Abstract
Cowhage is routinely used in human studies of histamine-independent itch. As such, it is of interest to understand its mechanism of action. The active component of cowhage is a cysteine protease called mucunain. As serine proteases can activate members of the protease-activated receptor (PAR) family to induce itch and pain, the mechanism of action of mucunain and cathepsin S, a human cysteine protease, were likewise thought to be through activation of PARs. The demonstration that cathepsin S activates members of the Mas-related G-protein coupled receptor family to induce itch led us to evaluate the activity of mucunain on these receptors. We find that mucunain activates the human receptors MRPGRX1 and MRGPRX2 and induces degranulation of human mast cells. These findings indicate that Mrgprs may be involved in itch induced by cowhage.
Keywords: itch, pruritus, protease, Mas-related G-protein coupled receptor, protease-activated receptor, Mrgpr, 7TM receptor
Cowhage is the term applied to the itch-inducing spicules or trichomes that cover the seedpods of the tropical plant Mucuna pruriens. Botany purists often prefer the term Stizolobium pruriens. There are numerous terms, based on the local customs and language, equivalent to cowhage. Itching powder is one of the colloquial terms in the English language that refers to the spicules. Cowhage can also refer to the active component in the spicules, a protease, also known as mucunain.
Cowhage and histamine are the two most widely used substances employed to measure itch in human psychophysical studies. Cowhage activates mechanically-sensitive sensory nerve fibers whereas histamine activates mechanically-insensitive(Schmelz et al., 2000). The intensity of cowhage-induced itch is significantly higher than histamine-induced itch(Papoiu et al., 2011). Cowhage-induced itch is mediated by pathways that are independent of histamine(Davidson et al., 2007, Kosteletzky et al., 2009). The majority of itches encountered in the clinic have a limited response to antihistamines, highlighting the importance of histamine-independent pathways. The molecular mechanism of cowhage-induced itch is thus of interest and importance with respect to understanding itch and the development of therapeutics.
In the mid-1950s, proteins, not genes, were the rage in biomedical research. Proteases, which are proteins with the capacity to cut molecules, were part of this phenomenon. Consistent with the times, it was suggested in 1955 that a protease may be the active pruritogen in cowhage, and it was given the name mucunain(Shelley and Arthur, 1955). This prescient observation was confirmed in 2008 with the isolation and identification of a cysteine protease from cowhage(Reddy et al., 2008). Heat-inactivated spicules reconstituted with the plant protease induced itch in humans akin to native cowhage spicules. In addition, an irreversible inhibitor of cysteine proteases, E64, blocked the sensations induced both by native cowhage and the spicules reconstituted with mucunain(Reddy et al., 2008).
Mucunain was shown to activate the protease-activated receptors (PAR) 2 and 4, implicated previously in itch and pain(Reddy et al., 2008). Further studies revealed that additional plant proteases known to induce itch are also cysteine proteases and activate PAR2 and 4(Reddy and Lerner, 2010). An endogenous cysteine protease, cathepsin S, which shares active site sequence homology with mucunain(Reddy et al., 2010) was also found to activate PARs and induce itch, pricking and burning sensations similar to cowhage(Reddy et al., 2010). Transgenic mice in which cathepsin S was overexpressed, spontaneously developed an intensely pruritic disorder resembling atopic dermatitis(Kim et al., 2012). Cathepsin S levels were also found to be elevated in psoriatic keratinocytes, consistent with its disease association with itch(Schonefuss et al., 2010).
There was however, an inconsistency. The data on the role of PAR4 in itch are limited and contradictory(Asfaha et al., 2007, Tsujii et al., 2008). On one hand, the expression of PAR2 on keratinocytes and dorsal root ganglion neurons (DRGs), and the previously proposed roles for this receptor in itch and pain(Steinhoff et al., 2003), supported a role for PAR2 in cathepsin S and cowhage-induced itch. On the other hand, cathepsin S-induced itch remains intact in PAR2 knock out mice(Reddy et al., 2015).
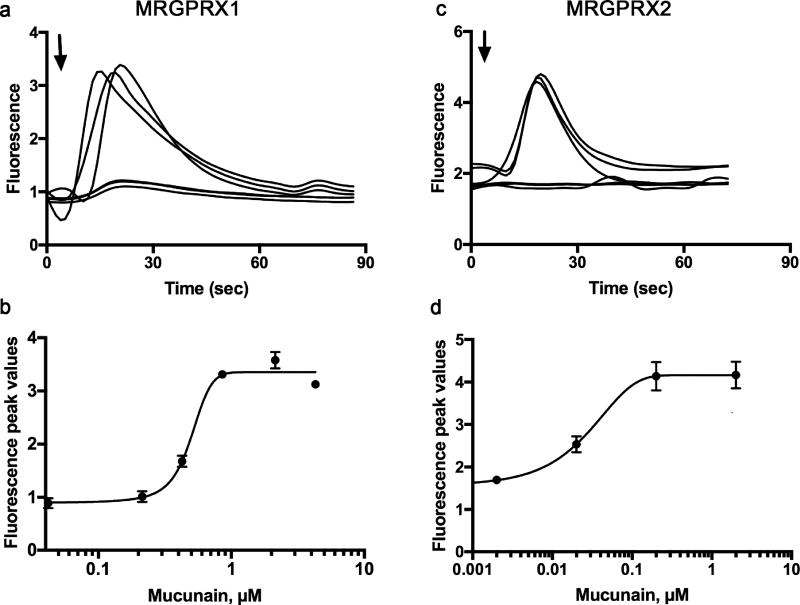
The key to solving the problem of cowhage and cathepsin S-induced itch, and potentially conditions associated with histamine-independent itch, is a family of receptors known as Mas-related G-protein-coupled receptors or Mrgprs. Mrgprs were identified in 2001 and their role in histamine-independent itch is now clear(Dong et al., 2001). Further studies revealed that cathepsin S activates human MRGPRX2 and, as opposed to PAR2 knock out mice, cathepsin S-induced itch is significantly decreased in Mrgpr cluster knock out mice(Reddy et al., 2015). Here we report that mucunain activates human MRGPRX1 and MRGPRX2 (Figure 1).
