Abstract
ApoA-I activates LCAT that converts lipoprotein cholesterol to cholesteryl ester (CE). Molecular dynamic simulations suggested earlier that helices 5 of two antiparallel apoA-I molecules on discoidal HDL form an amphipathic tunnel for migration of acyl chains and unesterified cholesterol to the active sites of LCAT. Our recent crystal structure of Δ(185–243)apoA-I showed the tunnel formed by helices 5/5, with two positively charged residues arginine 123 positioned at the edge of the hydrophobic tunnel. We hypothesized that these uniquely positioned residues Arg123 are poised for interaction with fatty acids produced by LCAT hydrolysis of the sn-2 chains of phosphatidylcholine, thus positioning the fatty acids for esterification to cholesterol. To test the importance of Arg123 for LCAT phospholipid hydrolysis and CE formation, we generated apoA-I[R123A] and apoA-I[R123E] mutants and made discoidal HDL with the mutants and WT apoA-I. Neither mutation of Arg123 changed the particle composition or size, or the protein conformation or stability. However, both mutations of Arg123 significantly reduced LCAT catalytic efficiency and the apparent Vmax for CE formation without affecting LCAT phospholipid hydrolysis. A control mutation, apoA-I[R131A], did not affect LCAT phospholipid hydrolysis or CE formation. These data suggest that Arg123 of apoA-I on discoidal HDL participates in LCAT-mediated cholesterol esterification.
Keywords: cholesterol/metabolism, fatty acid/transferase, high density lipoprotein/metabolism, high density lipoprotein/structure, lipid and lipoprotein metabolism, catalytic efficiency, mutant proteins, protein conformation.
ApoA-I is the principle protein component of HDL and a key element of the reverse cholesterol transport pathway (1–3). Reverse cholesterol transport involves esterification of free cholesterol after its efflux from peripheral cells to lipid-poor apoA-I and nascent discoidal HDL. The esterification of cholesterol is catalyzed by the enzyme LCAT that enables the transfer of a fatty acid present in phosphatidylcholine to the hydroxyl group of cholesterol (4). The reaction consists of two steps (5, 6). The first step is mediated by a phospholipase A2 activity of LCAT and involves the cleavage of the sn-2 ester bond of phosphatidylcholine and the release of a fatty acid and lysophosphatidylcholine. The second step includes the transacylation of the fatty acid to the 3-hydroxyl group of cholesterol, thereby forming cholesteryl ester (CE). LCAT binds reversibly to HDL particles, and apoA-I on HDL is the principal physiological activator of the enzyme (4, 7). CE molecules formed by the LCAT-catalyzed reaction partition into the hydrophobic core of the particles, thus preventing cholesterol from returning to cells by diffusion. Cholesterol esterification on HDL leads to conversion of nascent discoidal HDL into mature spherical particles (2, 8). Uptake of CE from the core of mature HDL by the scavenger receptor class B type 1 receptor in the liver leads to net efflux of cholesterol from the circulation (1–3, 8). Thus, LCAT-mediated esterification of cholesterol is a central step in HDL maturation and reverse cholesterol transport. Yet, the molecular details of LCAT activation by apoA-I are not fully understood.
ApoA-I (243 amino acids, 28 kDa) has a flexible and adaptable structure that enables the protein to exchange among lipid-free, lipid-poor, and various lipid-bound states during reverse cholesterol transport (9, 10). The first 43 residues of apoA-I form a G-helix, and the rest of the protein consists of 10 tandem 11/22-residue repeats that have high propensity to form amphipathic α-helices. To identify the specific part of apoA-I involved in LCAT activation, a number of groups have investigated the effects of apoA-I mutation on LCAT-mediated cholesterol esterification in reactions carried out in vitro (11–18). Several studies concluded that apoA-I helix 6 alone (residues 143−164) plays a main role in LCAT activation (11–14, 17). Analysis of apoA-I natural mutations associated with the reduced LCAT activity suggested that a more extended region between residues 110 and 160 is involved in LCAT activation (18), while immunochemical studies indicated that the apoA-I region between residues 95 and 121 is likely important for LCAT activation (19). Taken together, the majority of the studies, although not all (15), implicated the central helices 4−6 (residues 99−164) of apoA-I in LCAT activation. It was established that the correct secondary conformation of apoA-I, including the proper alignment and orientation of the hydrophobic face of the central α-helices, is important for LCAT activation (12–14, 16, 17, 20).
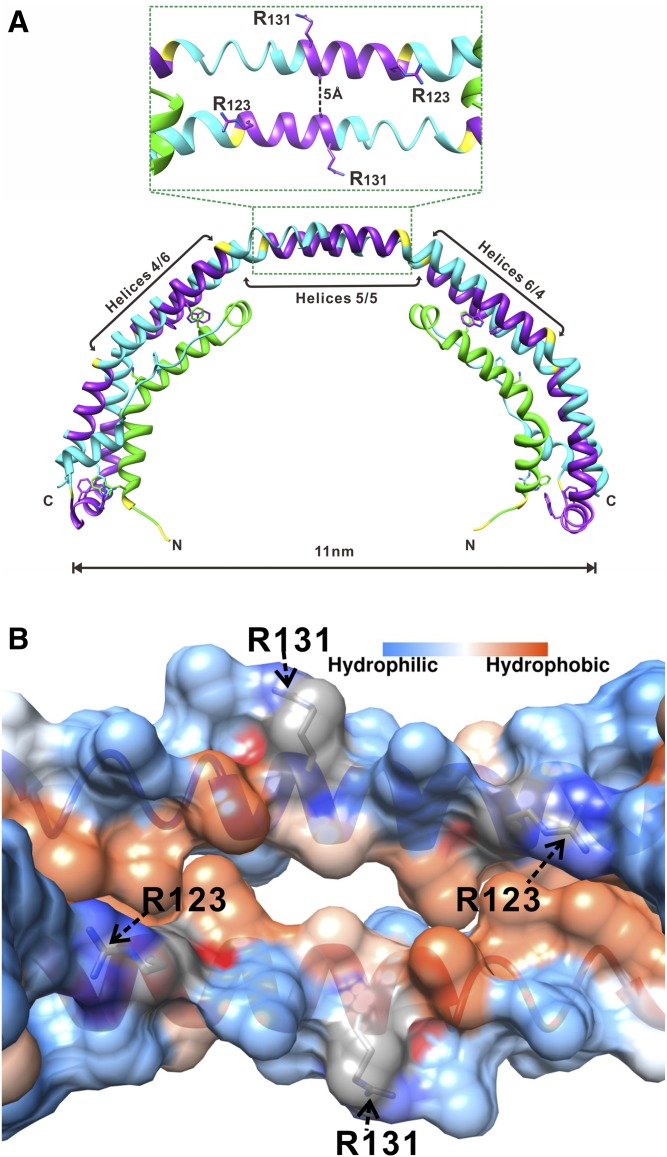
Deeper insights into the function of apoA-I as a LCAT activator were gained by more detailed understanding of the structural organization of apoA-I on HDL. Structure of discoidal (nascent) HDL particles, which are the preferred LCAT substrate, has been well-studied using reconstituted discoidal HDL (rHDL) produced in vitro. Despite some debates on details of certain regions of apoA-I in discoidal HDL, the majority of experimental and theoretical data support the “double belt” organization of apoA-I (16, 21–28). In this model, amphipathic α-helices from two antiparallel apoA-I molecules wrap around the perimeter of the discs to form a closed double belt, with helices 5/5 of the two apoA-I molecules being in register. Molecular dynamics simulations of discoidal HDL suggested that when LCAT binds to discoidal HDL, the helix 5/5 domain of the antiparallel apoA-I molecules forms an amphipathic presentation tunnel for migration of hydrophobic acyl chains and amphipathic cholesterol from the lipid bilayer to the active sites of LCAT (27). Our 2.2 Å-resolution X-ray crystal structure of the lipid-free C-terminal truncated Δ(185–243)apoA-I (Protein Data Bank entry 3R2P) (29) supports the double belt model and substantiates the tunnel formed by helices 5/5 of two antiparallel apoA-Is. Although lipid-free, the Δ(185–243)apoA-I forms a curved crystallographic dimer with a degree of curvature similar to that of the double belt on rHDL with 10.5 nm diameter (Fig. 1A). The key conformational elements of the central part (residues 79–178) of the crystallographic dimer resemble those of the full-length apoA-I double belt on discoidal HDL, as shown by experimental studies and molecular dynamic simulations (reviewed in Refs. 30–32). The crystal structure clearly shows that helices 5/5 of the two antiparallel apoA-I molecules form a central hole (tunnel) of ∼5 Å diameter (Fig. 1). Lipid binding is expected to rotate helices resulting in enlargement of the tunnel and rearrangements of some salt bridges (30). The tunnel seen in the crystal structure has a strongly charged outside surface and a hydrophobic inside surface (Fig. 1B), with only two positively charged residues Arg123 (one from each antiparallel apoA-I molecule) on the edge of the hydrophobic face that project toward the hydrophobic region at each end of the tunnel. We propose that these uniquely positioned residues Arg123 are poised for interaction with fatty acids produced by LCAT hydrolysis of the sn-2 chain of phospholipids, thus positioning the fatty acids for esterification to cholesterol.
