraphen A showed a decrease of saturated fatty acids (Fig. 2C) and a strong reduction of PG levels (Fig. 2B), we analyzed the ability of stearic acid (18:0) and of PG (in the form of DOPG), respectively, to compensate the inhibition of ACC in EC migration assays. Stearic acid (1 μM, 24 h) did not influence the undirected migration of ECs (Fig. 6C). Due to the low solubility of DOPG, liposomes (multilaminar vesicles) were prepared. The successful incorporation of DOPG into the membrane phospholipids was confirmed via determining the total amount of PG (Fig. 6D). The total amount of other phospholipids, exemplarily characterized for PC, was not significantly affected (Fig. 6D). As shown before (Fig. 2B), soraphen A reduced the amount of PG, whereas the addition of DOPG restored the levels of PG after soraphen A treatment (Fig. 6D). Both HUVECs and HMECs were treated with DOPG (10 and 100 μM, 24 h) and soraphen A (30 μM, 24 h). DOPG completely rescued the antimigratory effect of ACC inhibition in HUVECs and HMECs (Fig. 6E). Our findings provide evidence that the antimigratory action of ACC inhibition is causally linked to the reduction of the phospholipid subgroup PG.
DISCUSSION
The fatty acid metabolism has been increasingly recognized as an interesting target system to treat various metabolic pathologies like insulin resistance, hepatic steatosis, dyslipidemia, obesity, metabolic syndrome, and nonalcoholic fatty liver disease. Moreover, alterations in the fatty acid metabolism are a hallmark of cancer cell proliferation (42, 43). Compared with healthy cells, many tumor cells synthesize fatty acids at higher rates, which results in an elevated dependence on lipogenesis (44). Also the fatty acid metabolism of the endothelium has gained attention. Although ECs are continually exposed to fatty acids from the blood, de novo lipogenesis has been suggested to be of importance for the maintenance and regulation of physiological as well as pathophysiological endothelial functions, such as endothelial nitric oxide synthase activity (45), endothelial permeability (46), or angiogenesis-associated proliferation and sprouting (47).
The inhibition of ACC offers two attractive options to interfere with fatty acid metabolism: It stimulates the fatty acid oxidation in oxidative tissues and, concurrently, inhibits the de novo lipogenesis in lipogenic tissues (2, 48). Recent studies confirm ACC as a valuable drug target for the treatment of the metabolic syndrome and obesity, cancer, and, of note, also autoimmune diseases. For instance, Harriman et al. (9) reported that the ACC inhibitor ND-630, which was administered to rats in a model of diet-induced obesity, reduced hepatic steatosis and weight gain and improved insulin sensitivity and dyslipidemia. In an in vivo model of non-small-cell lung cancer, the ACC inhibitor ND-646 succeeded in blocking tumor growth (49). Moreover, soraphen A decreased the formation of pro-inflammatory TH17 cells, stimulated the development of anti-inflammatory Treg cells, and attenuated the symptoms in a murine model of multiple sclerosis (14).
Despite this knowledge, the role of ACC and the impact of ACC inhibition on EC functions have not been investigated so far. Our study provides the first link between ACC/fatty acid metabolism and migration in ECs. We demonstrated that ACC1 is the predominant isoform both in macrovascular HUVECs and microvascular HMECs. ACC inhibition led to reduced levels of malonyl-CoA and, consequently, to a marked shift in the phospholipid composition. As a result, membrane fluidity was reduced. These changes were associated with a reduced amount of filopodia in migrating ECs (without affecting the cytoskeleton in confluent cells) and a strongly lowered migratory capacity. The antimigratory action of ACC inhibition depended on increased concentrations of PUFAs and, most importantly, decreased levels of a distinct subgroup of membrane lipids, namely PG.
