Abstract
Objectives
Plexiform neurofibromas (PN) are complex, histologically benign peripheral nerve sheath tumors that are challenging to measure by simple line measurements. Computer-aided volumetric segmentation of PN has become the recommended method to assess response in clinical trials directed at PN. Different methods for volumetric analysis of PN have been developed. The goal of this study is to test the level of agreement in volume measurements and in interval changes using two separate methods of volumetric MRI analysis.
Methods
Three independent volume measurements were performed on 15 PN imaged at three time-points using 3DQI software at Massachusetts General Hospital (MGH) and National Cancer Institute (NCI) and MEDx software at NCI.
Results
Median volume differences at each time-point comparing MGH-3DQI and NCI-3DQI were −0.5, −4.2, −19.9 ml; comparing NCI-3DQI and NCI-MEDx −21.0, −47.0, −21.0 ml; comparing MGH-3DQI and NCI-MEDx −10.0, −70.3, −29.9 ml. Median differences in percentage change over time comparing MGH-3DQI and NCI-3DQI were −1.7, 1.1, −1.0%; comparing NCI-3DQI and NCI-MEDx −2.3, 3.3, −1.1%; comparing MGH-3DQI and NCI-MEDx −0.4, 2.0, −1.5%. Volume differences were < 20% of the mean of the two measurements in 117 of 135 comparisons (86.7%). Difference in interval change was < 20% in 120 of the 135 comparisons (88.9%), while disease status classification was concordant in 115 of 135 comparisons (85.2%).
Conclusions
The volumes, interval changes, and progression status classifications were in good agreement. The comparison of two volumetric analysis methods suggests no systematic differences in tumor assessment. A prospective comparison of the two methods is planned.
Keywords: Neurofibromatosis type 1, Plexiform neurofibroma, Tumor Response Evaluation, Volumetric MRI
Introduction
Plexiform neurofibromas (PN) are histologically benign nerve sheath tumors typically associated with neurofibromatosis type-1 (NF1). These tumors develop along multiple branches of peripheral nerves and can be large, or irregularly shaped, making standard linear measurements unreliable. PNs are well visualized by magnetic resonance imaging (MRI) using Short TI Inversion Recovery (STIR) sequence, and can be contoured using computer-aided volumetric lesion segmentation methods1–3. Volumetric evaluation has become the method of choice to determine tumor response and time to disease progression in recent clinical trials for NF1-related PNs4–9.
In the phase 1 trial of the MEK inhibitor selumetinib 71% of patients with inoperable PN experienced at least 20% volume reduction, and in some cases improvement in clinical symptoms4. If the ongoing phase 2 trial confirms a similar response rate, and proves that the moderate size decreases are indeed associated with clinical improvement, selumetinib may become the first medical therapy approved by the US Food and Drug Administration (FDA) for the treatment of PN.
In order to facilitate drug development, NF researchers organized the Response Evaluation in Neurofibromatosis and Schwannomatosis (REiNS) International Collaboration with the goal of evaluating and standardizing clinical trial endpoints10. The REiNS consensus recommendations have been discussed with representatives of the FDA to ensure that selected endpoints and outcome measures would meet standards requested by the agency for drug approval. The FDA had no objection to evaluating treatment response in NF1 PN using volumetric MRI analysis, however recommended the testing of agreement by independent readers and different measurement tools in measuring volume change, which has not been done to date. The FDA also emphasized the need for a volumetric method, which could be utilized by multiple sites with similar results.
There are two independently developed volumetric methods optimized for PN measurement. The MEDx software used at the National Cancer Institute (NCI) performs a slice-by-slice histogram analysis of selected areas to identify the signal intensity threshold between tumor and normal tissues2. The 3DQI method developed at Massachusetts General Hospital (MGH) generates a three-dimensional rendering of the image data, with the tumor surface identified by the dynamic-threshold level set method starting with a seed initiation within the lesion and propagating shell expanding to the boundary1. Both methods employ various editing tools to finalize the tumor contour to user specifications. The end results are highly reproducible and closely resemble manually placed outlines. Reliably detecting volume change on serial scans of complex tumors requires identifying and consistently measuring all parts of the target lesion and can be challenging.
We tested the agreement between PN volumes and classification of progression status over time as determined by two different volumetric methods (3DQI and MEDx) and different users (NCI and MGH investigator).
Methods
Patients
Study subjects were selected by E.D. among participants of the NCI NF1-Natural History study to be representative of clinical trial patients. PN of different sizes, locations (orbit, face, neck, chest, abdomen, pelvis, extremities), levels of complexity, and imaging characteristics (diffuse or well circumscribed) were included. Patient identifiers were removed from the MRI data.
The MRIs and volumetric analyses were performed under IRB approved protocols (NCI NF1-Natural History - NCT00924196, and MGH Tumor Imaging Metrics Core).