Fig. 1.
Location of Arg123 and Arg131 in the structure of apoA-I. A: X-ray crystal structure of Δ(185−243)apoA-I (Protein Data Bank entry 3R2P) showing a crystallographic dimer with the central opening (amphipathic tunnel), ∼5 Å in diameter, between the antiparallel helices 5/5 (29). B: Inside view of the amphipathic central tunnel formed by the central antiparallel helices 5/5 with the surface colored according to the residue hydrophobicity or charge. The central tunnel has a strongly charged outside surface and a hydrophobic inside surface. The two positively charged Arg123 residues, one from each apoA-I antiparallel molecule, are projecting toward the hydrophobic region at each end of the tunnel. Two Arg131 residues, one from each antiparallel molecule, are located on the outside surface of the tunnel.
To test this hypothesis, we generated apoA-I mutations, R123A and R123E, that ablate and reverse, respectively, the positive charge of Arg123. We investigated the effects of these mutations on the conformation and stability of apoA-I in solution and in rHDL and on the ability of apoA-I to promote LCAT phospholipid hydrolysis and cholesterol esterification. We compared mutation of Arg123 to mutation of another arginine, Arg131, that is also located in helix 5, but is not projecting toward the hydrophobic inside of the tunnel (Fig. 1). Our data demonstrate that Arg123, but not Arg131, is essential for LCAT-mediated cholesterol esterification, while neither of these two arginine residues seem to be important for LCAT-mediated phospholipid hydrolysis.
MATERIALS AND METHODS
Materials
POPC was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). The 8-anilino-2-naphthalene-sulfonate (ANS), cholesterol, sodium cholate, and fatty-acid free BSA were from Sigma-Aldrich. The 1,2-[3H]cholesterol was purchased from PerkinElmer Life Sciences Inc. Purified recombinant human LCAT was a generous gift of Dr. John S. Parks (Wake Forest University). All chemicals were of highest purity analytical grade.
Plasmid construction, expression, purification, and preparation of proteins
WT apoA-I, apoA-I[R123A], apoA-I[R123E], and apoA-I[R131A] were expressed in the His6-MBP-TEV expression system in Escherichia coli and purified using the techniques and materials described previously (29, 33). The purified proteins had a single glycine at the N terminus derived from the TEV cleavage site. Protein purity was analyzed by 12% SDS-PAGE followed by staining with Coomassie blue R259 and by Western blotting using monoclonal antibodies against human apoA-I. The proteins with purity higher than 95% were frozen with liquid nitrogen and then stored at −80°C until use. Before the experiments, the proteins were freshly refolded by dialysis against 4 M guanidine hydrochloride (GdnHCl) followed by extensive dialysis against TBS buffer, pH 7.6. The protein samples prepared for the circular dichroism (CD) experiments were dialyzed against 10 mM sodium phosphate, pH 7.6. Protein concentration was determined by Lowry protein assay.
Preparation and characterization of rHDL
rHDL particles comprised of POPC, cholesterol, and either WT apoA-I or one of the apoA-I mutants were prepared by the sodium cholate dialysis method (34) at starting molar ratio POPC:cholesterol:apoA-I of 80:8:1, as previously described (35). rHDL particles used in the CE formation assays were made with traces of radiolabeled [1,2-3H]cholesterol, to get 30,000 dpm per 1 μg of cholesterol. Formation of rHDL particles was verified by nondenaturing gradient (8–25%) gel electrophoresis and electron microscopy; electron micrographs of negatively stained samples of rHDL were taken and the major diameter of the particles was determined as described (36). The final composition of rHDL was determined by Phospholipids C and Free Cholesterol E assays (Wako Diagnostics, Richmond, VA) and modified Lowry protein assay in the presence of 1% SDS.
CD spectroscopy: thermally induced and denaturant-induced unfolding
CD measurements were performed on AVIV 62DS or AVIV 215 spectropolarimeters (AVIV Associates, Inc.) at protein concentrations of 25–80 μg/ml, as described (36, 37). Lipid-free apoA-I is monomeric in this range of protein concentrations under the conditions used in our experiments (37). Far-UV spectra were recorded at 25°C at several protein concentrations and then normalized to mean residue ellipticity, [Θ]. The α-helical content was determined from the mean residue ellipticity at 222 nm, [Θ222] (38).
Stability of lipid-free apoA-I was assessed by monitoring protein thermal unfolding by ellipticity at 222 nm. Proteins were heated in the cuvette of the spectropolarimeter from 3°C to 98°C with 1 degree increments at several different heating rates between 0.4 and 0.7°C/min. The melting (midpoint) temperature, Tm, and van’t Hoff enthalpy, ΔHv, were determined from van’t Hoff analysis of the melting curves (37). Stability of rHDL was assessed from thermally and GdnHCl-induced unfolding of apoA-I in the particles. Thermal unfolding was monitored by ellipticity at 222 nm, while the particles were heated from 5°C to 98°C with 1 degree increments at the constant rate (0.3°C/min). Midpoint of thermal unfolding, T1/2, at which denaturation of apoA-I was half-completed, was determined from the thermal unfolding curves. For GdnHCl-induced unfolding of apoA-I on rHDL, aliquots of rHDL at protein concentrations of 15–135 μg/ml were preincubated with various concentrations of GndHCl, ranging from 0 to 5 M, at 4°C for 72 h, to allow apoA-I to achieve complete denaturation; then ellipticity at 222 nm was recorded at 25°C. The conformational stability, ΔGDo, the midpoint of denaturation, D1/2, and m parameter, which reflects the steepness of the denaturation curve in the transition region, were determined by a linear extrapolation method, as previously described (36).