We clearly showed that ACC inhibition caused reduced formation of filopodia. We suggest that this reduction could be the mechanistic basis for the inhibition of EC migration. Filopodia are known to play an important role in cell migration and wound healing. They serve as “antennae” of cells to sense their microenvironment and to set the direction of cell migration (40). Integrins in filopodia promote cell adhesion and migration (50). Accordingly, a loss of filopodia leads to a reduced undirected and directed migration capacity, as shown in our experiments. This specific effect was also represented by the strongly decreased FMI:Y, whereas ACC inhibition had only a weak effect on the velocity of cell migration. With a reduced amount of filopodia, the cell loses its ability to sense the environment and, hence, FMI:Y is decreased. A study by Scott et al. (35) corroborates the essential role of ACC/fatty acid metabolism in mediating the distribution of F-actin: Pharmacological inhibition of ACC in a Src-transformed fibroblast cell line led to reduced formation of invadopodia. Invadopodia are actin-rich protrusions of the plasma membrane in invasive cancer cells, which are utilized to invade the extracellular matrix (51).
An increase in the degree of desaturation of fatty acids is expected to increase membrane fluidity, because unsaturated fatty acids cannot be packed as densely as saturated ones. In contrast, a shift toward longer fatty acid chains should decrease membrane fluidity due to intensified van der Waals forces. As a matter of fact, soraphen A decreased membrane fluidity. In accordance, soraphen A treatment augmented the fatty acid chain length of PC. However, soraphen A concurrently raised the portion of unsaturated fatty acids and increased the amount of shorter fatty acid chains within the phospholipid subgroup PG. We speculate that the increase in the fatty acid chain length in PC overcompensates the increase in desaturation and, thus, dominates the effect of soraphen A on membrane fluidity, in particular because PC is much more abundant in membranes than PG. An influence of soraphen A on another crucial regulator of membrane fluidity, cholesterol, was excluded. In accordance with our results, Braig et al. (52) also found that soraphen A treatment of MDA-MB-231 mammary cancer cells increased the grade of desaturation and the chain length of phospholipid fatty acids, augmented membrane rigidity, and inhibited the migratory capacity of these cells. In contrast to our findings, however, ACC inhibition did not lead to changes in the total amount of phospholipid subgroups. This difference might be due to the usage of different cell types, treatment times, and concentrations of soraphen A.
A very important finding of our study is that PGs as well as PUFAs are critical modulators of membrane fluidity and migration of ECs. PUFAs mimicked the antimigratory action of soraphen A. Saturated fatty acids did not prevent the effect of soraphen A on EC migration, whereas an addition of PG fully rescued the effect of ACC inhibition. This indicates that the antimigratory action of soraphen A is linked to the amount of PG present in EC membranes. The role of PG in modulating cell migration was unknown in the literature so far.
Our study reveals for the first time a link between migration and an altered phospholipid composition in ECs. On the one hand, soraphen A represents a useful chemical tool to investigate the role of fatty acid metabolism in ECs. On the other hand, the inhibition of ACC offers a new and valuable therapeutic perspective for the treatment of EC migration-related diseases.
Supplementary Material
Acknowledgments
The authors thank Dr. Mukul Ashtikar for the professional assistance in preparing multilaminar vesicles and Mareike Lang for the support in Western blot analysis.