MR Image Acquisition
Regional or whole-body MRIs were performed on Philips Medical Systems Achieva or GE Genesis Signa scanners at 1.5 tesla magnetic field strength and included axial and coronal STIR sequences with consistent imaging parameters between time-points (Field of view: 15 to 50 cm; Matrix: 224×224 to 512×512; TI: 150–180; TR: 4000–6350; TE: 12–36; Slice thickness: 4–10mm).
Image Analysis
The 3DQI and MEDx volumetric methods were applied to 15 PN, each imaged at 3 time-points (45 MRI studies).
In order to evaluate the effect of different volumetric analysis systems as well as user variability, three independent measurements were performed on each MRI; the MGH analyst used 3DQI, and the NCI analyst used both 3DQI and MEDx systems for analyses. All measurements were done on anonymized MRIs for the purpose of this comparison; the two analysts agreed on the target lesions and used identical MRI slices for volumetric analyses. There was a six months interval between the 3DQI and MEDx evaluations at the NCI.
To classify disease status, we defined progressive disease as ≥ 20% volume increase, partial response as ≥20% volume decrease, and stable disease as <20% change between time-points, in agreement with reported clinical trial practices11.
Statistical Analysis
The Wilcoxon signed rank test was used to statistically compare the numerical difference between the lesion volumes as well as the difference between the percent changes in volumes over time. The Jonckheere-Terpstra trend test was used to establish the degree of significance of the association in disease response status classification; small p-values (p<0.05 for example), would suggest that the two measures provided similar classification. For completeness, the McNemar test for paired categorical data (for two categories) or an exact marginal homogeneity test (for 3 ordered categories) was used to demonstrate the degree of balance in the discordant results, after establishing the overall degree of agreement; p<0.05 would suggest strong imbalances in the response assessment between methods but such findings were not anticipated given the strong concordances identified. Finally, a 95% confidence interval was calculated on the fraction of concordant results. In view of the small numbers of subjects evaluated, these results should be considered primarily hypothesis generating.
All p-values are two-tailed and presented without any adjustment for multiple comparisons.
Results
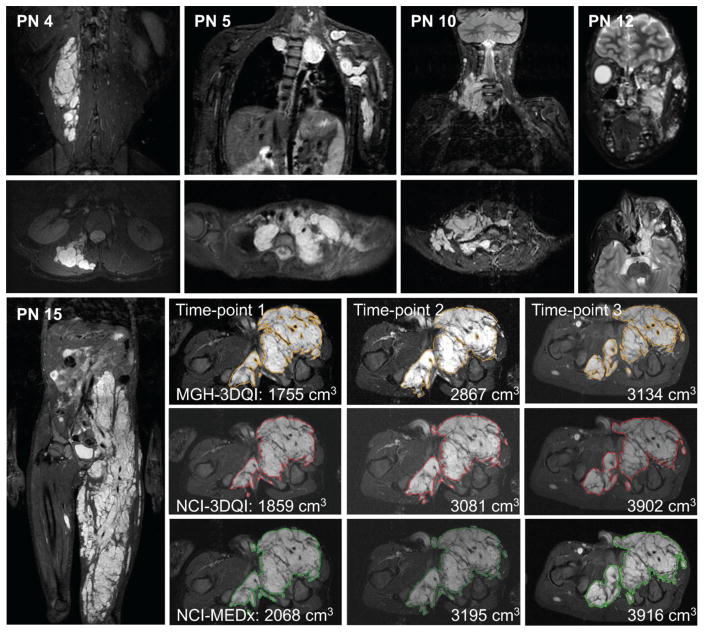
Representative images of the complex PN included in the study are shown in Figure 1. The MGH-3DQI, NCI-3DQI and NCI-MEDx analyses resulted in similar lesion contours (Figure 1), comparable PN volumes (Table 1), and similar growth trends over time (Supplementary Figure 1). Examples of the complete volume segmentations can be reviewed at the REiNS collaboration website (https://ccrod.cancer.gov/confluence/display/REINS/Presentations).
Figure 1. Examples of plexiform neurofibromas included in the study.
Coronal (top row), and axial (second row) STIR MR images of PN included in the study. PN 4: small, well defined PN in the right lumbar paraspinal muscle layer. PN 5: Large and highly complex PN in the upper chest, left shoulder, and arm. PN 10: medium sized right neck PN with moderately complex shape. PN 12: medium sized PN in left face, infiltrating the orbit, and facial muscles. PN 15: Coronal overview and segmentation contours at comparable levels in the axial plane from the three volumetric analyses, as labelled.
Table 1.