ANS fluorescence measurements
Fluorescence of ANS (0.25 mM) in the presence of 0.05 mg/ml of lipid-free apoA-I was recorded as previously described (36, 39). Fluorescent measurements were performed at 25°C on a FluoroMax-2 fluorescence spectrometer (Instruments S.A., Inc.), with 395 nm excitation wavelength and the emission spectra scanned from 400 to 560 nm. The wavelength of maximum fluorescence (WMF) and fluorescence intensity at the WMF, I, were measured from the spectra after subtraction of the buffer baseline.
Assays for CE formation
LCAT assays were performed in duplicate in 0.5 ml TBS. Various amounts of rHDL containing traces of 3H-cholesterol were preincubated with 0.4% BSA (fatty acid-free) and 2 mM β-mercaptoethanol for 20 min at 37°C, then purified recombinant LCAT was added to initiate the reaction. Concentrations of rHDL cholesterol (reaction substrate) in the incubation mixtures ranged between 0 and 16 μM and the amount of LCAT ranged between 11 and 130 ng, depending on the type and amount of rHDL. Tubes were purged with N2 and the reactions were carried out at 37°C for 15–25 min. Control incubations contained all components except LCAT. The reaction was stopped by the addition of chloroform:methanol (2:1, volume ratio). The lipids were extracted from each incubation mixture and cholesterol and cholesterol ester were separated by thin-layer chromatography, as described (40). The bands corresponding to cholesterol and CE were quantified by scintillation counting. To maintain first-order kinetics, the extent of cholesterol esterification in the reactions was kept below 15% by adjusting the incubation time and the amount of LCAT, which resulted in various values for the rHDL/LCAT molar ratio in the incubation mixtures. The cholesterol esterification rate was expressed in nanomoles of CE formed per hour per milliliter of LCAT and plotted against the substrate (rHDL cholesterol) concentrations. The apparent rate constants, Km and Vmax, were determined from Lineweaver-Burk double reciprocal plots.
Assay for phospholipid hydrolysis
The effect of the apoA-I mutations on LCAT phospholipid hydrolysis was studied using a fluorometric assay kit for measuring phospholipase A2 activity of LCAT (STA-615; Cell Biolabs, Inc., San Diego, CA). The approach utilizes a fluorogenic dual-labeled POPC with fluorophores attached to the terminal ends of both acyl chains, similar to the fluorescence assays developed for measuring the phospholipase A2 activity of LCAT (41) and other enzymes (42, 43). The fluorophores are in a quenched state when the phospholipid is uncleaved. Upon hydrolysis of POPC at the sn-2 position by phospholipase A2 activity of LCAT, fluorescent monomer chains are produced, which are measured in a fluorescence reader. The assays were performed in duplicate in 0.13 ml buffer containing rHDL substrate (15 μg of POPC) with 1 μM fluorogenic dual-labeled POPC, 0.4% BSA, 2 mM β-mercaptoethanol, and 15–20 ng of purified recombinant LCAT. All constituents, except LCAT, were preincubated for 20 min at 37°C, and then the reaction was initiated by adding LCAT. Each assay included two types of control incubations, with all constituents except LCAT and with all constituents except rHDL. The reactions were carried out in tubes purged with N2 at 37°C for 15 min to 1 h. Control experiments performed with rHDL containing 3H-cholesterol showed that CE formation at these experimental conditions was linear up to 1 h and the extent of CE formation was <20%. The reaction was quenched with methanol at a final concentration of 25%, and the fluorescence of the samples was measured in 96-well microplates at 342 nm excitation and 400 nm emission wavelengths using an Infinite M1000 PRO microplate reader (Tecan Group Ltd.). The difference between the fluorescent signal in the absence of rHDL and in the presence of rHDL, normalized by the incubation time and the amount of LCAT added, was used as a relative measure for LCAT phospholipid hydrolysis. The method was validated in a separate set of incubations with increasing concentrations of rHDL, which resulted in proportionally increasing values of the relative measure for LCAT phospholipid hydrolysis, expressed in the relative fluorescence intensity, I400, per hour of incubation per milliliter of LCAT.
RESULTS
Effect of the mutations on the conformation and stability of lipid-free apoA-I
Parameters determined from CD analysis of the lipid-free apoA-I forms are listed in Table 1. The α-helical content did not differ between the WT and the mutant forms of apoA-I, suggesting that none of the mutations alter the secondary conformation of lipid-free apoA-I. The thermal unfolding curves for WT apoA-I and the three mutant apoA-I forms normalized to molar residue ellipticity, [Θ222], overlapped or were close to each other (not shown). The melting temperature, Tm, and van’t Hoff enthalpy, ΔHv, determined from the melting curves indicate that none of the mutations affected the stability or unfolding cooperativity of lipid-free apoA-I.
TABLE 1.
α-Helical content, thermodynamic parameters, and ANS fluorescence for lipid-free WT apoA-I and the apoA-I mutant forms
| α-Helixa (%) | Tmb (°C) | ΔHvb (kcal/mol) | Ic (relative units) | WMFc (nm) | |
| WT apoA-I | 56 ± 2 | 62 ± 1 | 41 ± 2 | 1.0 | 474 |
| apoA-I[ R123A] | 58 ± 3 | 63 ± 1 | 45 ± 2 | 0.8 | 477 (+3) |
| apoA-I[R123E] | 59 ± 2 | 64 ± 1 | 43 ± 2 | 0.8 | 476 (+2) |
| apoA-I[R131A] | 57 ± 2 | 64 ± 1 | 44 ± 2 | 1.0 | 474 |
The values are mean ± SD from at least three experiments.
Estimated from the [Θ222] at 25°C.
The melting temperature, Tm, and van’t Hoff enthalpy, ΔHv, were determined from van’t Hoff analysis of thermal unfolding curves monitored by CD.
Parameters of ANS fluorescence were determined in the presence of WT apoA-I or one of the apoA-I mutants. I, ANS fluorescence intensity in relative units compared with the fluorescence in the presence of WT apoA-I.
Fluorescence of the amphipathic dye, ANS, in the presence of WT apoA-I or one of the apoA-I mutants was measured to investigate the effect of the mutations on the tertiary folding of the lipid-free protein. The R123A and R123E mutations resulted in a slightly lower fluorescent intensity, I, and a 2–3 nm “red” shift of the ANS spectra (Table 1), suggesting that the substitutions of Arg123 for Ala or Glu lead to slightly lesser exposure of the hydrophobic surfaces of lipid-free apoA-I. ANS fluorescence in the presence of the apoA-I[R131A] was similar to that of WT apoA-I, suggesting that the mutation of Arg131 did not affect the tertiary folding of the lipid-free protein.
Effect of the mutations on the rHDL characteristics
Electron micrographs of rHDL made with WT apoA-I or one of the apoA-I mutants showed discoidal complexes that are stacked on edge. For all types of rHDL, the major diameter determined from electron micrographs as the average of more than 200 particles was between 7.8 and 9.2 nm, with the standard deviation ±2 nm, indicating no significant difference between sizes of rHDL formed by WT apoA-I or any of the apoA-I mutants. Assays for phospholipid, cholesterol, and protein content indicated that all the four types of rHDL particles were similar in composition (Table 2).
TABLE 2.