Footnotes
Abbreviations:
- ACC
- acetyl-CoA carboxylase
- Bkg
- background
- DOPG
- dioleoylphosphatidylglycerol
- EC
- endothelial cell
- FMI:Y
- forward migration index toward the Y-axis direction
- Gf
- grating factor
- HMEC
- human microvascular endothelial cell
- HUVEC
- human umbilical vein endothelial cell
- LDH
- lactate dehydrogenase
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- qPCR
- quantitative PCR
- TMA-DPH
- trimethylammonium diphenylhexatriene
- TOFA
- 5-tetradecyloxy-2-furonic acid
- UPLC
- ultra-performance LC
This work was supported by German Research Foundation (DFG-Research Group FOR 1406) Grants FU 691/9-2 (to R.F.) and WE 2260/11-2 (to O.W.). A.K. was supported by the German Research Council (GRK 1715) and the Phospholipid Research Center Heidelberg (Heidelberg, Germany). The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Tong L. 2005. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell. Mol. Life Sci. 62: 1784–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong L., and Harwood H. J. Jr. 2006. Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J. Cell. Biochem. 99: 1476–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakil S. J., Stoops J. K., and Joshi V. C.. 1983. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 52: 537–579. [DOI] [PubMed] [Google Scholar]
- 4.Abu-Elheiga L., Matzuk M. M., Abo-Hashema K. A., and Wakil S. J.. 2001. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 291: 2613–2616. [DOI] [PubMed] [Google Scholar]
- 5.Schreurs M., Van Dijk T., Gerding A., Havinga R., Reijngoud D. J., and Kuipers F.. 2009. Soraphen, an inhibitor of the acetyl-CoA carboxylase system, improves peripheral insulin sensitivity in mice fed a high-fat diet. Diabetes Obes. Metab. 11: 987–991. [DOI] [PubMed] [Google Scholar]
- 6.Harwood H. J., Jr 2004. Acetyl-CoA carboxylase inhibition for the treatment of metabolic syndrome. Curr. Opin. Investig. Drugs. 5: 283–289. [PubMed] [Google Scholar]
- 7.Harwood H. J., Jr 2005. Treating the metabolic syndrome: acetyl-CoA carboxylase inhibition. Expert Opin. Ther. Targets. 9: 267–281. [DOI] [PubMed] [Google Scholar]
- 8.Stiede K., Miao W., Blanchette H. S., Beysen C., Harriman G., Harwood H. J. Jr., Kelley H., Kapeller R., Schmalbach T., and Westlin W. F.. 2017. Acetyl-CoA carboxylase inhibition reduces de novo lipogenesis in overweight male subjects: a randomized, double-blind, crossover study. Hepatology. 66: 324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harriman G., Greenwood J., Bhat S., Huang X., Wang R., Paul D., Tong L., Saha A. K., Westlin W. F., Kapeller R., et al. 2016. Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats. Proc. Natl. Acad. Sci. USA. 113: E1796–E1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swinnen J. V., Brusselmans K., and Verhoeven G.. 2006. Increased lipogenesis in cancer cells: new players, novel targets. Curr. Opin. Clin. Nutr. Metab. Care. 9: 358–365. [DOI] [PubMed] [Google Scholar]
- 11.Milgraum L. Z., Witters L. A., Pasternack G. R., and Kuhajda F. P.. 1997. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin. Cancer Res. 3: 2115–2120. [PubMed] [Google Scholar]
- 12.Beckers A., Organe S., Timmermans L., Scheys K., Peeters A., Brusselmans K., Verhoeven G., and Swinnen J. V.. 2007. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 67: 8180–8187. [DOI] [PubMed] [Google Scholar]
- 13.Yahagi N., Shimano H., Hasegawa K., Ohashi K., Matsuzaka T., Najima Y., Sekiya M., Tomita S., Okazaki H., Tamura Y., et al. 2005. Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur. J. Cancer. 41: 1316–1322. [DOI] [PubMed] [Google Scholar]
- 14.Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C. N., Bahre H., et al. 2014. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20: 1327–1333. [DOI] [PubMed] [Google Scholar]
- 15.Kim K-H. 1997. Regulation of mammalian acetyl-coenzyme A carboxylase. Annu. Rev. Nutr. 17: 77–99. [DOI] [PubMed] [Google Scholar]
