Abstract
Serum amyloid A (SAA) is a family of acute-phase reactants. Plasma levels of human SAA1/SAA2 (mouse SAA1.1/2.1) can increase ≥1,000-fold during an acute-phase response. Mice, but not humans, express a third relatively understudied SAA isoform, SAA3. We investigated whether mouse SAA3 is an HDL-associated acute-phase SAA. Quantitative RT-PCR with isoform-specific primers indicated that SAA3 and SAA1.1/2.1 are induced similarly in livers (∼2,500-fold vs. ∼6,000-fold, respectively) and fat (∼400-fold vs. ∼100-fold, respectively) of lipopolysaccharide (LPS)-injected mice. In situ hybridization demonstrated that all three SAAs are produced by hepatocytes. All three SAA isoforms were detected in plasma of LPS-injected mice, although SAA3 levels were ∼20% of SAA1.1/2.1 levels. Fast protein LC analyses indicated that virtually all of SAA1.1/2.1 eluted with HDL, whereas ∼15% of SAA3 was lipid poor/free. After density gradient ultracentrifugation, isoelectric focusing demonstrated that ∼100% of plasma SAA1.1 was recovered in HDL compared with only ∼50% of SAA2.1 and ∼10% of SAA3. Thus, SAA3 appears to be more loosely associated with HDL, resulting in lipid-poor/free SAA3. We conclude that SAA3 is a major hepatic acute-phase SAA in mice that may produce systemic effects during inflammation.
Keywords: adipose tissue, animal models, inflammation, liver
Acute-phase serum amyloid A (SAA) is a family of acute-phase proteins that have been evolutionarily conserved for approximately 500 million years (1, 2). Teleologically, this conservation suggests an important role for SAA in host defense. The precise functions of the SAA family have not been defined, although it is suggested that SAA has antibacterial properties (3) and can serve as a chemoattractant for monocytes and neutrophils (4). SAA can also promote inflammation by inducing inflammatory cytokines, such as IL-1β, TNF-α, and IL-6, as well as metalloproteinases in smooth muscle cells and macrophages (5–9). SAA may act as a danger-associated molecular pattern molecule by interacting with multiple pattern recognition receptors, such as Toll-like receptors, CD36, and FPRL1 (1). The pro-inflammatory functions of SAA should be seen in the context of recent data that SAA is an innate immune molecule capable of activating the NLR family pyrin domain containing 3 (NLRP3) inflammasome, although the precise molecular events underlying this activation are not completely understood (10–13). SAA has a strong association with cardiovascular disease, where it can serve as a predictor of mortality (14). The key question is whether SAA serves as more than a marker, but rather as a mediator of acute vascular syndromes (15).
The SAA family comprises at least four isoforms that likely arose from gene duplication. In humans, two SAA isoforms (SAA1 and SAA2) are highly induced during an acute-phase response, with plasma levels increasing up to 1,000-fold or more. SAA1 and SAA2 are predominantly expressed in the liver, but also in extrahepatic tissues, and are found in plasma associated with HDL (16). The mouse homologs (SAA1.1 and SAA2.1) are similarly induced and share more than 90% amino acid identity (2). The SAA4 isoform is an HDL-associated apolipoprotein that is ∼55% homologous to SAA1/2 and constitutively expressed at relatively low levels in both human and mouse liver. To date, no known immune or inflammatory activities have been ascribed to SAA4 (17, 18) and its function is largely unknown.
Mice express a third acute-phase SAA isoform, SAA3, which is not expressed in humans due to a premature stop codon in the human Saa3 gene (19). Mouse SAA3 is 69% homologous to SAA1.1/2.1 (2) and is induced in the liver as well as extrahepatic tissues (20), particularly adipocytes, and to a lesser extent in macrophages in inflammatory settings. Prior studies have also reported SAA3 expression in testis, spleen, intestine, and kidney in acute inflammatory states (21), as well as in intestinal epithelial cells (22), lymphocytes, lymphoid follicles, and plasma cells of many normal tissues (23). In general, SAA3 is considered to be the “extrahepatic” SAA, exerting similar pro-inflammatory activities in mouse macrophages (24) and adipose tissue (21, 25) as human SAA1 and SAA2. Whether SAA3 contributes to systemic SAA levels in mice has been the focus of a limited number of studies, with equivocal results. Whereas SAA3 was determined to be a circulating SAA bound to HDL in mice undergoing an acute inflammatory response to lipopolysaccharaide (LPS) injection (24, 26), it was not detected in HDL in plasma from ob/ob mice with chronic low-grade inflammation (25). The goals of the present study were to carefully define the degree to which SAA3 contributes to circulating SAA during an acute-phase response, the extent to which SAA3 is associated with HDL, and its cellular sources.
MATERIALS AND METHODS
Animals
C57BL/6 mice were obtained from Jackson Laboratory, Bar Harbor, ME. Mice deficient in SAA1.1 and SAA2.1 were bred to obtain a >99.9% C57BL/6 background (27). Mice deficient in SAA3 have been described (28). For simplicity, C57BL/6 mice, mice lacking SAA1.1/SAA2.1, and mice lacking SAA3 are referred to as SAA-WT, SAA1.1/2.1-DKO, and SAA3-KO, respectively. An acute-phase response was elicited by intraperitoneal injection of 25 or 100 μg LPS (Escherichia coli 0111:B4; Sigma Chemical Co.), then mice were humanely euthanized and plasma and tissue were obtained for analyses and preparation of HDL (LPS dose and duration for each experiment are indicated in the figure legends). Control animals were not injected. All studies were performed with the approval of the University of Kentucky or University of Washington Institutional Animal Care and Use Committee.
Primary hepatocyte cultures
Primary hepatocytes were isolated from SAA-WT and SAA1.1/2.1-DKO mice 6 h after injection with 25 μg LPS using a two-step perfusion method (29). Briefly, the liver was first perfused with Ca+2/Mg+2-free HBSS containing 10 mM glucose, 10 mM HEPES, and 0.3 mM EDTA and then with HBSS containing 0.05% collagenase type IV (C5138; Sigma), 1.3 mM CaCl2, 0.5 mM MgCl2, 10 mM glucose, and 10 mM HEPES. Hepatocytes were washed by repeated low speed centrifugation (50 g for 2 min) and cell viability (normally 90–95%) was assessed by trypan blue exclusion. Cells were plated onto 12-well plates precoated with rat tail collagen (354236; BD Biosciences, Bedford, MA) at a density of 2 × 105 cells/well and cultured at 37°C under 5% CO2 in Williams’ Medium E (GIBCO) containing 10% fetal bovine serum (GIBCO), 2% penicillin-streptomycin, 1% sodium pyruvate, 1% l-glutamine, and 1% insulin-transferrin-selenium (GIBCO). Cells were cultured for 18 h after plating and then collected for RNA isolation.