Composition of rHDL made with WT apoA-I or one of the mutant apoA-I and the conformational and stability characteristics of apoA-I in the rHDL
| POPC:Cholesterol:apoA-Ia (M:M:M) | α-Helixb (%) | T1/2c (°C) | ΔGDood (kcal/mol) | md [kcal/(moles of apoA-I × moles of GdnHCl)] | D1/2d (M) | |
| WT apoA-I | (76 ± 7):(7 ± 0.8):1 | 79 ± 2 | 78 ± 0.5 | 3.8 ± 0.3 | 1.4 ± 0.1 | 2.5 ± 0.1 |
| apoA-I[R123A] | (68 ± 5):(6 ± 0.7):1 | 77 ± 3 | 79 ± 1.0 | 3.4 ± 0.2 | 1.3 ± 0.1 | 2.6 ± 0.1 |
| apoA-I [R123E] | (71 ± 6):(7 ± 0.8):1 | 76 ± 1 | 79 ± 1.0 | 3.4 ± 0.2 | 1.4 ± 0.1 | 2.5 ± 0.0 |
| apoA-I [R131A] | (77 ± 4):(8 ± 0.5):1 | 79 ± 2 | 79 ± 0.5 | 3.9 ± 0.2 | 1.6 ± 0.1 | 2.4 ± 0.1 |
The values are mean ± SD from at least three experiments.
Molar composition of rHDL (determined in three independent reconstitution experiments).
Estimated from the [Θ222] at 25°C.
The midpoint of thermal unfolding, T1/2, at which half of the total change in the CD signal is observed, was determined from the thermal unfolding curves for rHDL.
The conformational stability, ΔGDo, m value, and midpoint of denaturation, D1/2, were determined by the linear extrapolation method from the chemical unfolding curves for rHDL.
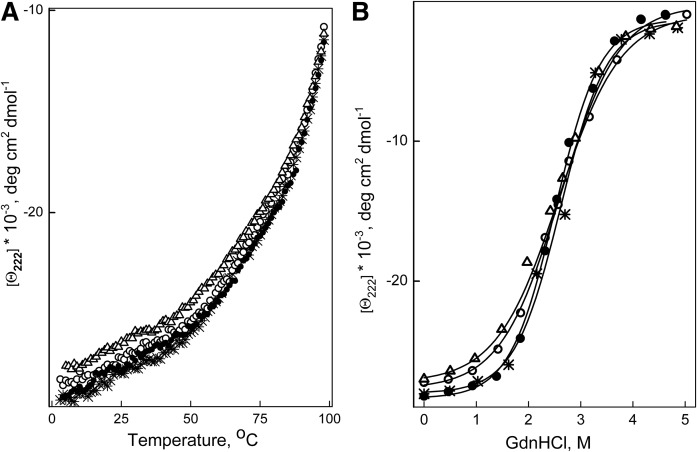
Far-UV CD analysis showed that the α-helical content of apoA-I in rHDL was not altered by any of the mutations (Table 2). The thermal unfolding curves for each type of rHDL were close to each other (Fig. 2A), and the midpoint of thermal denaturation determined from the curves was almost identical for all types of rHDL (Table 2). The denaturant-unfolding curves for all types of rHDL were also close to each other (Fig. 2B) and the conformational stability, the midpoint of denaturation, and the m value did not differ significantly between all types of rHDL (Table 2). These data indicate that the stability of apoA-I in rHDL was not affected by the mutations of Arg123 or Arg131.
Fig. 2.
Unfolding of apoA-I in rHDL monitored by the ellipticity at 222 nm. rHDLs were made with WT apoA-I (*), apoA-I[R123A] (open circle), apoA-I[R123E] (open triangle), or apoA-I[R131A] (closed circle). A: Thermal unfolding was induced by heating the proteins from 2°C to 98°C in the cuvette within the CD spectrometer holder. B: Chemical unfolding was induced by incubation of aliquots of rHDL with various concentrations of GndHCl, ranging from 0 to 5 M, at 4°C for 72 h, and then ellipticity at 222 nm was recorded at 25°C.
CE formation by LCAT
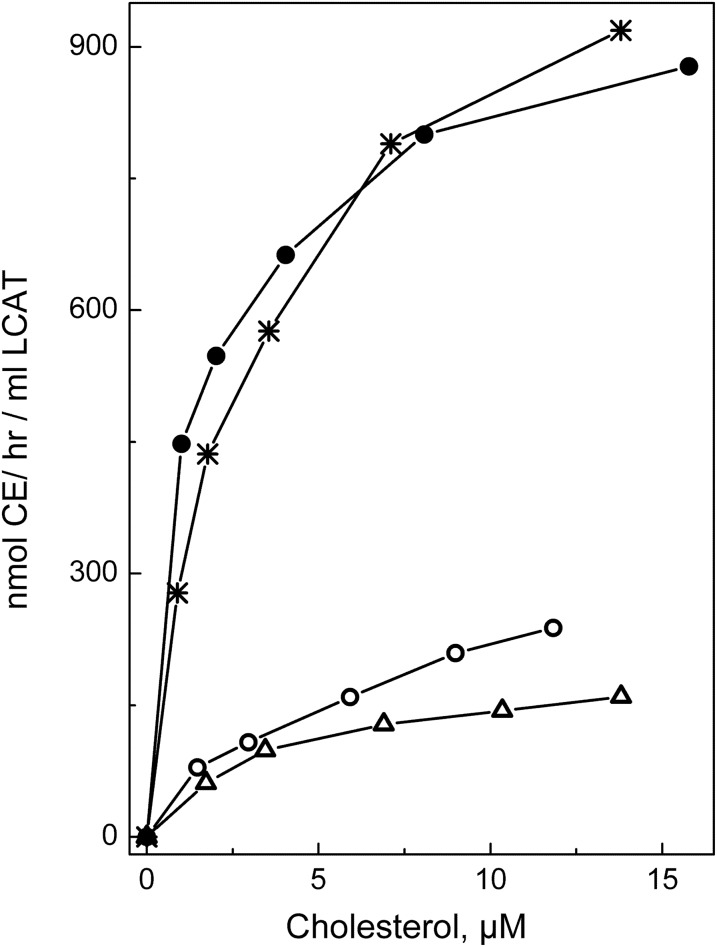
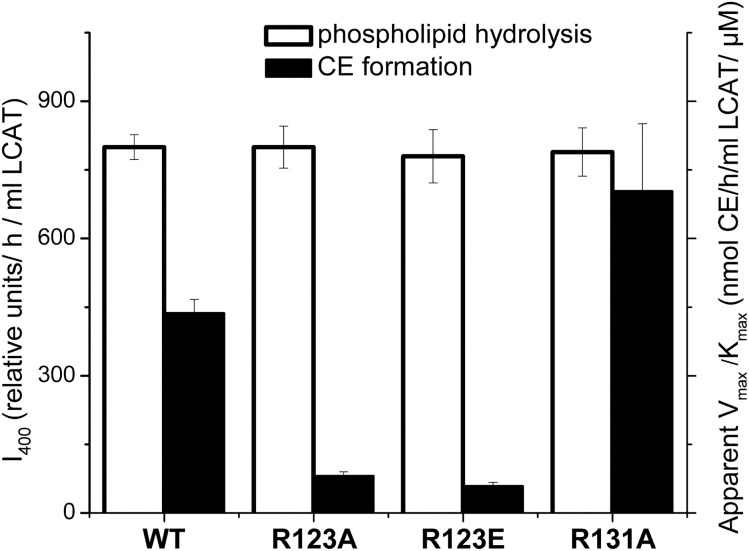
After establishing that the conformation and stability of apoA-I in rHDL was not affected by any of the three mutations, functional assays were performed. To assess the effect of the apoA-I mutations on the LCAT-mediated cholesterol esterification, rHDL particles made with WT apoA-I or mutant apoA-I were used as a substrate in the CE formation assay. Figure 3 shows the cholesterol esterification rate for each type of rHDL as a function of the substrate (rHDL cholesterol) concentrations. The apparent kinetic constants for the LCAT reaction are listed in Table 3. The apparent Michaelis constant, Km, was slightly (∼1.7 times) increased for the apoA-I[R123E], but not significantly changed for the apoA-I[R123A]. However, the apparent maximum reaction rate, Vmax, was 3.6 and 4.1 times lower and the apparent catalytic efficiency, Vmax/Km, was 5.4 and 7.4 times lower for rHDL made with apoA-I[R123A] and with apoA-I[R123E], respectively, compared with rHDL made with WT apoA-I. The data show that both mutations of Arg123 significantly reduced the ability of apoA-I to promote LCAT-mediated cholesterol esterification. In contrast, the R131A mutation was associated with a slight, but not statistically significant, increase in the catalytic efficiency of LCAT, indicating that the mutation of Arg131 did not significantly change the ability of apoA-I to activate LCAT. The effect of the apoA-I mutations on the apparent catalytic efficiency of LCAT is shown graphically in Fig. 4.