Three independent volumetric MRI measurements of 15 PN imaged at three different time-points
| ID | PN Location | Time-point 1 MRI | Time-point 2 MRI | Time-point 3 MRI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MGH-3DQI | NCI-3DQI | NCI-MEDx | MGH-3DQI | NCI-3DQI | NCI-MEDx | MGH-3DQI | NCI-3DQI | NCI-MEDx | ||
| 1 | Abdomen | 2568.7 | 3013 | 3302 | 4204.5 | 4127 | 4533 | 4724.3 | 4930 | 5150 |
| 2 | Neck, chest | 588.2 | 599 | 647 | 682.7 | 674 | 753 | 743.6 | 746 | 818 |
| 3 | Neck, chest | 893.0 | 824 | 892 | 1112.1 | 1087 | 1244 | 1399.5 | 1380 | 1425 |
| 4 | Back | 129.4 | 126 | 147 | 137.3 | 135 | 150 | 151.7 | 149 | 170 |
| 5 | Brachial plexus | 359.0 | 444 | 591 | 421.1 | 514 | 659 | 527.6 | 657 | 771 |
| 6 | Chest | 1861.3 | 1856 | 1838 | 1938.9 | 1964 | 2030 | 2253.1 | 2273 | 2283 |
| 7 | Chest, left arm | 1243.0 | 1426 | 1564 | 1377.9 | 1690 | 1820 | 1880.8 | 2060 | 2287 |
| 8 | Face | 180.1 | 155 | 177 | 175.7 | 173 | 181 | 204.8 | 194 | 201 |
| 9 | Lower leg | 255.4 | 285 | 270 | 299.1 | 329 | 328 | 270.5 | 303 | 290 |
| 10 | Neck | 152.0 | 151 | 162 | 175.8 | 180 | 188 | 257.7 | 256 | 288 |
| 11 | Orbit | 91.8 | 88.3 | 87.8 | 82.2 | 96.7 | 106 | 63.0 | 76.7 | 79.3 |
| 12 | Orbit, face | 175.1 | 180 | 168 | 211.4 | 201 | 194 | 241.9 | 200 | 194 |
| 13 | Pelvis | 987.5 | 988 | 970 | 993.8 | 1086 | 1133 | 1098.5 | 1146 | 1196 |
| 14 | Abdomen, pelvis | 1021.2 | 868 | 836 | 1634.6 | 1311 | 1282 | 2864.1 | 2949 | 2835 |
| 15 | Pelvis, thigh | 1755.2 | 1859 | 2068 | 2867.2 | 3081 | 3195 | 3134.1 | 3902 | 3916 |
The customary reporting style at MGH is to give all values with decimals, while at NCI volumes over 100 ml are rounded to the nearest whole. The smallest volume in the dataset is 63.0 ml (PN11, MGH-3DQI, time-point 3), and the largest volume is 5150 ml (PN1, NCI-MEDx, time-point 3). Volume trends by age are provided in supplementary figure 1.
Pairwise comparison of the volume differences among the three sets of analyses showed variable agreement (Table 2A and Supplementary Figure 2).
Table 2.
Summary of volume measurement differences
| A. VOLUMES | Time-point 1 MRI | Time-point 2 MRI | Time-point 3 MRI |
|---|---|---|---|
| MGH-3DQI - NCI-3DQI | |||
| Volume difference: Median (range) – ml* |
−0.5 (−444.3 to 153.2) P=0.45 |
−4.2 (−312.1 to 323.6) P=0.42 |
−19.9 (−767.9 to 41.9) P=0.035 |
| *Corresponding relative volume difference - % of mean (PN ID) | 0.1 (PN13) 15.9 (PN1); 16.2 (PN14) |
2.4 (PN10) 20.3 (PN7); 22.0 (PN14) |
0.9 (PN6) 21.8 (PN15); 19.0 (PN12) |
| Relative volume difference Median (range) - % of mean** |
5.7 (0.1 to 21.2) | 5.0 (1.3 to 22.0) | 4.3 (0.3 to 21.8) |
| **Corresponding absolute volume difference - ml (PN ID) | 103.8 (PN15) 0.5 (PN13); 85.0 (PN5) |
10.4 (PN12) 8.7 (PN2); 323.6 (PN14) |
205.7 (PN1) 2.4 (PN2); 767.9 (PN15) |
| Number of PN with <20% relative volume difference | 14 | 13 | 13 |
| NCI-3DQI - NCI-MEDx | |||
| Volume difference: Median (range) – ml* |
−21.0 (−289.0 to 32.0) P=0.071 |
−47.0 (−406.0 to 29.0) P=0.0024 |
−21.0 (−227.0 to 114.0) P=0.018 |
| *Corresponding relative volume difference - % of mean (PN ID) | 15.4 (PN4) 9.2 (PN1); 3.8 (PN14) |
4.2 (PN13) 24.7 (PN1); 2.2 (PN14) |