RNA isolation and RT-PCR
Total RNA was isolated from mouse livers and hepatocytes using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA from epididymal fat was isolated by RNeasy lipid tissue mini kit (7484; Qiagen). RNA samples (9 μg) were treated with DNase 1 (TURBO DNA-free™ kit (AM1907; Invitrogen) for 30 min at 37°C. RNA from liver, epididymal fat (0.5 μg), or cultured cells (0.1 μg) was reverse transcribed into cDNA using the reverse transcription system (4368814; Applied Biosystems). After 4-fold dilution, 5 μl were used as a template for real-time RT-PCR. Amplification was done for 40 cycles using Power SYBR Green PCR Master Mix kit (4367659; Applied Biosystems). Quantification of mRNA was performed using the ΔΔCT method and normalized to GAPDH for liver and hepatocytes or 36B4 for epididymal fat. Primer sequences are as follows: GAPDH (accession number NM_008084) 5′-CTC ATG ACC ACA GTC CAT GCC A-3′, 5′-GGA TGA CCT TGC CCA CAG CCT T-3′ 36B4 (accession number NM_007475) 5′-AAG CGC GTC CTG GCA TTG TCT-3′, 5′-CCG CAG GGG CAG CAG TGG T-3′ SAA1.1/2/1 (accession number NM_009117.3) 5′-CTC CTA TTA GCT CAG TAG GTT GTG-3′, 5′-CAC TTC CAA GTT TCT GTT TAT TAC CC-3; SAA3 (accession number NM_011315.3) 5′-TTT CTC TTC CTG TTG TTC CCA GTC-3′, 5′- TCA CAA GTA TTT ATT CAG CAC ATT GGG A-3′.
Electrofocusing and immunochemical analysis
Aliquots of plasma (7 μl) or HDL (13.5 μg) isolated from LPS-injected mice were subjected to isoelectric focusing (IEF), as previously described (26), using an Ampholine gradient consisting of 20% (v/v) Ampholine (pH 3–10), 40% (v/v) Ampholine (pH 4–6.5), and 40% (v/v) Ampholine (pH 7–9) (Pharmacia LKB Biotechnology Inc.). Electrofocused samples were subjected to immunochemical analysis, as described (26). The SAA isoforms were identified by immunochemical staining utilizing rabbit anti-mouse SAA antibody to identify SAA1.1 and SAA2.1 and rabbit anti-mouse SAA3 (a gift from Dr. Phillip Scherer, University of Texas Southwestern) to identify SAA3.
Plasma lipoprotein analysis and SAA measurement
Lipoproteins in 50 μl of plasma pooled from six mice injected with 100 μg LPS were separated by fast protein (FP)LC utilizing a Superose 6 column (17-5172-01; GE Healthcare, Uppsala, Sweden). The cholesterol content of 0.5 ml fractions was determined. SAA1.1/2.1 and SAA3 in plasma and FPLC fractions were determined by a mouse SAA ELISA kit (TP802M; Tridelta Development Ltd., Ireland) and a mouse SAA3 ELISA kit (EZMSAA3-12K; EMD Millipore, Danvers, MA), respectively.
Mass spectrometric analysis
HDL from mice that were injected with 25 μg LPS were subjected to SDS-PAGE or IEF. Coomassie-stained bands corresponding to the apparent molecular mass of SAA separated by SDS-PAGE (∼12 kDa) or the isoelectric point (pI) of SAA3 separated by IEF (∼9.2) were excised, washed, and subjected to in-gel trypsin digestion (30). The resulting tryptic peptides were extracted and subjected to LC-MS/MS using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). The LC-MS/MS results were searched against the mouse portion of the UniProt database using a local Mascot server (version 2.3; Matrix Science) for protein identification, including methionine oxidation and cysteine carbamidomethylation as allowed side chain modifications. A false discovery rate of 1% was used in decoy search for the high-confidence peptides. Proteins with a score of at least 30 for single high-confidence peptides were considered positive identifications. UniProt protein names and identifier numbers are used throughout this work.
Immunohistochemistry
Paraffin-embedded liver sections (8 μm thick) were fixed in 4% paraformaldehyde and incubated with 3% H2O2 for 10 min to quench endogenous peroxidases. After blocking endogenous biotin using an avidin/biotin blocking kit (Vector Laboratories, Burlingame, CA), slides were incubated overnight at 4°C with rabbit anti-mouse SAA (26) (1:2,000 dilution). Sections were subsequently incubated with biotinylated anti-rabbit IgG and avidin-peroxidase according to the vendor’s instructions (Vectastain Elite ABC kit; Vector Laboratories). Immunoreactivity was visualized using a DAB kit (K3468; Dako North America, Inc., Carpinteria, CA), followed by counterstaining with hematoxylin. Immunoreactivity was visualized using the red chromogen, 3-amino-9-ethyl carbazole (Vector Laboratories). Images were captured on a Nikon ECLIPSE 80i microscope with the aid of NIS-Elements BR 4.00.08 software.
In situ hybridization
In situ hybridization was performed essentially as described (31). For PCR amplification of liver cDNA, the following SAA gene-specific primers were used: murine SAA1.1/2.1 (NM_009117/NM_011314) forward primer: 5′-ctcctattagctcagtaggttgtg-3′, reverse primer: 5′-cacttccaagtttctgtttattaccc-3′ and murine SAA3 (NM_011315) forward primer: 5′-tttctcttcctgttgttcccagtc-3′, reverse primer: 5′-tcacaagtatttattcagcacattggg-3′. PCR was performed using 5 μl cDNA in a total volume of 50 μl containing 0.2 μM primer mix, 1× PCR buffer, 3.0 mM MgCl2, 200 μM dNTP mixture, and 1 U GoTaq Flexi DNA polymerase (Promega) and the following program: 2 min at 95°C, 40 cycles at (45 s at 95°C, 45 s at 56°C, and 45 s at 72°C), and a 10 min extension at 72°C. The PCR products were subcloned into pGEM-T (Promega) and their sequence identity was confirmed by double-stranded DNA sequencing. Plasmid constructs were linearized by SphI and NdeI for SAA1.1/2.1 and SAA3. Riboprobes in antisense and sense orientation were generated from linearized plasmid constructs by in vitro transcription using the appropriate RNA polymerases and 35S-UTP in a volume of 10 μl containing 1 μg of linearized plasmids. After 90 min incubation at 37°C, 10 U of RNase-free DNase I were added to the reactions and incubated for another 15 min. After adding RNase-free water to 20 μl and 20 μl sodium carbonate buffer [80 mM NaHCO3 and 120 mM Na2CO3 (pH 10.2)] for limited hydrolysis, the reaction was incubated at 60°C for the appropriate time [t = (Lo – Lf)/K × Lo × Lf, where Lo is the cDNA length, Lf is the expected length of the probes (i.e., 250 bp), and K is 0.11], as described (31). The reaction was stopped by adding 2 μl of 10% acetic acid. The probes were purified using Micro Bio-Spin P-30 columns (Bio-Rad). For in situ hybridization histochemistry, frozen 8 μm-thick liver sections were fixed in 4% paraformaldehyde for 60 min. After three washes in PBS and incubation in 0.4% Triton X-100 for 10 min, the slides were rinsed in deionized water and transferred to 0.1 M triethanolamine (pH 8.0). After 1 min incubation, acetic anhydride was added under strong stirring to a final concentration of 0.25% (v/v) and further incubated for 10 min. The slides were washed for 10 min in PBS and rinsed in deionized water prior to dehydration in 50% and 70% ethanol. Radioactive probes were diluted to 5 × 104 dpm/μl in hybridization solution [600 mM NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA-Na2, 0.05% tRNA, 20 mM dithiothreitol, 1× Denhardt’s solution, 10% dextran sulfate, 100 μg/ml sonicated salmon sperm DNA, and 50% formamide]. Prehybridized sections were incubated with 100 μl hybridization solution containing radioactive RNA probes at 60°C in a formamide humid chamber box for 16 h. After a series of washes, the sections were dehydrated in 50% and 70% ethanol. For autoradiographic detection, slides were coated with Kodak NTB-2 nuclear emulsion (Eastman Kodak, Rochester, NY) and exposed at 4°C for 5–30 days in the dark. Slides were developed in Kodak D19 solution and counterstained with cresyl violet. Photographic documentation was performed under bright-field illumination using a Nikon ECLIPSE 55i microscope with the aid of NIS-Elements BR 4.13.04 software.