Fig. 3.
Cholesterol esterification by LCAT. rHDL made with WT apoA-I (*), apoA-I[R123A] (open circle), apoA-I[R123E] (open triangle), or apoA-I[R131A] (closed circle) and containing traces of [3H]cholesterol were used as a substrate in the CE formation assays. Increasing concentrations of rHDL were incubated with purified LCAT to get initial rate constants for CE formation, as described in the Materials and Methods. The cholesterol esterification rate is expressed in nanomoles of CE formed per hour of incubation per milliliter of LCAT and plotted against the substrate (rHDL cholesterol) concentrations. All points represent an average of two to three individual experiments of duplicate determinations for each cholesterol concentration.
TABLE 3.
Apparent kinetic constants for the reaction of LCAT with rHDL made with WT apoA-I or one of the apoA-I mutant forms
| Apparent Km (μM) | Apparent Vmax (nmol CE/h/ml LCAT) | Apparent Vmax/Km (nmol CE/h/ml LCAT/μM) | |
| WT apoA-I | 2.3 ± 0.4 | 977 ± 46 | 437 ± 30 |
| apoA-I[R123A] | 3.3 ± 0.6 | 270 ± 18a | 81 ± 9a |
| apoA-I[R123E] | 4.1 ± 0.5b | 235 ± 33a | 59 ± 8a |
| apoA-I[R131A] | 1.3 ± 0.3 | 923 ± 61 | 704 ± 147 |
The values represent the mean ± SD of at least three independent measurements. The apparent LCAT kinetic parameters were determined from the CE formation assays using Lineweaver-Burk plots (double reciprocal plots) of reaction velocity versus rHDL cholesterol concentration as described in the Material and Methods.
P < 0.0005 compared with the values for WT apoA-I.
P < 0.05 compared with the values for WT apoA-I.
Fig. 4.
Effect of the apoA-I mutations on LCAT phospholipid hydrolysis and CE formation. rHDLs made with WT apoA-I or one of the apoA-I mutants were used as a substrate for the assays to assess phospholipid hydrolysis and CE formation by LCAT. Values were calculated as the mean ± SD of at least three independent measurements. Phospholipid hydrolysis was assessed by measuring fluorescence of monomeric labeled fatty acids produced by hydrolysis of dual-labeled POPC (1 μM) following the incubation of rHDL (15 μg of POPC) with purified LCAT, as described in the Materials and Methods. The relative values for LCAT phospholipid hydrolysis (white bars) are expressed in the relative fluorescence intensity at 400 nm, I400, normalized by the incubation time and the amount of LCAT (left y axis). CE formation was measured in the assays as described for Fig. 3. The values for the apparent catalytic efficiency for CE formation, Vmax/Km, (listed in Table 3) were determined from Lineweaver-Burk double reciprocal plots generated from the assay data. The values for the apparent Vmax/Km (black bars) are expressed in nanomoles of CE formed per hour of incubation per milliliter of LCAT per μM of substrate cholesterol (right y axis).
Phospholipid hydrolysis by LCAT
The effect of the apoA-I mutations on LCAT-mediated phospholipid hydrolysis was assessed by measuring fluorescence of monomeric labeled fatty acids produced by hydrolysis of dual-labeled POPC following incubations of rHDL with LCAT. The values for the relative fluorescence intensity normalized by the incubation time and the amount of LCAT were similar for rHDL made with WT apoA-I and each of the mutant apoA-Is (Fig. 4). The data indicate that neither of the mutations of Arg123 nor the mutation of Arg131 affected the ability of LCAT to hydrolyze POPC and produce free fatty acids.
DISCUSSION
In this work, we tested the hypothesis that in the LCAT reaction on nascent (discoidal) HDL, Arg123 of apoA-I interacts with free fatty acids produced by LCAT hydrolysis of the sn-2 chains of phospholipids to position the fatty acids for esterification to cholesterol. The study was designed to investigate whether mutation of Arg123 and “control” mutation of Arg131 of apoA-I on rHDL affect LCAT-mediated CE formation and phospholipid hydrolysis on the rHDL. Figure 4 summarizes the observed effects of the apoA-I mutations on LCAT phospholipid hydrolysis and CE formation.
The findings that neither mutation of Arg123 affected LCAT phospholipid hydrolysis support the view that Arg123 plays no role in the first step of LCAT reaction that is hydrolysis of the sn-2 ester bond of phospholipid. The markedly reduced LCAT catalytic efficiency on rHDL containing apoA-I with mutated Arg123 supports the notion that Arg123 of apoA-I is involved in LCAT-mediated cholesterol esterification. The fact that both charge ablation and charge reversal of Arg123 reduced CE formation suggests that the interaction of Arg123 with free fatty acids may have an electrostatic component. The observed large reduction in the apparent catalytic efficiency, Vmax/Km, was due to the large reduction in the apparent Vmax, while the apparent Km was not significantly changed (for the R123A) or only slightly increased (for R123E). It was shown earlier that the substitution of the region Arg123−Tyr166 of apoA-I with the region Ser12−Ala75 of apoA-II also led to greatly reduced LCAT activity due to a reduction in Vmax with unaltered Km (44). Furthermore, it was shown that the presence of apoA-I in rHDL specifically increases the apparent Vmax for LCAT-mediated cholesterol esterification, when compared with rHDL made with other apolipoproteins, apoA-II (45) or apo E4 (46). The dissociation constants, Km, were of the same order of magnitude for rHDL made with either of these apolipoproteins, or even for pure lipid vesicles (45). The presence of Arg123 of apoA-I on rHDL may contribute to the relatively high values for the catalytic efficiency and the maximum reaction rate observed for apoA-I-containing rHDL.
Previous studies have shown that changes in the secondary conformation and stability of apoA-I on substrate rHDL, as well as the composition and size of the particles, may affect LCAT activity (13, 17, 20, 47). Therefore, to investigate the involvement of Arg123 in LCAT reaction, it was important to determine whether the generated mutations of apoA-I alter the conformation and stability of apoA-I and the characteristics of rHDL. We found that the substitutions of Arg123 for Ala or Glu did not change the secondary conformation or stability of the lipid-free apoA-I and only slightly reduced exposure of the protein hydrophobic surfaces to solution, suggesting that the mutations of Arg123 led to a slightly more compact folding of monomeric apoA-I in solution. However, none of the mutations of Arg123 affected the conformation or stability of apoA-I on rHDL. Also, the apoA-I[R123A]- and apoA-I[R123E]-containing rHDL particles were similar to WT apoA-I-containing rHDL particles in regard to the composition, morphology, and the average diameter. This indicates that the large reduction in the LCAT-activating ability of apoA-I resulting from mutation of Arg123 is not due to variations in the composition or size of the substrate rHDL or to changes in the apoA-I conformation or stability.