13.2 (PN4) 10.4 (PN7); 3.9 (PN14) |
| Relative volume difference Median (range) - % of mean** |
7.7 (0.6 to 28.4) | 4.5 (0.3 to 24.7) | 4.3 (0.4 to 16.0) |
| **Corresponding absolute volume difference - ml (PN ID) | 48.0 (PN2) 0.5 (PN11); 147.0 (PN5) |
8.0 (PN8) 1.0 (PN9); 145.0 (PN5) |
50.0 (PN13) 10.0 (PN6); 114.0 (PN5) |
| Number of PN with <20% relative volume difference | 14 | 14 | 15 |
| MGH-3DQI - NCI-MEDx | |||
| Volume difference: Median (range) – ml* |
−10.0 (−733.3 to 185.2) P=0.21 |
−70.3 (−442.1 to 352.6) P=0.015 |
−29.9 (−781.9 to 47.9) P=0.010 |
| *Corresponding relative volume difference - % of mean (PN ID) | 6.4 (PN10) 25.0 (PN1); 19.9 (PN14) |
9.8 (PN2) 27.6 (PN7); 24.2 (PN14) |
1.3 (PN6) 22.2 (PN15); 22.0 (PN12) |
| Relative volume difference Median (range) - % of mean** |
6.4 (0.1 to 48.8) | 9.8 (3.0 to 44.0) | 9.5 (1.0 to 37.5) |
| **Corresponding absolute volume difference - ml (PN ID) | 10.0 (PN10) 1.0 (PN3); 232.0 (PN5) |
70.3 (PN2) 5.3 (PN8); 237.9 (PN5) |
74.4 (PN2) 29.1 (PN14); 243.4 (PN5) |
| Number of PN with <20% relative volume difference | 12 | 11 | 11 |
| B. INTERVAL CHANGES | Time-point 1 to 2 | Time-point 2 to 3 | Time-point 1 to 3 |
|---|---|---|---|
| MGH-3DQI - NCI-3DQI | |||
| Difference in % change Median (range) - % |
−1.7 (−20.0 to 26.7) P=0.56 |
1.1 (−49.7 to 14.9) P=0.80 |
−1.0 (−59.3 to 27.0) P=0.27 |
| Largest discrepancy in interval change - % (PN ID) | 63.7 vs. 37.0 (PN1) | 75.2 vs. 124.9 (PN14) | 180.5 vs. 239.7 (PN14) |
| Number of PN with <20% difference in interval change | 13 | 14 | 11 |
| Number of PN with concordant disease status assessment | 14 | 14 | 12 |
| NCI-3DQI - NCI-MEDx | |||
| Difference in % change Median (range) - % |
−2.3 (−11.2 to 11.2) P=0.60 |
3.3 (−11.0 to 12.4), P=0.095 |
−1.1 (−8.2 to 20.5) P=0.60 |
| Largest discrepancy in interval change - % (PN ID) | 65.7 vs. 54.5 (PN15) | 27.0 vs. 14.5 (PN3) | 109.9 vs. 89.4 (PN15) |
| Number of PN with <20% difference in interval change | 15 | 15 | 14 |
| Number of PN with concordant disease status assessment | 13 | 13 | 13 |
| MGH-3DQI - NCI-MEDx | |||
| Difference in % change Median (range) - % |
−0.4 (−31.2 to 26.4) P=0.68 |
2.0 (−45.9 to 14.4) P=0.39 |
−1.5 (−58.7 to 28.0) P=0.64 |
| Largest discrepancy in interval change - % (PN ID) | −10.5 vs. 20.7 (PN11) | 75.2 vs. 121.1 (PN14) | 180.5 vs. 239.1 (PN14) |
| Number of PN with <20% difference in interval change | 13 | 14 | 11 |
| Number of PN with concordant disease status assessment | 12 | 12 | 12 |
In large tumors, absolute volume differences tend to be greater, but in proportion to their size smaller lesions can have more measurement discrepancy, therefore we provide both the numeric and relative differences. The volume differences were calculated by subtracting one volume from the matching other, as listed in the header, and ranged from −781.9 ml (PN15, MGH3DQI vs NCI-MEDx, time-point 3) to 406.0 ml (PN1, MGH-3DQI vs. NCI-3DQI, time-point 2). Relative volume differences were calculated using the absolute volume differences between the matching pairs (irrespective of negative or positive values), and comparing to the mean of the same pairs. PN ID refers to the lesion identification as listed in Table 1.