Statistical analysis
Data are expressed as mean ± SD. Results for Figs. 2 and 3B were analyzed by one-way ANOVA followed by a Bonferroni posttest. Statistical analysis for Fig. 4D was performed using the two-tailed Student’s t-test for unpaired data. P < 0.05 was considered statistically significant. All statistical analyses were carried out using GraphPad Prism 5.
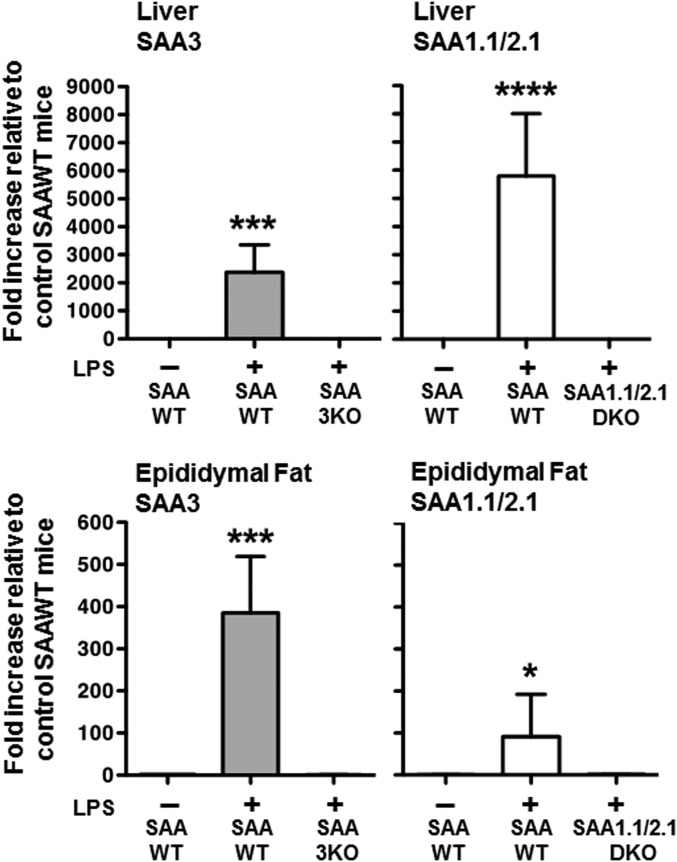
Fig. 2.
SAA 1.1/2.1 and SAA3 expression in mouse liver and adipose tissue. Semi-quantitative RT-PCR using isoform-specific primers was performed using cDNA prepared from 0.5 μg liver RNA (top panel) and 0.5 μg epididymal fat RNA (bottom panel), isolated from control SAA-WT or LPS-injected (100 μg LPS) SAA-WT, SAA1.1/2.1-KO, and SAA3-KO mice. Values are the mean ± SD, n = 3–6. *P < 0.05; ***P < 0.001; ****P < 0.0001.
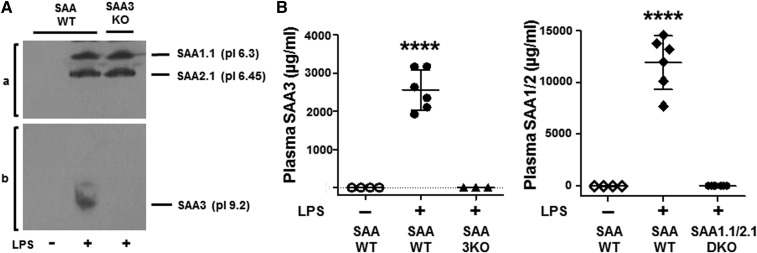
Fig. 3.
SAA protein in plasma. A. SAA isoforms in plasma of untreated SAA-WT and LPS-injected (100 μg LPS) SAA-WT and SAA3-KO mice were separated by IEF utilizing an Ampholine gradient, as described in the Materials and Methods, and pressure-blotted onto nitrocellulose. The SAA isoforms were identified by immunochemical staining utilizing rabbit anti-mouse SAA antibody to identify SAA1.1 and SAA2.1 (a) and rabbit anti-mouse SAA3 to identify SAA3 (b). B: SAA3 and SAA1.1/2.1 in the plasma of control SAA-WT and LPS-injected (100 μg LPS) SAA-WT, SAA3-KO, and SAA1.1/2.1-DKO mice were measured by ELISA utilizing kits specific for the mouse SAA isoforms. Values are the mean ± SD, n = 3–6. ****P < 0.0001.
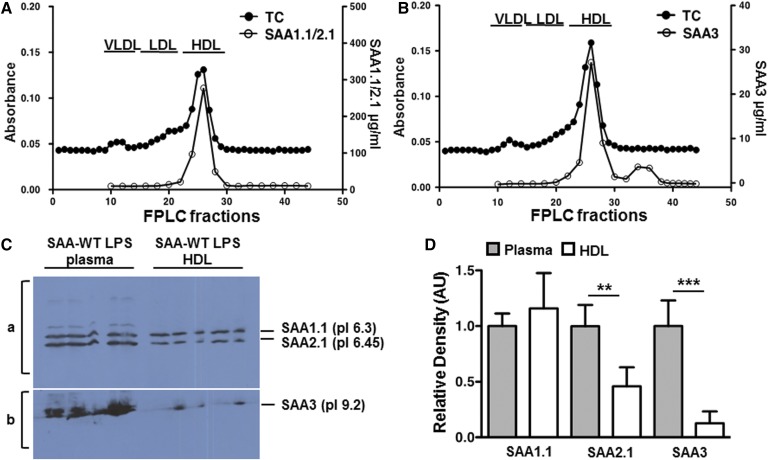
Fig. 4.
SAA3 association with HDL. Pooled plasma from six SAA-WT mice injected with 100 μg LPS was separated by FPLC and the total cholesterol (TC) content of eluted fractions was measured to determine the lipoprotein profile (y axis of panels A and B). The SAA1.1/2.1 content of FPLC fractions (A) and SAA3 content of FPLC fractions (B) were measured by isoform-specific ELISAs. C: HDL (d 1.063–1.21) was isolated by sequential density gradient ultracentrifugation from the plasma of SAA-WT mice injected with 25 μg LPS. Aliquots of plasma (7 μl) and HDL (13.5 μg) were subjected to IEF utilizing an Ampholine gradient, as described in the Materials and Methods, to separate the SAA isoforms, followed by pressure-blotting onto nitrocellulose. Equivalent amounts of HDL-cholesterol were loaded in each lane. SAA1.1 and SAA2.1 were detected with a rabbit anti-SAA antibody (a) and SAA3 was detected by an anti-mouse SAA3 antibody (gift of Dr. Phillip Scherer, University of Texas Southwestern) (b). D: Immunopositive bands from C were quantified by densitometric scanning and the values for isolated HDL were expressed relative to the corresponding values for plasma, which were arbitrarily given a value of 1. **P < 0.01, ***P < 0.001.