Figure 1. Mucunain activates the human receptors MRGPRX1 and human MRGPRX2.
(a, c) Responses of individual cells to 300 nM mucunain added at the arrow. (b, d) Concentration effect curves for mucunain on MRGPRX1 (EC50 0.50 µM) and MRGPRX2 (EC50 0.11) respectively. For (a) and (b), HEK-293 cells were transiently transfected with a cDNA encoding the human receptor MRGPRX1. For (b) and (d), a HEK-293 cell line was engineered to stably express a cDNA encoding the human receptor MRGPRX2. Intracellular calcium [Ca2+]i was determined by ratiometric Fura-2 imaging as an indicator of receptor activation following the addition of mucunain.
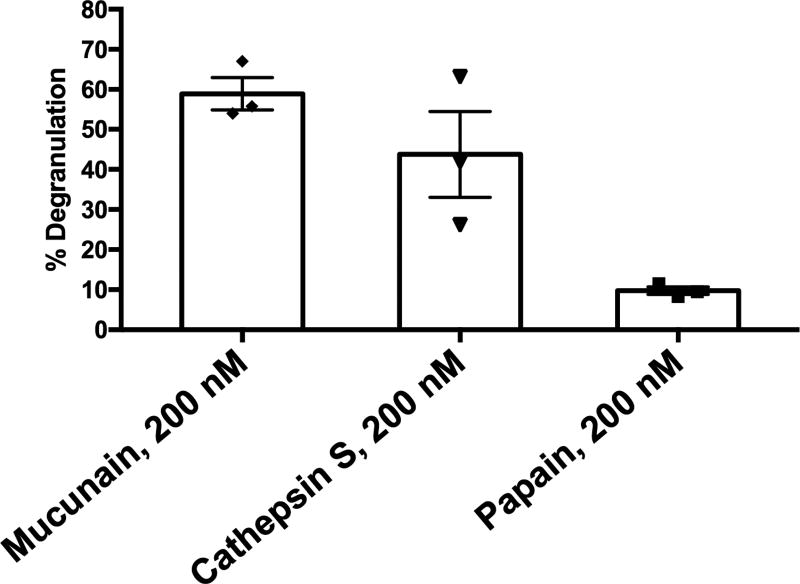
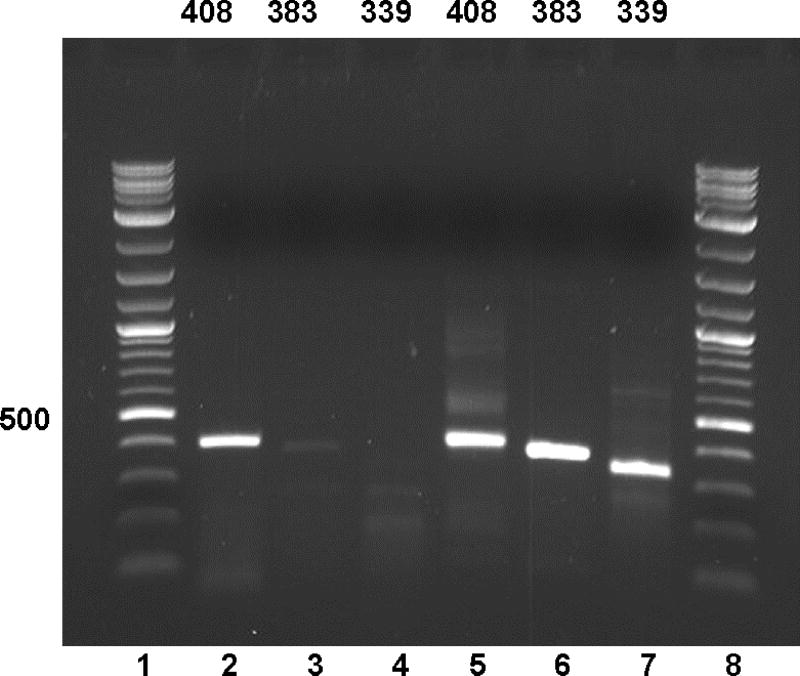
The mechanism of cowhage-induced itch may now be explained as follows: MRGPRX1 is expressed on human DRG neurons. Chloroquine and BAM8-22, in addition to cowhage, activate this receptor to induce histamine-independent itch(Liu et al., 2009, Sikand et al., 2011). MRGPRX2 is expressed on human mast cells(McNeil et al., 2014) and mediates IgE-independent mast cell degranulation induced by a number of medications and basic secretagogues(Azimi et al., 2016, McNeil et al., 2014). Mucunain induces mast cell degranulation (Figure 2), likely via activation of MRGPRX2, and may release histamine (McNeil et al., 2014). MRGPRX2, in addition to MRGPRX1, is transcribed (Figure 3) in and found on human DRG neurons(Robas et al., 2003, Tatemoto et al., 2006, Zhang et al., 2005) allowing for cowhage to induce itch by multiple mechanisms. We acknowledge as a possible limitation that MRGPRX2 expression on neurons could result from mast cell contamination.. We are also aware of the report in which transcriptional profiling from human DRG neurons did not identify MRGPRX2(Flegel et al., 2015) but these data were generated without amplification and do not take into account the possibility that pruritogenic or inflammatory stimuli could contribute to expression.