The median difference (range) between MGH-3DQI and NCI-3DQI volumes at time-point one was −0.5 ml (−444.3 to 153.2 ml), at time-point two −4.2 ml (−312.1 to 323.6 ml), and at time-point three −19.9 ml (−767.9 to 41.9 ml) (P=0.45; 0.42; 0.035 respectively). The largest volume differences between MGH-3DQI and NCI-3DQI volumes at each time-point correspond to 15.9% (444.3 ml in PN1), 22.0% (323.6 ml in PN 14), and 21.8% (767.9 ml in PN 15) of the mean of the two volume measurements. In proportion to the mean of the MGH-3DQI and NCI-3DQI measurements the volume difference was less than 20% in 40 of the 45 volume pairs (14 of 15 at time-point 1; 13 of 15 at time-points 2 and 3). The median difference (range) between NCI-3DQI and NCI-MEDx volumes at time-point one was −21.0 ml (−289.0 to 32.0 ml), at time-point two −47.0 ml (−406.0 to 29.0 ml), and at time-point three −21.0 ml (−227.0 to 114.0 ml) (P=0.071; 0.0024; 0.018 respectively). The largest volume differences between NCI-3DQI and NCI-MEDx volumes at each time-point correspond to 9.2% (289 ml in PN 1), 9.4% (406 ml in PN 1), and 10.4% (227 ml in PN 7) of the mean of the two volume measurements. In proportion to the mean of the NCI-3DQI and NCI-MEDx measurements the volume difference was less than 20% in 43 of the 45 volume pairs (14 of 15 at time-points 1 and 2; 15 of 15 at time-point 3). The median difference (range) between MGH-3DQI and NCI-MEDx volumes at time-point one was −10.0 ml (−733.3 to 185.2 ml), at time-point two −70.3 ml (−442.1 to 352.6 ml), and at time-point three −29.9 ml (−781.9 to 47.9 ml) (P=0.21; 0.015; 0.010 respectively). The largest volume differences between MGH-3DQI and NCI-MEDx volumes at each time-point correspond to 25% (733.3 ml in PN1), 27.6% (442.1 ml in PN 7), and 22.2% (781.9 ml in PN 15) of the mean of the two volume measurements. In proportion to the mean of the MGH-3DQI and NCI-MEDx measurements the volume difference was less than 20% in 34 of the 45 volume pairs (12 of 15 at time-point 1; 11 of 15 at time-points 2 and 3).
Of the total of 135 volume pairs compared in 117 (86.7%) the volume differences accounted to less than 20% of the mean of the two measurements (Table 2A). Overall there was a general tendency for the MEDx method to result in larger volumes, which sometimes reached statistical significance, and the closest agreement was observed between the different users (MGH vs. NCI) of the 3DQI method. Over time, as most tumors grew larger, volume measurement variability appears somewhat increased.
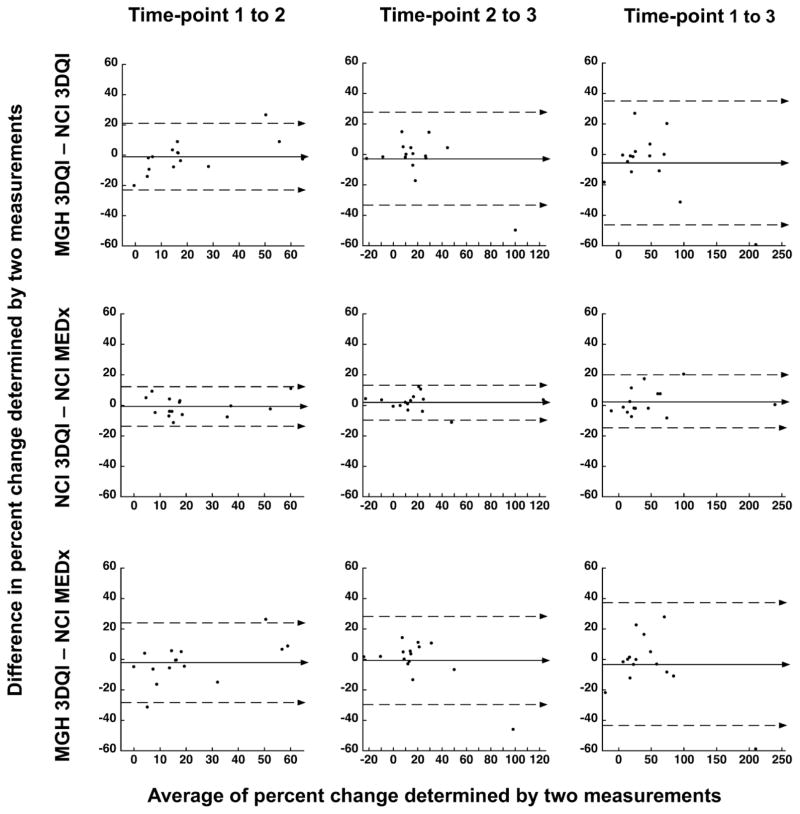
More relevant for clinical trials than agreement in absolute volumes, percent volume changes between time-points were remarkably consistent (Table 2B and Figure 2).
Figure 2. Bland-Altman plots of percent change differences between time-points.
Bland-Altman plots of percent change differences from time-point 1 to 2 (left panels), from time-point 2 to 3 (middle panels), and from time point 1 to 3 (right panels). Measurement differences are plotted against the averages of the two measurements. The top row compares the results of 3DQI method at MGH to 3DQI method at NCI (same method, different user), middle row compares the results of 3DQI method at NCI to MEDx method at NCI (different method, same user), and bottom row compares the results of 3DQI method at MGH to MEDx method at NCI (different method, different user).