In the crystal structure of the Δ(185–243)apoA-I dimer (29), there is a very weak and unusual intramolecular salt bridge between Arg123 and Glu120, with Pro121 breaking the helical conformation between the two residues. Replacing Arg123 by Glu or Ala in the crystallographic dimer eliminates this weak salt bridge, but is not expected to affect other salt bridge interactions, lead to formation of new salt bridges, or alter the conformation or stability of the dimer. Indeed, the intramolecular salt bridge between Arg123 and Glu120 may not exist in the presence of lipids. Nevertheless, the substitution of Arg123 for Glu or Ala would eliminate this interaction, but is unlikely to change existing stabilizing salt bridge interactions or lead to formation of new stabilizing or destabilizing interactions in apoA-I, given the unaltered conformation and stability characteristics of the mutant apoA-I forms on rHDL. These data are consistent with the notion that a free fatty acid produced by LCAT phospholipid hydrolysis interacts with Arg123 of apoA-I on rHDL (replacing the weak interaction with Glu120, if it exists), but cannot interact with negatively charged Glu or neutral Ala at position 123 in the apoA-I[R123E] or apoA-I[R123A], resulting in the reduced LCAT-activating ability of these mutant proteins.
Noticeably, a number of naturally occurring human apoA-I mutations in the central part of apoA-I (helices 4−7) are associated with low plasma HDL levels and abnormal LCAT activation (18, 48, 49). While none of these mutations involves Arg123 directly, these mutations alter the conformation and/or stability of apoA-I on rHDL (18, 33, 35, 39, 48, 50, 51) and thereby may affect the structure of the helix 5/5 domain and the position of Arg123 in this structure. The natural mutation, L141R (apoA-IPisa), located in helix 5 and associated with reduced LCAT activity (52), was shown to destabilize apoA-I (35) and is also expected to disturb the hydrophobic face of the helix 5/5 domain. Several naturally occurring apoA-I mutations associated with low LCAT activity are clustered in helix 6 and two mutations, K107del and E110K, are located in helix 4 (48, 49). These mutations may disturb salt bridge interactions of helix 6 with its pairwise mate, helix 4 of the antiparallel apoA-I molecule on discoidal HDL (22, 27, 29). Because the pairwise interaction between helices 4 and 6 occurs on both sides of the helix 5/5 domain (Fig. 1A), the natural mutations in helices 4 and 6 may have a major effect on the structure of the helix 5/5 domain and thus affect LCAT activity.
Our data show that, similar to the mutations of Arg123, the “control” mutation of Arg131 in helix 5 did not change the conformation or stability of apoA-I either in solution or in rHDL, nor did it affect LCAT phospholipid hydrolysis. However, in contrast to mutation of Arg123, mutation of Arg131 did not affect any of the catalytic parameters of LCAT-mediated CE formation, indicating that Arg131 of apoA-I is not essential for LCAT activity on rHDL. These observations are consistent with the position of Arg131 on the outside surface of the tunnel (Fig. 1), and support the concept that the position of Arg123 on the inside surface of the tunnel allows this residue to be involved in the transacylation activity of LCAT.
Previous studies have shown that three other arginine residues of apoA-I, R149, R153, and R160, located in helix 6, are critical for LCAT activity (16). These three arginine residues form a positively charged cluster on the outer surface of discoidal HDL particles that is believed to participate in LCAT binding. This is consistent with the location of R149, R153, and R160 on the outside surface of the dimer backbone in the crystal structure of Δ(185–243)apoA-I that potentially enables polar interactions of these residues with LCAT. Arginine residues in other parts of apoA-I are likely involved in salt bridge interactions stabilizing the double belt structure, as suggested by the double belt model of apoA-I (22, 27) and demonstrated in the crystal structure of the Δ(185–243)apoA-I dimer (29). Therefore, we did not consider mutations of other arginine residues of apoA-I as “control” mutations in our study, because they are expected to alter the conformation and/or stability of apoA-I (36) and affect LCAT activation. In fact, the naturally occurring mutations involving arginine residues in apoA-I, including R10L, R27T, R151C, R153P, R160L, and R173C, are associated with low plasma HDL and abnormal LCAT activity (48, 49).
Jauhiainen and Dolphin (5) proposed a mechanism of LCAT reaction that involves transacylation of free fatty acids produced by the phospholipase A2 activity of LCAT to serine residues of LCAT followed by transesterification of the fatty acids to LCAT cysteine residues. The resulting thioester is believed to subsequently donate the fatty acids to the 3-hydroxyl group of cholesterol. In the same study, the authors hypothesized that serine residues of apoA-I may act as transient acceptors of free fatty acids following phospholipid hydrolysis. The findings did not confirm the latter hypothesis (5), thus offering no mechanism for the LCAT-activation function of apoA-I. It is possible that the interaction of free fatty acids with serine residues of LCAT, proposed by Jauhiainen and Dolphin (5), is preceded by the interaction of the free fatty acids with Arg123 of apoA-I inside the presentation tunnel. Jones et al. (27) suggested that when substrate for LCAT reaction is in the tunnel, apoA-I becomes integrated with the LCAT molecule and the tunnel connects to the active site of LCAT.
Among the studies supporting the double belt organization of apoA-I on discoidal HDL, there are several studies suggesting that particular parts of apoA-I molecules on the discs may form flexible loops protruding into solution that causes a localized opening between the parallel belts (25, 26, 28). Wu et al. (26) suggested, based on hydrogen/deuterium exchange experiments, that residues 159–180 of apoA-I on discoidal HDL form a flexible loop that interacts with and activates LCAT. However, a more recent study by Chetty et al. (28) using a similar experimental approach demonstrated that residues 159–180 do not form a loop, while the region spanning residues 125–158 (or residues 115–158) of one or both apoA-I molecules in the double belt can form either helix or a partially disordered loop on discoidal HDL (reviewed in Ref. 32). The latter findings are consistent with our earlier studies (35) and the partially unfolded helices 5/5 in the crystal structure of Δ(185–243)apoA-I (Fig. 1A) and are in line with our results suggesting the involvement of Arg123 of apoA-I in LCAT reaction on discoidal HDL. Because the double belt model is believed to be a common organizational motif for apoA-I in both discoidal and spherical HDL of various sizes (31, 53, 54), it is possible that Arg123 of apoA-I may be involved in LCAT reaction on spherical HDL also. Cross-linking and mass spectrometry studies of spherical reconstituted and human plasma HDL (53, 54) led to a cage-like trefoil model, in which three apoA-I molecules are positioned on spherical HDL with helices of each apoA-I molecule involved in the same salt bridge interactions as in the double belt model. In this trefoil model, each apoA-I molecule is bent in the middle of helix 5, thus creating an opening between helices 5. According to a model proposed by Gursky (31), adaptation of apoA-I conformation to variable sizes and shapes of HDL involves rearrangements of the N- and C-terminal parts of the molecule, while the central half of the double belt, including helices 4–6, remains structurally conserved and resembles the corresponding part of the crystal structure of the Δ(185–243)apoA-I dimer. According to this model, the structure of the helix 5/5 domain, including the position of Arg123, remains “constant” in discoidal and spherical HDL of various sizes.