- 16.McGarry J. D., Mannaerts G., and Foster D. W.. 1977. A possible role for malonyl-CoA in the regulation of hepatic fatty acid oxidation and ketogenesis. J. Clin. Invest. 60: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saggerson D. 2008. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu. Rev. Nutr. 28: 253–272. [DOI] [PubMed] [Google Scholar]
- 18.Sumpio B. E., Riley J. T., and Dardik A.. 2002. Cells in focus: endothelial cell. Int. J. Biochem. Cell Biol. 34: 1508–1512. [DOI] [PubMed] [Google Scholar]
- 19.Pearson J. D. 2000. Normal endothelial cell function. Lupus. 9: 183–188. [DOI] [PubMed] [Google Scholar]
- 20.Lamalice L., Le Boeuf F., and Huot J.. 2007. Endothelial cell migration during angiogenesis. Circ. Res. 100: 782–794. [DOI] [PubMed] [Google Scholar]
- 21.Michaelis U. R. 2014. Mechanisms of endothelial cell migration. Cell. Mol. Life Sci. 71: 4131–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y., Volrath S. L., Weatherly S. C., Elich T. D., and Tong L.. 2004. A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product. Mol. Cell. 16: 881–891. [DOI] [PubMed] [Google Scholar]
- 23.Cho Y. S., Lee J. I., Shin D., Kim H. T., Jung H. Y., Lee T. G., Kang L-W., Ahn Y-J., Cho H-S., and Heo Y-S.. 2010. Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK. Biochem. Biophys. Res. Commun. 391: 187–192. [DOI] [PubMed] [Google Scholar]
- 24.Gerth K., Pradella S., Perlova O., Beyer S., and Müller R.. 2003. Myxobacteria: proficient producers of novel natural products with various biological activities—past and future biotechnological aspects with the focus on the genus Sorangium. J. Biotechnol. 106: 233–253. [DOI] [PubMed] [Google Scholar]
- 25.Ades E. W., Candal F. J., Swerlick R. A., George V. G., Summers S., Bosse D. C., and Lawley T. J.. 1992. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J. Invest. Dermatol. 99: 683–690. [DOI] [PubMed] [Google Scholar]
- 26.Nicoletti I., Migliorati G., Pagliacci M. C., Grignani F., and Riccardi C.. 1991. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods. 139: 271–279. [DOI] [PubMed] [Google Scholar]
- 27.Koeberle A., Shindou H., Koeberle S. C., Laufer S. A., Shimizu T., and Werz O.. 2013. Arachidonoyl-phosphatidylcholine oscillates during the cell cycle and counteracts proliferation by suppressing Akt membrane binding. Proc. Natl. Acad. Sci. USA. 110: 2546–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harada N., Oda Z., Hara Y., Fujinami K., Okawa M., Ohbuchi K., Yonemoto M., Ikeda Y., Ohwaki K., and Aragane K.. 2007. Hepatic de novo lipogenesis is present in liver-specific ACC1-deficient mice. Mol. Cell. Biol. 27: 1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savage D. B., Choi C. S., Samuel V. T., Liu Z-X., Zhang D., Wang A., Zhang X-M., Cline G. W., Yu X. X., and Geisler J. G.. 2006. Reversal of diet-induced hepatic steatosis and hepatic insulin resistance by antisense oligonucleotide inhibitors of acetyl-CoA carboxylases 1 and 2. J. Clin. Invest. 116: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fullerton M. D., Galic S., Marcinko K., Sikkema S., Pulinilkunnil T., Chen Z. P., O’Neill H. M., Ford R. J., Palanivel R., O’Brien M., et al. 2013. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat. Med. 19: 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vahlensieck H. F., Pridzun L., Reichenbach H., and Hinnen A.. 1994. Identification of the yeast ACC1 gene product (acetyl-CoA carboxylase) as the target of the polyketide fungicide soraphen A. Curr. Genet. 25: 95–100. [DOI] [PubMed] [Google Scholar]
- 33.Gerth K., Bedorf N., Irschik H., Höfle G., and Reichenbach H.. 1994. The soraphens: a family of novel antifungal compounds from Sorangium cellulosum (Myxobacteria). J. Antibiot. (Tokyo). 47: 23–31. [DOI] [PubMed] [Google Scholar]
- 34.Baron A., Migita T., Tang D., and Loda M.. 2004. Fatty acid synthase: a metabolic oncogene in prostate cancer? J. Cell. Biochem. 91: 47–53. [DOI] [PubMed] [Google Scholar]