The median difference (range) between the percent volume changes determined by MGH-3DQI and NCI-3DQI analyses from time-point one to time-point two was −1.7% (−20.0% to 26.7%), from time-point two to time-point three 1.1% (−49.7% to 14.9%), and from time-point one to time-point three −1.0% (−59.3% to 27.0%) (P=0.56; 0.80; 0.27 respectively). The median difference (range) between the percent volume changes determined by NCI-MEDx and NCI-3DQI analyses from time-point one to time point-two was −2.3% (−11.2% to 11.2%), from time-point two to time-point three 3.3% (−11.0% to 12.4%), and from time-point one to time-point three −1.1% (−8.2% to 20.5%) (P=0.60; 0.095; 0.60 respectively). The median difference (range) between the percent volume changes determined by MGH-3DQI and NCI-MEDx analyses from time-point one to time-point two was −0.40% (−31.2% to 26.4%), from time-point two to time-point three 2.0% (−45.9% to 14.4%), and from time-point one to time-point three −1.5% (−58.7% to 28.0%) (P=0.68; 0.39; 0.64 respectively).
Less than 20% difference in interval percentage change was recorded in 120 of the total of 135 comparisons (88.9%).
Disease status classification between time-points was concordant for 12–14 (80–93.3%) of the 15 cases in the nine sets of comparisons (Supplementary Figure 3), or 115 of the combined 135 comparisons (85.2%). These results suggest moderately strong to strong agreement in disease status classification, with no evidence of significant imbalance in the direction of classification. The 95% confidence interval for 12 of 15 concordant cases is 51.5–95.7%; for 13 of 15 concordant cases 59.5–98.3%; and for 14 of 15 concordant cases 68.1–99.8%
Discussion
While standard solid tumor response criteria continue to be based on uni-, or bi-dimensional tumor measurements12, 13, volumetric lesion assessment has become increasingly utilized in recent years, and most of the commonly used medical image-viewing systems now offer lesion-contouring options. Volume measurements are especially helpful in capturing subtle size changes in slow growing or complex shaped tumors, such as PN.
The success of drug development for PN depends in part on the ability to objectively and sensitively assess tumor response in clinical trials. There is consensus among researchers in the NF field, that linear measurements of PN are not sufficient for imaging response evaluation, and volumetric analysis has become the standard method in clinical trials11.
The level of agreement between different volumetric techniques and different observers has not been tested to date. In this study, we compared sequential volume measurements on 15 complex PN that were generated by two independently developed volumetric methods, and two independent observers. There were no systematic differences in tumor volume approximation, and the absolute volumes, percent changes between time-points, and progression status classifications were in good agreement. Some level of discrepancy between the results is expected due to the difficulty of the analysis.
Variability in size measurements and calculations of interval change are unavoidable, no matter what assessment methods are used. It is generally accepted, that experienced radiologists can measure the longest diameter of the majority of lesions with less than 5% variation from the same scan, but clearly there are exceptions. Zhao et al. propose, that changes of 8% or greater in unidimensional lesion size exceed the measurement variability when measured on chest CT, an imaging modality known to be well reproducible14. The range of discrepancies in linear measurements performed on CT scans of variable body areas is well summarized in a meta-analysis by Yoon et al.15. In pooled estimates the 95% limit of agreement of relative measurement differences was −17.8 to 16.1% for the same observers (5 studies, 648 lesions measured), −22.1 to 25.4% between two observers (8 studies, 1878 lesions measured), and −31.3 to 30.3% in calculating the interval change by different observers (3 studies, 575 lesions measured). Some of the discrepant cases thus exceeded the threshold of significant difference in disease classification. Another study compared CT scans acquired less than 15 minutes apart and found significant differences when measuring the longest diameter of the same lesions side by side16. Given the limitations of the standard measurement tools, it is not rare that investigator reported objective response rates are not confirmed by independent review17. Compared to line measurements, variability may be reduced when using a volumetric tool18–20.
In our series, the volume difference was less than 20% in 117 of 135 volume pairs that were compared. The largest difference in volume measurements was 48.8% (PN 5 at time-point 1: MGH-3DQI=359 ml versus NCI-MEDx=591 ml). 48.8% volume difference between two spheres would be equivalent to about 15% difference in their longest diameters, thus the variation in volumetric size estimates compares favorably with what is reported for standard linear measurements. Similarly, the difference in interval volume change was less than 20% in 120 of the 135 comparisons, while the disease status classification was concordant in 115 of 135 comparisons.
Some of the measurement variations are explained by inherent sources of ambiguity in the medical image data. MRI provides an imperfect pixelated representation of the body. The image units at the edge of a tumor are subject to so called partial volume-averaging, meaning that they represent a mixture of tumor and healthy tissue with intermediate signal intensity, and the actual lesion contour is hidden within that gray zone. Large body segments are typically imaged with in-plane resolution of 0.5–1.5 mm, leading to a natural uncertainty in contour placement. Just 1 mm shift in the outline of a 10-cm spherical lesion will result in 7% difference between the corresponding volumes. With increasing surface to volume ratios in complex-shaped, or very small tumors the variation can be even larger. Using image-sharpening tools can reduce this type of variability; however post-acquisition processing will never reveal as much detail, as images acquired at higher resolution, and some ambiguity in localizing the exact tissue boundary will remain. Importantly, different edge finding algorithms may handle the partial volume-averaging zone differently.