In conclusion, the data reported here support the role of Arg123 of apoA-I in transacylation of fatty acids produced by LCAT phospholipid hydrolysis to cholesterol. This novel mechanistic view on the function of apoA-I as LCAT activator on nascent (discoidal) HDL may be helpful for better understanding the mechanisms of disorders involving “dysfunctional” HDL and abnormal LCAT activity (1, 8, 55, 56).
Acknowledgments
The authors are indebted to Dr. John S. Parks and Abraham K. Gebre of Wake Forest School of Medicine, Winston-Salem, NC, for the generous gift of pure recombinant LCAT and advice. The authors gratefully acknowledge Donald L. Gantz for the expert help with electron microscopy and Dr. Olga Gursky for helpful discussions.
Footnotes
Abbreviations:
- ANS
- 8-anilino-2-naphthalene-sulfonate
- CD
- circular dichroism
- CE
- cholesteryl ester
- GdnHCl
- guanidine hydrochloride
- rHDL
- reconstituted discoidal HDL
- WMF
- wavelength of maximum fluorescence
This work was supported by National Institutes of Health Grant R01HL116518. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Rye K. A., and Barter P. J.. 2014. Cardioprotective functions of HDLs. J. Lipid Res. 55: 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothblat G. H., and Phillips M. C.. 2010. High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr. Opin. Lipidol. 21: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenson R. S., Brewer H. B. Jr., Davidson W. S., Fayad Z. A., Fuster V., Goldstein J., Hellerstein M., Jiang X. C., Phillips M. C., Rader D. J., et al. 2012. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 125: 1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fielding C. J., and Fielding P. E.. 1995. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 36: 211–228. [PubMed] [Google Scholar]
- 5.Jauhiainen M., and Dolphin P. J.. 1986. Human plasma lecithin-cholesterol acyltransferase. an elucidation of the catalytic mechanism. J. Biol. Chem. 261: 7032–7043. [PubMed] [Google Scholar]
- 6.Wang J., Gebre A. K., Anderson R. A., and Parks J. S.. 1997. Amino acid residue 149 of lecithin:cholesterol acyltransferase determines phospholipase A2 and transacylase fatty acyl specificity. J. Biol. Chem. 272: 280–286. [DOI] [PubMed] [Google Scholar]
- 7.Jonas A. 1998. Regulation of lecithin-cholesterol acyltransferase activity. Prog. Lipid Res. 37: 209–234. [DOI] [PubMed] [Google Scholar]
- 8.Rye K. A., and Barter P. J.. 2014. Regulation of high-density lipoprotein metabolism. Circ. Res. 114: 143–156. [DOI] [PubMed] [Google Scholar]
- 9.Nolte R. T., and Atkinson D.. 1992. Conformational analysis of apolipoprotein A-I and E-3 based on primary sequence and circular dichroism. Biophys. J. 63: 1221–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., and Anantharamaiah G. M.. 1992. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33: 141–166. [PubMed] [Google Scholar]
- 11.Sorci-Thomas M., Kearns M. W., and Lee J. P.. 1993. Apolipoprotein A-I domains involved in lecithin-cholesterol acyltransferase activation. Structure:function relationships. J. Biol. Chem. 268: 21403–21409. [PubMed] [Google Scholar]
- 12.Sorci-Thomas M. G., Curtiss L., Parks J. S., Thomas M. J., and Kearns M. W.. 1997. Alteration in apolipoprotein A-I 22-mer repeat order results in a decrease in lecithin:cholesterol acyltransferase reactivity. J. Biol. Chem. 272: 7278–7284. [DOI] [PubMed] [Google Scholar]
- 13.Sorci-Thomas M. G., Curtiss L., Parks J. S., Thomas M. J., Kearns M. W., and Landrum M.. 1998. The hydrophobic face orientation of apolipoprotein A-I amphipathic helix domain 143–164 regulates lecithin:cholesterol acyltransferase activation. J. Biol. Chem. 273: 11776–11782. [DOI] [PubMed] [Google Scholar]
- 14.Sviridov D., Hoang A., Sawyer W. H., and Fidge N. H.. 2000. Identification of a sequence of apolipoprotein A-I associated with the activation of lecithin:cholesterol acyltransferase. J. Biol. Chem. 275: 19707–19712. [DOI] [PubMed] [Google Scholar]
- 15.Scott B. R., McManus D. C., Franklin V., McKenzie A. G., Neville T., Sparks D. L., and Marcel Y. L.. 2001. The N-terminal globular domain and the first class A amphipathic helix of apolipoprotein A-I are important for lecithin:cholesterol acyltransferase activation and the maturation of high density lipoprotein in vivo. J. Biol. Chem. 276: 48716–48724. [DOI] [PubMed] [Google Scholar]
- 16.Roosbeek S., Vanloo B., Duverger N., Caster H., Breyne J., De Beun I., Patel H., Vandekerckhove J., Shoulders C., Rosseneu M., et al. 2001. Three arginine residues in apolipoprotein A-I are critical for activation of lecithin:cholesterol acyltransferase. J. Lipid Res. 42: 31–40. [PubMed] [Google Scholar]
- 17.Alexander E. T., Bhat S., Thomas M. J., Weinberg R. B., Cook V. R., Bharadwaj M. S., and Sorci-Thomas M.. 2005. Apolipoprotein A-I helix 6 negatively charged residues attenuate lecithin-cholesterol acyltransferase (LCAT) reactivity. Biochemistry. 44: 5409–5419. [DOI] [PubMed] [Google Scholar]
- 18.Hoang A., Huang W., Sasaki J., and Sviridov D.. 2003. Natural mutations of apolipoprotein A-I impairing activation of lecithin:cholesterol acyltransferase. Biochim. Biophys. Acta. 1631: 72–76. [DOI] [PubMed] [Google Scholar]
- 19.Banka C. L., Bonnet D. J., Black A. S., Smith R. S., and Curtiss L. K.. 1991. Localization of an apolipoprotein A-I epitope critical for activation of lecithin-cholesterol acyltransferase. J. Biol. Chem. 266: 23886–23892. [PubMed] [Google Scholar]
- 20.Sparks D. L., Frank P. G., and Neville T. A.. 1998. Effect of the surface lipid composition of reconstituted LPA-I on apolipoprotein A-I structure and lecithin: cholesterol acyltransferase activity. Biochim. Biophys. Acta. 1390: 160–172. [DOI] [PubMed] [Google Scholar]
- 21.Borhani D. W., Rogers D. P., Engler J. A., and Brouillette C. G.. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94: 12291–12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., and Harvey S. C.. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274: 31755–31758. [DOI] [PubMed] [Google Scholar]
- 23.Li H., Lyles D. S., Thomas M. J., Pan W., and Sorci-Thomas M. G.. 2000. Structural determination of lipid-bound ApoA-I using fluorescence resonance energy transfer. J. Biol. Chem. 275: 37048–37054. [DOI] [PubMed] [Google Scholar]
- 24.Silva R. A., Hilliard G. M., Li L., Segrest J. P., and Davidson W. S.. 2005. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 44: 8600–8607. [DOI] [PubMed] [Google Scholar]
- 25.Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., and Oda M. N.. 2006. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J. Biol. Chem. 281: 20418–20426. [DOI] [PubMed] [Google Scholar]
- 26.Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M. I. I. I., Smith J. D., Gogonea V., and Hazen S. L.. 2007. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14: 861–868. [DOI] [PubMed] [Google Scholar]