RESULTS
SAA1.1/2.1 and SAA3 expression in mouse liver
The classical murine acute-phase reactants, SAA1.1 and SAA2.1, are expressed primarily in the livers of mice and are dramatically upregulated following an inflammatory stimulus (20). In contrast to SAA1.1 and SAA2.1, SAA3 is generally thought to be mainly expressed extra-hepatically, predominantly by adipocytes, but also by macrophages (21, 24). However, the relative expression of the various mouse acute-phase SAA isoforms in tissues and plasma has not been adequately documented, partly due to the lack of validated reagents that distinguish SAA1.1/2.1 and SAA3. The recent development of mice deficient in SAA1.1/2.1 (27) or SAA3 (28) allowed us to confirm the isoform specificity of the reagents used in our study to distinguish SAA3 from the other two acute-phase SAA isoforms. As expected, SAA1.1/2.1 mRNA was readily detected by RT-PCR in the liver of a mouse injected with LPS, but not a control mouse liver (Fig. 1A). Notably, SAA3 mRNA appeared to be similarly induced by LPS in SAA-WT mouse liver. The specificity of the primers used to amplify SAA1.1/2.1 and SAA3 cDNAs was demonstrated by the absence of RT-PCR products in LPS-injected SAA1.1/2.1-DKO and SAA3-KO mouse livers, respectively (Fig. 1A).
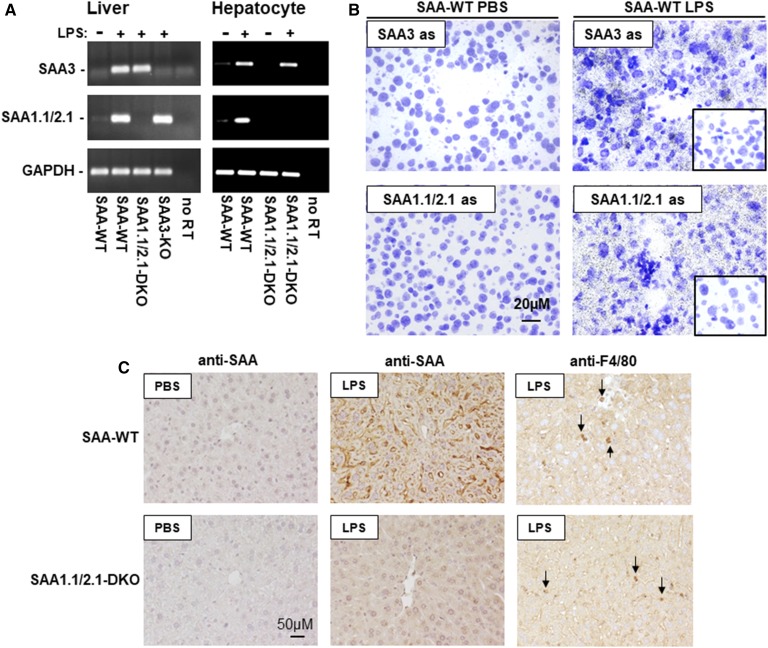
Fig. 1.
SAA 1.1/2.1 and SAA3 in mouse liver. A: Total RNA was isolated from the livers of noninjected SAA-WT mice, and SAA-WT, SAA1.1/2.1-DKO, and SAA3-KO mice injected with 100 μg LPS, as well as primary hepatocytes isolated from SAA-WT and SAA1.1/2.1-DKO mice injected with 25 μg LPS or PBS, as indicated. RT-PCR was performed using primers specific for SAA3 and SAA1.1/2.1. B: High-resolution bright-field images of emulsion-coated liver sections from PBS- or LPS-injected SAA-WT mice hybridized with SAA3- or SAA1.1/2.1-anti-sense probes. Inserts in the right-hand panels represent hybridization of SAA3- or SAA1.1/2.1-sense probes to liver sections of an LPS-injected SAA-WT mouse. Magnification, 40× scale bar, 20 μM; exposure times, 5 days for SAA1.1/2.1 and 30 days for SAA3. C: Immunostaining of liver sections from a SAA-WT or SAA1.1/2.1-DKO mice injected with either PBS or LPS (25 μg), as indicated, using antibodies that detect SAA1.1/2.1 and SAA3 (anti-SAA) or macrophages (anti-F4/80). Images were photographed under 20× objective magnification (scale bar, 50 μM). Arrows in the right panels indicate immunopositive staining for F4/80 (red/brown chromogen).
It was of interest to determine whether SAA3 expressed in acute-phase mouse liver was due to induction in hepatocytes or other hepatic cell types, such as resident macrophages. As a first approach to address this question, primary hepatocytes were isolated from SAA-WT and SAA1.1/2.1-DKO mice 6 h after injection with 25 μg LPS, and then cultured for 18 h prior to RNA isolation. Results from RT-PCR indicated that hepatocytes were induced to express SAA3 during an acute-phase response (Fig. 1A). We next performed in situ hybridization as another approach to investigate which cell type(s) express SAA3 mRNA in acute-phase mouse liver (Fig. 1B). Cresyl violet-stained liver sections showed dense signals for SAA mRNA in liver sections of LPS-injected SAA-WT mice, but not PBS-injected mice (Fig. 1B). Control experiments using sense-strand riboprobes demonstrated the specificity of the hybridization signals (inserts on right panels, Fig. 1B). Moreover, the broad distribution of the hybridization signal throughout the tissue section suggested that SAA3 mRNA expression is not limited to resident macrophages (i.e., Kuppfer cells) in acute-phase mouse liver, and that hepatocytes are likely an important source of SAA3 expressed in the liver during an inflammatory response. This conclusion was supported by results from immunohistochemical staining using a polyclonal anti-SAA antibody (Fig. 1C). As expected, robust staining for SAA was observed in liver sections from LPS-injected SAA-WT mice, with a tissue distribution consistent with expression by hepatocytes. Similarly distributed immunopositive staining was also evident in liver sections of an LPS-injected SAA1.1/2.1-DKO mouse, albeit at less intensity compared with staining in LPS-injected SAA-WT mice. Immunoreactivity was negligible in control SAA-WT and SAA1.1/2.1-DKO mouse liver, strongly suggesting that the signal observed in LPS-injected SAA1.1/2.1-DKO mouse liver represents SAA3. Moreover, the diffuse immunopositive staining for SAA1.1/2.1-DKO sections stained with anti-SAA was distinct from the signal observed for anti-F4/80, indicating that Kupffer cells are not likely to be the only source of SAA3 in mouse liver (Fig. 1C). Taken together, our data indicate that SAA3 is a highly inducible acute-phase reactant in mouse hepatocytes.