For example, MEDx places the segmentation contour at the outer edge of the boundary zone, while 3DQI on the internal side, and therefore we find a general tendency for the MEDx method to give slightly larger volumes. In most cases the differences remained similar across time-points, and there was good agreement between the changes over time. However, this finding supports our recommendation that the same measurement methodology should be used for the entire duration of a clinical trial11.
The type of lesion selected for contouring also has an impact on test-retest variation. PNs with well-defined borders are easier to outline and can be more consistently measured, as opposed to diffuse, infiltrative PNs. In addition, there may be ambiguity in distinguishing tumor tissue from adjacent structures with similar signal characteristics, such as bowel content, pulmonary atelectasis, cysts or lymph nodes, and can be interpreted differently by different users or even by the same user on repeated analyses.
When comparing two sets of images there can be apparent changes in tumor size without any real structural change. There may be regions where the tumor is not clearly distinguishable from surrounding tissues, and the non-tumor related bright signals could change considerably between MRIs. If direct comparison is not possible between two scans, for example when the body position is changed, it is harder to verify that the same areas are included in the analysis. If there are technical differences between the images that affect the appearance of the tumor edge a consistent contour placement becomes difficult. With simple anatomic MR sequences, temporary swelling in the tumor is indistinguishable from active tumor growth. An experienced user might be able to recognize and compensate for some of these extraneous factors.
The final step of each volumetric analysis is to scrutinize the details and make manual corrections. When visually obvious changes occur, there may be a tendency to neglect the fine-tuning of final contours. This is the likely explanation for the largest discrepancy in interval change in our series (PN 14, time-point 1 to 3, MGH3DQI 1021.2 ml to 2864.2 ml 180.5% increase versus NCI-MEDx 868 ml 2949 ml 239.7% increase). The overall trends are still in very good agreement. In this study both 3DQI and MEDx performed well, and both have some advantages and disadvantages. The MEDx methodology is robust, uncomplicated, and has been in use for over 10 years, but each MRI slice of the slice-by-slice analysis takes about a minute, adding up to an hour or more for complex cases. In contrast, 3DQI processing on the entire three-dimensional image data is more time efficient and on average takes 10–20 minutes. The use of 3DQI is currently limited to researchers at MGH, but the imaging core facility offers processing for a fee for outside institutes. Further development and validation of the 3DQI software is under way with support from the Children’s Tumor Foundation, and the method is intended to be available for broader use in the future. MEDx no longer has technical support by the developing company, however licenses can still be obtained. For ongoing treatment trials, the NCI will continue to utilize MEDx to ensure the continuity of data analysis.
Limitations of our study include the potential bias in the selection of PN for analysis and the retrospective study design. Our focus was to evaluate methodological and user dependent sources of measurement variability. To limit other sources of variance, we selected consistently high-quality MR images from a representative cohort of clinical trial candidates with measurable complex tumors, rather than randomly assigning patients for the study. In a real-life clinical trial, there may be even more challenging or sub-optimally imaged cases.
In a retrospective study having access to images from subsequent time-points helps to resolve some of the structural ambiguities and potentially reduce the discrepancy between different observers. The results reported here might not be fully predictive of the level of agreement in a prospective setting.
In conclusion, our study demonstrated good agreement in percent volume changes between time-points, and progression status classifications. A prospective real-time response evaluation using both methods in a clinical trial is planned. The ultimate goal is to provide options for validated volumetric analysis methods for use in NF1-PN clinical trials and NF clinics worldwide. Reliability, ease of use, and processing speed are critical factors in adapting these technologies from the clinical trials research domain into clinical practice.
Supplementary Material
Acknowledgments
Funding sources: This research was supported in part by the National Cancer Institute Center for Cancer Research Intramural Research Program, by the National Institutes of Health Small Business Technology Transfer (STTR) grant number 1R41CA192600 to Massachusetts General Hospital through a subcontract from IQ Medical Imaging LLC., and by the Children’s Tumor Foundation.
Footnotes
Declaration of interest:
E. Dombi Conflicts of interest: none.
W. Cai receives research support from NCI, American Cancer Society, and the Children’s Tumor Foundation. He serves on the editorial boards of the International Journal of Intelligent Information Processing, World Journal of Radiology, International Journal of Radiology, and Advances in Modern Oncology Research.
SM. Steinberg Conflicts of interest: none.
MA. Bredella Conflicts of interest: none.
GM. Basinsky Conflicts of interest: none.
B. Somarouthu Conflicts of interest: none.
SR. Plotkin receives research support from the Children’s Tumor Foundation, NIH, and the Department of Defense Neurofibromatosis Clinical Trials Consortium.
J. Solomon Conflicts of interest: none.
BC. Widemann receives research support through the NCI CCR Intramural Research Program.