- 35.Scott K. E., Wheeler F. B., Davis A. L., Thomas M. J., Ntambi J. M., Seals D. F., and Kridel S. J.. 2012. Metabolic regulation of invadopodia and invasion by acetyl-CoA carboxylase 1 and de novo lipogenesis. PLoS One. 7: e29761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swinnen J. V., Van Veldhoven P. P., Timmermans L., De Schrijver E., Brusselmans K., Vanderhoydonc F., Van de Sande T., Heemers H., Heyns W., and Verhoeven G.. 2003. Fatty acid synthase drives the synthesis of phospholipids partitioning into detergent-resistant membrane microdomains. Biochem. Biophys. Res. Commun. 302: 898–903. [DOI] [PubMed] [Google Scholar]
- 37.Yeagle P. L. 1985. Cholesterol and the cell membrane. Biochim. Biophys. Acta. 822: 267–287. [DOI] [PubMed] [Google Scholar]
- 38.Kuhry J-G., Fonteneau P., Duportail G., Maechling C., and Laustriat G.. 1983. TMA-DPH: a suitable fluorescence polarization probe for specific plasma membrane fluidity studies in intact living cells. Cell Biophys. 5: 129–140. [DOI] [PubMed] [Google Scholar]
- 39.Byfield F. J., Aranda-Espinoza H., Romanenko V. G., Rothblat G. H., and Levitan I.. 2004. Cholesterol depletion increases membrane stiffness of aortic endothelial cells. Biophys. J. 87: 3336–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattila P. K., and Lappalainen P.. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9: 446–454. [DOI] [PubMed] [Google Scholar]
- 41.Faix J., and Rottner K.. 2006. The making of filopodia. Curr. Opin. Cell Biol. 18: 18–25. [DOI] [PubMed] [Google Scholar]
- 42.Baenke F., Peck B., Miess H., and Schulze A.. 2013. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 6: 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mounier C., Bouraoui L., and Rassart E.. 2014. Lipogenesis in cancer progression. Int. J. Oncol. 45: 485–492. [DOI] [PubMed] [Google Scholar]
- 44.Liu H., Liu J-Y., Wu X., and Zhang J-T.. 2010. Biochemistry, molecular biology, and pharmacology of fatty acid synthase, an emerging therapeutic target and diagnosis/prognosis marker. Int. J. Biochem. Mol. Biol. 1: 69–89. [PMC free article] [PubMed] [Google Scholar]
- 45.Wei X., Schneider J. G., Shenouda S. M., Lee A., Towler D. A., Chakravarthy M. V., Vita J. A., and Semenkovich C. F.. 2011. De novo lipogenesis maintains vascular homeostasis through endothelial nitric-oxide synthase (eNOS) palmitoylation. J. Biol. Chem. 286: 2933–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patella F., Schug Z. T., Persi E., Neilson L. J., Erami Z., Avanzato D., Maione F., Hernandez-Fernaud J. R., Mackay G., Zheng L., et al. 2015. Proteomics-based metabolic modeling reveals that fatty acid oxidation (FAO) controls endothelial cell (EC) permeability. Mol. Cell. Proteomics. 14: 621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoors S., De Bock K., Cantelmo A. R., Georgiadou M., Ghesquiere B., Cauwenberghs S., Kuchnio A., Wong B. W., Quaegebeur A., Goveia J., et al. 2014. Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metab. 19: 37–48. [DOI] [PubMed] [Google Scholar]
- 48.Wakil S. J., and Abu-Elheiga L. A.. 2009. Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 50: S138–S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svensson R. U., Parker S. J., Eichner L. J., Kolar M. J., Wallace M., Brun S. N., Lombardo P. S., Van Nostrand J. L., Hutchins A., Vera L., et al. 2016. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22: 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galbraith C. G., Yamada K. M., and Galbraith J. A.. 2007. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 315: 992–995. [DOI] [PubMed] [Google Scholar]
- 51.Seano G., and Primo L.. 2015. Podosomes and invadopodia: tools to breach vascular basement membrane. Cell Cycle. 14: 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braig S., Schmidt B. S., Stoiber K., Händel C., Möhn T., Werz O., Müller R., Zahler S., Koeberle A., and Käs J. A.. 2015. Pharmacological targeting of membrane rigidity: implications on cancer cell migration and invasion. New J. Phys. 17: 083007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.