SAA1.1/2.1 and SAA3 expression in mouse adipose tissue
To address the widely held concept that SAA3 is the predominant isoform expressed in extra-hepatic tissues, we next determined the extent to which each of the three acute-phase SAA isoforms were induced above baseline in liver and epididymal fat of mice 19 h after injection with 100 μg LPS (Fig. 2). The values for LPS-treated mice are shown relative to those of control (non-LPS injected) SAA-WT mice, which were arbitrarily given a value of 1. SAA3 and SAA1.1/2.1 expression was significantly upregulated in the livers of LPS-treated SAA-WT mice (P = 0.0004 and P < 0.0001, respectively), and not detected in their corresponding LPS-treated KO controls. The increase in expression of SAA3 (∼2,500-fold) was somewhat less robust compared with SAA1.1/2.1 (∼6,000-fold). In contrast to liver, SAA3 was the more inducible SAA in epididymal fat, increasing ∼400-fold in epididymal fat of LPS-treated SAA-WT (P = 0.0002), compared with ∼100-fold induction of SAA1.1/2.1 (P = 0.047). Thus, while SAA3 appears to be a major SAA isoform in fat during inflammation, its expression in adipose tissue appears to be greatly overshadowed by a much more robust induction of SAA3 in hepatocytes.
SAA3 protein in plasma
According to a previous report, SAA3 is induced in adipose tissue of obese mice, but is not detected on circulating HDL (25). Thus, it was of interest to determine whether SAA3 is present in plasma of mice undergoing an acute-phase response. Plasma was collected from untreated SAA-WT mice and SAA-WT and SAA3-KO mice after LPS injection and separated by IEF. The Ampholine gradient for IEF was optimized to maximize the separation of SAA1.1 (pI 6.3), SAA2.1 (pI 6.45), and the more basic SAA3 (pI 9.2) (26). This technique confirmed the presence of SAA1.1/2.1 in the plasma of LPS-treated SAA-WT and SAA3-KO mice, but not in the plasma of untreated SAA-WT mice (Fig. 3A). An immunopositive band that migrated at the expected pI of SAA3 was also detected in LPS-injected SAA-WT mice, but not in SAA3-KO mice or untreated SAA-WT mice (Fig. 3A). The relative plasma concentrations of SAA isoforms in acute-phase SAA-WT mice were estimated by ELISA using two commercially available kits that were confirmed to be specific for either SAA1.1/2.1 or SAA3 using plasma from LPS-treated SAA1.1/2.1-DKO and SAA3-KO mice, respectively (Fig. 3B). Based on these assays, plasma SAA3 concentrations (∼2.5 mg/ml) corresponded to ∼20% of plasma SAA1.1/2.1 (∼12.0 mg/ml) in acute-phase mouse plasma.
SAA3 association with HDL
It is well-established that the vast majority of SAA1.1/2.1 in plasma is associated with HDL. SAA3 has also been reported to be on the HDL fraction after LPS injection (24, 26), but not in obese mice (25). To directly address whether there are differences in SAA1.1/2.1 and SAA3 association with HDL, plasma from LPS-injected SAA-WT mice was separated by FPLC and fractions were analyzed for SAA content. As expected, virtually all of the SAA1.1/SAA2.1 eluted in HDL-containing fractions (Fig. 4A). Interestingly, whereas the majority of plasma SAA3 was associated with the HDL peak, ∼15% eluted in fractions beyond the HDL peak (Fig. 4B), suggesting a portion of plasma SAA3 exists in a lipid-poor or lipid-free form. To investigate this finding further, HDL was isolated from acute-phase SAA-WT mouse plasma by density gradient ultracentrifugation and aliquots were subjected to SDS-PAGE or IEF. LC-MS/MS analysis of Coomassie-stained bands corresponding to acute-phase SAAs on SDS-PAGE (∼12 kDa) and SAA3 on IEF (pI ∼9.2) confirmed that the isolated HDL fraction did indeed contain SAA3 (data not shown). However, the recovery of each of the SAA isoforms during the ultracentrifugation steps was different, as evident in the results presented in Fig. 4C, D. For this experiment, aliquots of acute-phase plasma and HDL isolated from the plasma (d 1.063–1.21) were applied to an IEF gel and then immunoblotted; the amount of HDL analyzed was calculated to match the HDL-cholesterol in electrofocused plasma (Fig. 4C, D). Interestingly, based on this analysis, there appeared to be a selective loss of SAA2.1 and even greater loss of SAA3 in the isolated acute-phase HDL preparations compared with plasma, suggesting dissociation of SAA2.1 and, to a greater extent, SAA3 from HDL during ultracentrifugal isolation.
DISCUSSION
The mouse is a widely utilized animal model to investigate the pathogenic role of SAA in chronic diseases. The human Saa1 and Saa2 and the mouse Saa1.1 and Saa2.1 genes are thought to be evolutionary homologs based on their sequence conservation, relative map positions, and transcriptional orientations (2). Mouse SAA3 shares 69% amino acid identity with human SAA1 (2), the major human SAA isoform expressed in the liver and extrahepatic tissues, including adipocytes (32). However, Saa3 is a pseudogene in humans (19), which has limited the interest in studying SAA3 biology in mice. Moreover, given a prior publication indicating that adipocyte-derived SAA3 from ob/ob mice, which are characterized by chronic low grade inflammation, does not circulate (25), it has been suggested that SAA3 exerts only localized effects in tissues and does not represent a systemic acute-phase SAA (28, 33). Results from the current study in which stimulation of SAA3 was more intense as a result of LPS injection indicate that the role of SAA3 in acute systemic inflammation in mice should be reassessed. We show that SAA3 is a hepatocyte-expressed acute-phase reactant that comprises a substantial portion (15–20%) of the total plasma SAA in endotoxemic mice. Unlike SAA1.1/2.1, the other acute-phase mouse SAAs, a portion of plasma SAA3 is present in a lipid-poor form, not associated with HDL. These findings may have important implications for previous studies of SAA in mice that focused only on SAA1.1/2.1.
Our data, in which an acute-phase response was induced by the injection of LPS, contrast with previous studies that failed to detect SAA3 in the circulation during a chronic low-grade inflammatory response due to obesity (25). Thus, it is likely that the discrepant findings are due to variations in the ability of different inflammatory stimuli to induce SAA3. For example, it was previously published that casein injection was relatively ineffective in inducing SAA3 in livers of mice compared with SAA1.1/2.1 (20). In another study, the conclusion that SAA3 is not a circulating acute-phase SAA was based on mass spectrometric analysis of HDL isolated by sequential density ultracentrifugation from mice in which SAA3 was relatively mildly induced by obesity (25). Our data indicate that a portion of plasma SAA3 (∼15%) is not associated with HDL, and that the bulk of HDL-associated SAA3 is stripped off the particle during ultracentrifugation. Although the relative recovery of plasma SAA2.1 was also lower compared with SAA1.1 during HDL isolation, we were unable to detect lipid-poor SAA2.1 in acute-phase mouse plasma. Thus, our study indicates that SAA3 is a major acute-phase SAA isoform that differs from SAA 1.1/2.1 in that it seems to be more loosely associated with HDL. Differences in the sequence of the hydrophobic N terminus, the lipid associating domain of SAA, may account for differences in HDL association for SAA1.1/2.1 and SAA3 (34).