GJ. Harris receives research support from Children’s Tumor Foundation and NIH. He also serves on the scientific advisory board of Fovia, Inc. Member, IQ Medical Imaging LLC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cai W, Kassarjian A, Bredella MA, et al. Tumor burden in patients with neurofibromatosis types 1 and 2 and schwannomatosis: determination on whole-body MR images. Radiology. 2009;250:665–673. doi: 10.1148/radiol.2503080700. [DOI] [PubMed] [Google Scholar]
- 2.Solomon J, Warren K, Dombi E, Patronas N, Widemann B. Automated detection and volume measurement of plexiform neurofibromas in neurofibromatosis 1 using magnetic resonance imaging. Computerized medical imaging and graphics: the official journal of the Computerized Medical Imaging Society. 2004;28:257–265. doi: 10.1016/j.compmedimag.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Poussaint TY, Jaramillo D, Chang Y, Korf B. Interobserver reproducibility of volumetric MR imaging measurements of plexiform neurofibromas. AJR Am J Roentgenol. 2003;180:419–423. doi: 10.2214/ajr.180.2.1800419. [DOI] [PubMed] [Google Scholar]
- 4.Dombi E, Baldwin A, Marcus LJ, et al. Activity of Selumetinib in Neurofibromatosis Type 1-Related Plexiform Neurofibromas. N Engl J Med. 2016;375:2550–2560. doi: 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson KA, Nalepa G, Yang FC, et al. Imatinib mesylate for plexiform neurofibromas in patients with neurofibromatosis type 1: a phase 2 trial. Lancet Oncol. 2012;13:1218–1224. doi: 10.1016/S1470-2045(12)70414-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss B, Widemann BC, Wolters P, et al. Sirolimus for progressive neurofibromatosis type 1-associated plexiform neurofibromas: a neurofibromatosis Clinical Trials Consortium phase II study. Neuro-oncology. 2015;17:596–603. doi: 10.1093/neuonc/nou235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss B, Widemann BC, Wolters P, et al. Sirolimus for non-progressive NF1-associated plexiform neurofibromas: an NF clinical trials consortium phase II study. Pediatric blood & cancer. 2014;61:982–986. doi: 10.1002/pbc.24873. [DOI] [PubMed] [Google Scholar]
- 8.Widemann BC, Babovic-Vuksanovic D, Dombi E, et al. Phase II trial of pirfenidone in children and young adults with neurofibromatosis type 1 and progressive plexiform neurofibromas. Pediatric blood & cancer. 2014;61:1598–1602. doi: 10.1002/pbc.25041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Widemann BC, Dombi E, Gillespie A, et al. Phase 2 randomized, flexible crossover, double-blinded, placebo-controlled trial of the farnesyltransferase inhibitor tipifarnib in children and young adults with neurofibromatosis type 1 and progressive plexiform neurofibromas. Neuro-oncology. 2014;16:707–718. doi: 10.1093/neuonc/nou004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plotkin SR, Blakeley JO, Dombi E, et al. Achieving consensus for clinical trials: the REiNS International Collaboration. Neurology. 2013;81:S1–5. doi: 10.1212/01.wnl.0000435743.49414.b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dombi E, Ardern-Holmes SL, Babovic-Vuksanovic D, et al. Recommendations for imaging tumor response in neurofibromatosis clinical trials. Neurology. 2013;81:S33–40. doi: 10.1212/01.wnl.0000435744.57038.af. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47:207–214. doi: 10.1002/1097-0142(19810101)47:1<207::aid-cncr2820470134>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Zhao B, James LP, Moskowitz CS, et al. Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology. 2009;252:263–272. doi: 10.1148/radiol.2522081593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon SH, Kim KW, Goo JM, Kim DW, Hahn S. Observer variability in RECIST-based tumour burden measurements: a meta-analysis. Eur J Cancer. 2016;53:5–15. doi: 10.1016/j.ejca.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Oxnard GR, Zhao B, Sima CS, et al. Variability of lung tumor measurements on repeat computed tomography scans taken within 15 minutes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:3114–3119. doi: 10.1200/JCO.2010.33.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ford R, Schwartz L, Dancey J, et al. Lessons learned from independent central review. Eur J Cancer. 2009;45:268–274. doi: 10.1016/j.ejca.2008.10.031. [DOI] [PubMed] [Google Scholar]
- 18.Buckler AJ, Danagoulian J, Johnson K, et al. Inter-Method Performance Study of Tumor Volumetry Assessment on Computed Tomography Test-Retest Data. Acad Radiol. 2015;22:1393–1408. doi: 10.1016/j.acra.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dejaco D, Url C, Schartinger VH, et al. Approximation of head and neck cancer volumes in contrast enhanced CT. Cancer Imaging. 2015;15:16. doi: 10.1186/s40644-015-0051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino M, Guo M, Jackman DM, et al. CT tumor volume measurement in advanced non-small-cell lung cancer: Performance characteristics of an emerging clinical tool. Acad Radiol. 2011;18:54–62. doi: 10.1016/j.acra.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.