- 27.Jones M. K., Catte A., Li L., and Segrest J. P.. 2009. Dynamics of activation of lecithin:cholesterol acyltransferase by apolipoprotein A-I. Biochemistry. 48: 11196–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sevugan Chetty P., Mayne L., Kan Z. Y., Lund-Katz S., Englander S. W., and Phillips M. C.. 2012. Apolipoprotein A-I helical structure and stability in discoidal high-density lipoprotein (HDL) particles by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 109: 11687–11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei X., and Atkinson D.. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286: 38570–38582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segrest J. P., Jones M. K., Catte A., and Thirumuruganandham S. P.. 2012. Validation of previous computer models and MD simulations of discoidal HDL by a recent crystal structure of apoA-I. J. Lipid Res. 53: 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gursky O. 2013. Crystal structure of Δ(185-243)ApoA-I suggests a mechanistic framework for the protein adaptation to the changing lipid load in good cholesterol: from flatland to sphereland via double belt, belt buckle, double hairpin and trefoil/tetrafoil. J. Mol. Biol. 425: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips M. C. 2013. New insights into the determination of HDL structure by apolipoproteins: thematic review series: high density lipoprotein structure, function, and metabolism. J. Lipid Res. 54: 2034–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorshkova I. N., Mei X., and Atkinson D.. 2014. Binding of human apoA-I[K107del] variant to TG-rich particles: implications for mechanisms underlying hypertriglyceridemia. J. Lipid Res. 55: 1876–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matz C. E., and Jonas A.. 1982. Micellar complexes of human apolipoprotein A-I with phosphatidylcholines and cholesterol prepared from cholate-lipid dispersions. J. Biol. Chem. 257: 4535–4540. [PubMed] [Google Scholar]
- 35.Gorshkova I. N., Liu T., Zannis V. I., and Atkinson D.. 2002. Lipid-free structure and stability of apolipoprotein A-I: probing the central region by mutation. Biochemistry. 41: 10529–10539. [DOI] [PubMed] [Google Scholar]
- 36.Gorshkova I. N., Liu T., Kan H. Y., Chroni A., Zannis V. I., and Atkinson D.. 2006. Structure and stability of apolipoprotein A-I in solution and in discoidal high-density lipoprotein probed by double charge ablation and deletion mutation. Biochemistry. 45: 1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorshkova I. N., Liadaki K., Gursky O., Atkinson D., and Zannis V. I.. 2000. Probing the lipid-free structure and stability of apolipoprotein A-I by mutation. Biochemistry. 39: 15910–15919. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y. H., Yang J. T., and Martinez H. M.. 1972. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry. 11: 4120–4131. [DOI] [PubMed] [Google Scholar]
- 39.Chroni A., Kan H. Y., Kypreos K. E., Gorshkova I. N., Shkodrani A., and Zannis V. I.. 2004. Substitutions of Glu110 and Glu111 in the middle helix 4 of human ApoA-I by Alanine affect the structure and in vitro functions of apoA-I and induce severe hypertriglyceridemia in ApoA-I-deficient mice. Biochemistry. 43: 10442–10457. [DOI] [PubMed] [Google Scholar]
- 40.Miller K. R., Wang J., Sorci-Thomas M., Anderson R. A., and Parks J. S.. 1996. Glycosylation structure and enzyme activity of 1ecithin:cholesterol acyltransferase from human plasma, HepC2 cells, and baculoviral and Chinese hamster ovary cell expression systems. J. Lipid Res. 37: 551–561. [PubMed] [Google Scholar]
- 41.Bonelli F. S., and Jonas A.. 1992. Continuous fluorescence assay for lecithin:cholesterol acyltransferase using a water-soluble phosphatidylcholine. J. Lipid Res. 33: 1863–1869. [PubMed] [Google Scholar]
- 42.Keshavarz A., Zelaya L., Singh J., Ranganathan R., and Hajdu J.. 2017. Activity-based targeting of secretory phospholipase A2 enzymes: A fatty-acid-binding-protein assisted approach. Chem. Phys. Lipids. 202: 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darrow A. L., Olson M. W., Xin H., Burke S. L., Smith C., Schalk-Hihi C., Williams R., Bayoumy S. S., Deckman I. C., Todd M. J., et al. 2011. A novel fluorogenic substrate for the measurement of endothelial lipase activity. J. Lipid Res. 52: 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhoest A., Zhao Z., De Geest B., Deridder E., Sillen A., Engelborghs Y., Collen D., and Holvoet P.. 1997. Role of the Arg123-Tyr166 paired helix of apolipoprotein A-I in lecithin:cholesterol acyltransferase activation. J. Biol. Chem. 272: 15967–15972. [DOI] [PubMed] [Google Scholar]
- 45.Bolin D. J., and Jonas A.. 1994. Binding of lecithin:cholesterol acyltransferase to reconstituted high density lipoproteins is affected by their lipid but not apolipoprotein composition. J. Biol. Chem. 269: 7429–7434. [PubMed] [Google Scholar]
- 46.Vezeridis A. M., Chroni A., and Zannis V. I.. 2011. Domains of apoE4 required for the biogenesis of apoE-containing HDL. Ann. Med. 43: 302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parks J. S., and Gebre A. K.. 1997. Long-chain polyunsaturated fatty acids in the sn-2 position of phosphatidylcholine decrease the stability of recombinant high density lipoprotein apolipoprotein A-I and the activation energy of the lecithin:cholesterol acyltransferase reaction. J. Lipid Res. 38: 266–275. [PubMed] [Google Scholar]
- 48.Sorci-Thomas M. G., and Thomas M. J.. 2002. The effects of altered apolipoprotein A-I structure on plasma HDL concentration. Trends Cardiovasc. Med. 12: 121–128. [DOI] [PubMed] [Google Scholar]
- 49.Francheschini G. 2010. Human apolipoprotein A-I mutants. In High Density Lipoproteins, Dyslipidemia, and Coronary Heart Disease. E. J. Schaefer, editor. Springer, New York. 63–69. [Google Scholar]
- 50.Alexander E. T., Tanaka M., Kono M., Saito H., Rader D. J., and Phillips M. C.. 2009. Structural and functional consequences of the Milano mutation (R173C) in human apolipoprotein A-I. J. Lipid Res. 50: 1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gursky O., Jones M. K., Mei X., Segrest J. P., and Atkinson D.. 2013. Structural basis for distinct functions of the naturally occurring Cys mutants of human apolipoprotein A-I. J. Lipid Res. 54: 3244–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miccoli R., Zhu Y., Daum U., Wessling J., Huang Y., Navalesi R., Assmann G., and von Eckardstein A.. 1997. A natural apolipoprotein A-I variant, apoA-I (L141R)Pisa, interferes with the formation of alpha-high density lipoproteins (HDL) but not with the formation of pre beta 1-HDL and influences efflux of cholesterol into plasma. J. Lipid Res. 38: 1242–1253. [PubMed] [Google Scholar]
- 53.Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., and Davidson W. S.. 2008. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. USA. 105: 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang R., Silva R. A., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., and Davidson W. S.. 2011. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 18: 416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Eckardstein A., and Rohrer L.. 2016. HDLs in crises. Curr. Opin. Lipidol. 27: 264–273. [DOI] [PubMed] [Google Scholar]
- 56.Kunnen S., and Van Eck M.. 2012. Lecithin:cholesterol acyltransferase: old friend or foe in atherosclerosis? J. Lipid Res. 53: 1783–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]