The designation of SAA3 as the extrahepatic SAA appears to be a misnomer, given that the induction of Saa3 in mouse liver after LPS injection (∼2,500-fold) is of the same order of magnitude as Saa1.1/2.1 (∼6,000-fold). Because our oligonucleotide primers do not distinguish SAA1.1 and SAA2.1 mRNAs, RT-PCR quantification reflects the total abundance of both transcripts. Thus, it appears that the Saa1.1, Saa2.1, and Saa3 genes may be similarly induced in hepatocytes during an acute-phase response. Interestingly, the total amount of SAA1.1 and SAA2.1 protein detected in the plasma exceeded the amount of SAA3 by ∼5-fold. The modest discrepancy in mRNA and protein data may reflect ambiguities in assays, posttranscriptional differences in expression or secretion of the different SAA isoforms, or the susceptibility of lipid-poor SAA3 to be more rapidly cleared from the plasma. Whether SAA3 expressed in adipose or other extrahepatic tissues contributes to circulating SAA was not specifically addressed in this study. Our data are in agreement with other reports that SAA3 is more highly induced in adipose tissue compared with SAA1.1/2.1 (33). Nevertheless, it seems unlikely that adipose tissue-derived SAA3 makes a major contribution to total systemic SAA in endotoxemic mice, given the relative fold induction of SAA3 mRNA in fat (∼400-fold) compared with liver (∼2,500-fold).
The relative ability of different SAA isoforms to dissociate from HDL could hold functional implications. We and others previously reported that HDL-associated SAA, unlike lipid-poor SAA, does not activate pro-inflammatory signaling pathways (7, 35–37). Thus, in order to exert inflammatory effects, liver-derived SAA must dissociate from HDL in tissues. The mechanism by which this occurs is unknown, but may be mediated by interactions with proteoglycans, which are known to be involved in SAA deposition into amyloid fibrils (38, 39). Whether SAA3 may be particularly biologically active in vivo due to a higher propensity to spontaneously disassociate from HDL merits further study. Moreover, the respective contributions of hepatocyte-produced SAA (presumably associated with HDL) versus SAA derived from extrahepatic tissues (likely present in a lipid-poor form) in promoting inflammation also needs to be explored. As noted above, ∼15% of circulating SAA3 (∼2–3% of total SAA) in plasma of LPS-injected mice appears to be present in a lipid-poor form. Whether this minor component reflects SAA3 produced by extrahepatic tissues is not known, but it is notable that the fold-induction of SAA3 mRNA in adipose tissue corresponds to ∼15–20% of the fold-induction of SAA3 mRNA in the liver after LPS injection. It is possible that extrahepatic SAA (i.e., adipocyte- or macrophage-derived) produces inflammatory effects more potently than the abundant systemic SAA present on HDL particles. On the other hand, it is conceivable that HDL particles readily access extravascular spaces at sites of infection or tissue injury, and that SAA can be stripped off HDL to exert its biological actions.
As an innate immune molecule, SAA has been suggested to play a role in a significant number of chronic inflammatory diseases, including rheumatoid arthritis, cardiovascular disease, obesity, pulmonary diseases, and cancer (18). SAA has been shown to be a predictor of clinical outcomes in patients with acute coronary syndromes (14, 40, 41). Whether SAA plays a direct role in the pathogenesis of chronic inflammatory diseases, or is merely a marker of increased risk, has been the topic of intense investigations over the past several decades, and the recent development of mouse strains with altered SAA gene expression have provided important new insights. For example, we and others reported increased atherosclerosis development in apoE−/− mice overexpressing SAA1 (42, 43). However, deficiency of SAA1.1/2.1 has been shown to have no, or only modest, effects on atherosclerotic lipid deposition in apoE−/− and LDL receptor−/− mice, respectively (44, 45). The paradoxical findings may be due to the fact that the previous loss-of-function studies did not take into account the potential role of SAA3 in mouse atherosclerosis models. We recently determined that overexpression of SAA3 using an adeno-associated viral vector in apoE−/− mice resulted in a significant 4-fold increase in atherosclerosis lesion area compared with control apoE−/− mice, whereas anti-sense oligonucleotide-mediated suppression of SAA3 decreased atherosclerosis in apoE−/− mice deficient in SAA1.1/2.1 (46). On the other hand, recent studies from our laboratory determined that apoE−/− mice deficient in only SAA1.1/2.1 are protected from angiotensin II-induced abdominal aortic aneurysm (AAA) formation, suggesting a minor role for SAA3 in experimental AAA (6). In general, there is a lack of studies to address whether there are functional differences in SAA1.1, SAA2.1, and SAA3. Mouse SAA1.1 and SAA2.1 and human SAA1 and SAA2 are highly homologous (∼90% amino acid conservation) and coordinately regulated in cells, leading to the assumption that they are functionally redundant. However, differences in function have been identified. Most notably, SAA1/SAA1.1, and not SAA2/SAA2.1, has a propensity to be deposited extracellularly as insoluble amyloid fibrils (26). Modest differences in the capacity to promote cellular cholesterol efflux and interact with pattern recognition receptors have also been noted for SAA1.1 and SAA2.1 (47). Based on available data, it appears that SAA3 shares activities similar to SAA1.1/2.1, although more studies are needed. As noted above, our finding that lipid-poor SAA3 is present in plasma during the acute-phase response may have important functional implications. Several researchers have pointed to SAA’s ability to induce matrix-degrading enzymes as a central mechanism underlying its pathogenic effects in chronic inflammatory disease. In patients with rheumatoid arthritis or psoriatic arthritis, SAA induces matrix metalloproteinase in synovial fibroblasts and is associated with increased joint destruction (48). Similarly, SAA1.1/2.1 induces matrix metalloproteinases in the aortic vascular wall in a mouse model of angiotensin II-induced AAA formation (6). These data could suggest an explanation why the evolutionarily conserved pro-inflammatory SAA is beneficial in the innate immune context: by lessening connective tissue tension to promote cellular diapedesis and even accommodate edema formation. However, in “modern” chronic inflammatory diseases, this innate beneficial function can contribute to progression by negatively impacting connective tissue integrity.
In summary, these studies shed light on the previous misconceptions that mouse SAA3 is an extrahepatic SAA. Moreover, we show that SAA3 is present in the circulation bound to HDL. Unlike SAA1.1/2.1, a portion of SAA3 in plasma is present in a lipid-free form, a distinct feature that may have important functional implications. Additional studies are needed to address the respective activities of the acute-phase mouse SAAs, and how these activities relate to the human SAA isoforms. Ignoring SAA3 in mouse models of chronic inflammatory diseases may confound interpretation.
Footnotes
Abbreviations:
- AAA
- abdominal aortic aneurysm
- FPLC
- fast protein liquid chromatography
- IEF
- isoelectric focusing
- LPS
- lipopolysaccharide
- pI
- isoelectric point
- SAA
- serum amyloid A
This work was supported by the US Department of Veterans Affairs Awards CX000975 (L.R.T.) and CX000773 (N.R.W.) and National Institutes of Health Grants HL134731 (N.R.W., F.C.D.B.), HL092969 (A.C.), AT007177 (L.d.H.), and P20 GM103527 (support for used cores). Mass spectrometric analysis was performed at the University of Kentucky, Proteomics Core Facility. This core facility is supported in part by funds from the Office of the Vice President for Research. The content of this study is solely the responsibility of the authors and does not represent the official views of the US Department of Veterans Affairs, the National Institutes of Health, or the United States Government.
REFERENCES
- 1.Eklund K. K., Niemi K., and Kovanen P. T.. 2012. Immune functions of serum amyloid A. Crit. Rev. Immunol. 32: 335–348. [DOI] [PubMed] [Google Scholar]
- 2.Uhlar C. M., Burgess C. J., Sharp P. M., and Whitehead A. S.. 1994. Evolution of the serum amyloid A (SAA) protein superfamily. Genomics. 19: 228–235. [DOI] [PubMed] [Google Scholar]
- 3.Derebe M. G., Zlatkov C. M., Gattu S., Ruhn K. A., Vaishnava S., Diehl G. E., MacMillan J. B., Williams N. S., and Hooper L. V.. 2014. Serum amyloid A is a retinol binding protein that transports retinol during bacterial infection. eLife. 3: e03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badolato R., Wang J. M., Murphy W. J., Lloyd A. R., Michiel D. F., Bausserman L. L., Kelvin D. J., and Oppenheim J. J.. 1994. Serum amyloid A is a chemoattractant: induction of migration, adhesion, and tissue infiltration of monocytes and polymorphonuclear leukocytes. J. Exp. Med. 180: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furlaneto C. J., and Campa A.. 2000. A novel function of serum amyloid A: a potent stimulus for the release of tumor necrosis factor-alpha, interleukin-1beta, and interleukin-8 by human blood neutrophil. Biochem. Biophys. Res. Commun. 268: 405–408. [DOI] [PubMed] [Google Scholar]
- 6.Webb N. R., De Beer M. C., Wroblewski J. M., Ji A., Bailey W., Shridas P., Charnigo R. J., Noffsinger V. P., Witta J., Howatt D. A., et al. . 2015. Deficiency of endogenous acute-phase serum amyloid A protects apoE-/- mice from angiotensin II-induced abdominal aortic aneurysm formation. Arterioscler. Thromb. Vasc. Biol. 35: 1156–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song C., Shen Y., Yamen E., Hsu K., Yan W., Witting P. K., Geczy C. L., and Freedman S. B.. 2009. Serum amyloid A may potentiate prothrombotic and proinflammatory events in acute coronary syndromes. Atherosclerosis. 202: 596–604. [DOI] [PubMed] [Google Scholar]
- 8.Lee H. Y., Kim M. K., Park K. S., Bae Y. H., Yun J., Park J. I., Kwak J. Y., and Bae Y. S.. 2005. Serum amyloid A stimulates matrix-metalloproteinase-9 upregulation via formyl peptide receptor like-1-mediated signaling in human monocytic cells. Biochem. Biophys. Res. Commun. 330: 989–998. [DOI] [PubMed] [Google Scholar]
- 9.Migita K., Kawabe Y., Tominaga M., Origuchi T., Aoyagi T., and Eguchi K.. 1998. Serum amyloid A protein induces production of matrix metalloproteinases by human synovial fibroblasts. Lab. Invest. 78: 535–539. [PubMed] [Google Scholar]
- 10.Ather J. L., Ckless K., Martin R., Foley K. L., Suratt B. T., Boyson J. E., Fitzgerald K. A., Flavell R. A., Eisenbarth S. C., and Poynter M. E.. 2011. Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J. Immunol. 187: 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Migita K., Izumi Y., Jiuchi Y., Kozuru H., Kawahara C., Nakamura M., Nakamura T., Agematsu K., Masumoto J., Yasunami M., et al. . 2014. Serum amyloid A induces NLRP-3-mediated IL-1beta secretion in neutrophils. PLoS One. 9: e96703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niemi K., Teirila L., Lappalainen J., Rajamaki K., Baumann M. H., Oorni K., Wolff H., Kovanen P. T., Matikainen S., and Eklund K. K.. 2011. Serum amyloid A activates the NLRP3 inflammasome via P2X7 receptor and a cathepsin B-sensitive pathway. J. Immunol. 186: 6119–6128. [DOI] [PubMed] [Google Scholar]
- 13.Yu N., Liu S., Yi X., Zhang S., and Ding Y.. 2015. Serum amyloid A induces interleukin-1beta secretion from keratinocytes via the NACHT, LRR and PYD domains-containing protein 3 inflammasome. Clin. Exp. Immunol. 179: 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrow D. A., Rifai N., Antman E. M., Weiner D. L., McCabe C. H., Cannon C. P., and Braunwald E.. 2000. Serum amyloid A predicts early mortality in acute coronary syndromes: A TIMI 11A substudy. J. Am. Coll. Cardiol. 35: 358–362. [DOI] [PubMed] [Google Scholar]
- 15.Filep J. G., and El Kebir D.. 2008. Serum amyloid A as a marker and mediator of acute coronary syndromes. Future Cardiol. 4: 495–504. [DOI] [PubMed] [Google Scholar]
- 16.Coetzee G. A., Strachan A. F., van der Westhuyzen D. R., Hoppe H. C., Jeenah M. S., and de Beer F. C.. 1986. Serum amyloid A-containing human high density lipoprotein 3. Density, size, and apolipoprotein composition. J. Biol. Chem. 261: 9644–9651. [PubMed] [Google Scholar]
- 17.Whitehead A. S., de Beer M. C., Steel D. M., Rits M., Lelias J. M., Lane W. S., and de Beer F. C.. 1992. Identification of novel members of the serum amyloid A protein superfamily as constitutive apolipoproteins of high density lipoprotein. J. Biol. Chem. 267: 3862–3867. [PubMed] [Google Scholar]
- 18.De Buck M., Gouwy M., Wang J. M., Van Snick J., Opdenakker G., Struyf S., and Van Damme J.. 2016. Structure and expression of different serum amyloid A (SAA) variants and their concentration-dependent functions during host insults. Curr. Med. Chem. 23: 1725–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluve-Beckerman B., Drumm M. L., and Benson M. D.. 1991. Nonexpression of the human serum amyloid A three (SAA3) gene. DNA Cell Biol. 10: 651–661. [DOI] [PubMed] [Google Scholar]
- 20.Meek R. L., and Benditt E. P.. 1986. Amyloid A gene family expression in different mouse tissues. J. Exp. Med. 164: 2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benditt E. P., and Meek R. L.. 1989. Expression of the third member of the serum amyloid A gene family in mouse adipocytes. J. Exp. Med. 169: 1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reigstad C. S., Lunden G. O., Felin J., and Backhed F.. 2009. Regulation of serum amyloid A3 (SAA3) in mouse colonic epithelium and adipose tissue by the intestinal microbiota. PLoS One. 4: e5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urieli-Shoval S., Cohen P., Eisenberg S., and Matzner Y.. 1998. Widespread expression of serum amyloid A in histologically normal human tissues. Predominant localization to the epithelium. J. Histochem. Cytochem. 46: 1377–1384. [DOI] [PubMed] [Google Scholar]
- 24.Meek R. L., Eriksen N., and Benditt E. P.. 1992. Murine serum amyloid A3 is a high density apolipoprotein and is secreted by macrophages. Proc. Natl. Acad. Sci. USA. 89: 7949–7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiba T., Han C. Y., Vaisar T., Shimokado K., Kargi A., Chen M. H., Wang S., McDonald T. O., O’Brien K. D., Heinecke J. W., et al. . 2009. Serum amyloid A3 does not contribute to circulating SAA levels. J. Lipid Res. 50: 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Beer M. C., de Beer F. C., McCubbin W. D., Kay C. M., and Kindy M. S.. 1993. Structural prerequisites for serum amyloid A fibril formation. J. Biol. Chem. 268: 20606–20612. [PubMed] [Google Scholar]
- 27.de Beer M. C., Webb N. R., Wroblewski J. M., Noffsinger V. P., Rateri D. L., Ji A., van der Westhuyzen D. R., and de Beer F. C.. 2010. Impact of serum amyloid A on high density lipoprotein composition and levels. J. Lipid Res. 51: 3117–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.den Hartigh L. J., Wang S., Goodspeed L., Ding Y., Averill M., Subramanian S., Wietecha T., O’Brien K. D., and Chait A.. 2014. Deletion of serum amyloid A3 improves high fat high sucrose diet-induced adipose tissue inflammation and hyperlipidemia in female mice. PLoS One. 9: e108564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji A., Wroblewski J. M., Cai L., de Beer M. C., Webb N. R., and van der Westhuyzen D. R.. 2012. Nascent HDL formation in hepatocytes and role of ABCA1, ABCG1, and SR-BI. J. Lipid Res. 53: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamelgarn M., Chen J., Kuang L., Arenas A., Zhai J., Zhu H., and Gal J.. 2016. Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim. Biophys. Acta. 1862: 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Ji A., Weihe E., and Schafer M. K.. 2004. Cell-specific expression and lipopolysaccharide-induced regulation of tumor necrosis factor alpha (TNFalpha) and TNF receptors in rat dorsal root ganglion. J. Neurosci. 24: 9623–9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang R. Z., Lee M. J., Hu H., Pollin T. I., Ryan A. S., Nicklas B. J., Snitker S., Horenstein R. B., Hull K., Goldberg N. H., et al. . 2006. Acute-phase serum amyloid A: an inflammatory adipokine and potential link between obesity and its metabolic complications. PLoS Med. 3: e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han C. Y., Subramanian S., Chan C. K., Omer M., Chiba T., Wight T. N., and Chait A.. 2007. Adipocyte-derived serum amyloid A3 and hyaluronan play a role in monocyte recruitment and adhesion. Diabetes. 56: 2260–2273. [DOI] [PubMed] [Google Scholar]
- 34.Yu J., Zhu H., Guo J. T., de Beer F. C., and Kindy M. S.. 2000. Expression of mouse apolipoprotein SAA1.1 in CE/J mice: isoform-specific effects on amyloidogenesis. Lab. Invest. 80: 1797–1806. [DOI] [PubMed] [Google Scholar]
- 35.Franco A. G., Sandri S., and Campa A.. 2011. High-density lipoprotein prevents SAA-induced production of TNF-alpha in THP-1 monocytic cells and peripheral blood mononuclear cells. Mem. Inst. Oswaldo Cruz. 106: 986–992. [DOI] [PubMed] [Google Scholar]
- 36.Baranova I. N., Bocharov A. V., Vishnyakova T. G., Kurlander R., Chen Z., Fu D., Arias I. M., Csako G., Patterson A. P., and Eggerman T. L.. 2010. CD36 is a novel serum amyloid A (SAA) receptor mediating SAA binding and SAA-induced signaling in human and rodent cells. J. Biol. Chem. 285: 8492–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M. H., de Beer M. C., Wroblewski J. M., Webb N. R., and de Beer F. C.. 2013. SAA does not induce cytokine production in physiological conditions. Cytokine. 61: 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noborn F., Ancsin J. B., Ubhayasekera W., Kisilevsky R., and Li J. P.. 2012. Heparan sulfate dissociates serum amyloid A (SAA) from acute-phase high-density lipoprotein, promoting SAA aggregation. J. Biol. Chem. 287: 25669–25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J. P., Galvis M. L., Gong F., Zhang X., Zcharia E., Metzger S., Vlodavsky I., Kisilevsky R., and Lindahl U.. 2005. In vivo fragmentation of heparan sulfate by heparanase overexpression renders mice resistant to amyloid protein A amyloidosis. Proc. Natl. Acad. Sci. USA. 102: 6473–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosuge M., Ebina T., Ishikawa T., Hibi K., Tsukahara K., Okuda J., Iwahashi N., Ozaki H., Yano H., Kusama I., et al. . 2007. Serum amyloid A is a better predictor of clinical outcomes than C-reactive protein in non-ST-segment elevation acute coronary syndromes. Circ. J. 71: 186–190. [DOI] [PubMed] [Google Scholar]
- 41.Johnson B. D., Kip K. E., Marroquin O. C., Ridker P. M., Kelsey S. F., Shaw L. J., Pepine C. J., Sharaf B., Bairey Merz C. N., Sopko G., et al. . 2004. Serum amyloid A as a predictor of coronary artery disease and cardiovascular outcome in women: the National Heart, Lung, and Blood Institute-Sponsored Women’s Ischemia Syndrome Evaluation (WISE). Circulation. 109: 726–732. [DOI] [PubMed] [Google Scholar]
- 42.Dong Z., Wu T., Qin W., An C., Wang Z., Zhang M., Zhang Y., Zhang C., and An F.. 2011. Serum amyloid A directly accelerates the progression of atherosclerosis in apolipoprotein E-deficient mice. Mol. Med. 17: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J. C., Jayne C., Thompson J., Wilson P. G., Yoder M. H., Webb N., and Tannock L. R.. 2015. A brief elevation of serum amyloid A is sufficient to increase atherosclerosis. J. Lipid Res. 56: 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Beer M. C., Wroblewski J. M., Noffsinger V. P., Rateri D. L., Howatt D. A., Balakrishnan A., Ji A., Shridas P., Thompson J. C., van der Westhuyzen D. R., et al. . 2014. Deficiency of endogenous acute phase serum amyloid A does not affect atherosclerotic lesions in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 34: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishack P. A., Bhanvadia C. V., Lukens J., Sontag T. J., De Beer M. C., Getz G. S., and Reardon C. A.. 2015. Serum amyloid A facilitates early lesion development in Ldlr-/- mice. J. Am. Heart Assoc. 4: e001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J. C., Wilson P. G., Shridas P., Ji A., De Beer M. C., De Beer F. C., Webb N. R., and Tannock L. R.. 2017. Serum amyloid A3 is pro-atherogenic. Atherosclerosis. 268: 32–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tam S. P., Flexman A., Hulme J., and Kisilevsky R.. 2002. Promoting export of macrophage cholesterol: the physiological role of a major acute-phase protein, serum amyloid A 2.1. J. Lipid Res. 43: 1410–1420. [DOI] [PubMed] [Google Scholar]
- 48.Connolly M., Mullan R. H., McCormick J., Matthews C., Sullivan O., Kennedy A., FitzGerald O., Poole A. R., Bresnihan B., Veale D. J., et al. . 2012. Acute-phase serum amyloid A regulates tumor necrosis factor alpha and matrix turnover and predicts disease progression in patients with inflammatory arthritis before and after biologic therapy. Arthritis Rheum. 64: 1035–1045. [DOI] [PubMed] [Google Scholar]