Figure 2. Mucunain and cathepsin S degranulate human mast cells.
The level of degranulation in the LAD-2 mast cell line was assessed by the release of β-hexosaminidase as quantified by the level of its substrate p-nitrophenyl N-acetyl-β-d-glucosamide (PNAG) digested in a colorimetric assay. Human MRGPRX2 mediates IgE-independent mast cell degranulation. Mucunain and cathepsin S cause degranulation. In contrast, papain, another cysteine protease, does not, as it activates MRGPRX1, not present on LAD-2 cells.
Figure 3. MRGPRX2 appears to be transcribed in human dorsal root ganglia (DRG).
- F: TCAGCGGTCGTGTGTGTCCTGCTCTGGG
- R: GAAGAAGTAAATGATGGGGTTGGCAC
- X2INTF1: AATTCTGCACCCCCATGGAG
- X2INTR1: GCCTGCATTTGAACCCACAG
- X2INTF2: ATTAGCCAGTCGTGGTGGTG
- X2INTR2: CTCCATGGGGGTGCAGAATT
We propose that while cowhage primarily activates the histamine-independent pathway it can also induce the release of histamine from mast cells. This component of cowhage-induced itch was not previously appreciated. A question then arises: If mucunain induces mast cell degranulation, why is cowhage only sometimes associated with the typical wheal and flare response of histamine injection(Sikand et al., 2009)? The likely answer is that the intra epidermal nature of skin penetration by cowhage spicules combined with the limited amount of mucunain that spicules can carry and deliver is consistent with mast cells not being activated on a regular basis. In contrast, intradermal administration of histamine would allow its diffusion to reach mast cells leading to their activation. Indeed studies with histamine at concentrations equipotent to native cowhage spicules produce sensory qualities of similar magnitude and duration and a cowhage spicule reconstituted by histamine, is capable of inducing itch and nociceptive sensations in the absence of a visible flare(Sikand et al., 2009). This explanation is consistent with a study performed with plant proteases on 75 healthy subjects from the 1950s in which a variable degree of erythema and wheal formation was noted at the site of intradermal injection(Arthur and Shelley, 1955). However, when a single spicule was inserted in the epidermis, itch was produced in the absence of visible skin change(Arthur and Shelley, 1955).
In summary, we propose that MRGPRX1 and MRGPRX2 contribute to cowhage induced-itch. These receptors represent potential therapeutic targets for histamine-independent itch.
Acknowledgments
EA is the recipient of a grant from the National Eczema Association. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, R01AR057744 and R21AR067399 to EAL. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- DRG
dorsal root ganglion
- Mrgpr
mas-related G-protein coupled receptor
- PAR
protease-activated receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors state no conflict of interest.
References
- Arthur RP, Shelley WB. The role of proteolytic enzymes in the production of pruritus in man. The Journal of investigative dermatology. 1955;25(5):341–6. doi: 10.1038/jid.1955.138. [DOI] [PubMed] [Google Scholar]
- Asfaha S, Cenac N, Houle S, Altier C, Papez MD, Nguyen C, et al. Protease-activated receptor-4: a novel mechanism of inflammatory pain modulation. British journal of pharmacology. 2007;150(2):176–85. doi: 10.1038/sj.bjp.0706975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi E, Reddy VB, Shade KC, Anthony RM, Talbot S, Pereira PJ, et al. Dual action of neurokinin-1 antagonists on Mas-related GPCRs. JCI Insight. 2016;1(16):e89362. doi: 10.1172/jci.insight.89362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S, Zhang X, Yoon CH, Khasabov SG, Simone DA, Giesler GJ., Jr The itch-producing agents histamine and cowhage activate separate populations of primate spinothalamic tract neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27(37):10007–14. doi: 10.1523/JNEUROSCI.2862-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106(5):619–32. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- Flegel C, Schobel N, Altmuller J, Becker C, Tannapfel A, Hatt H, et al. RNA-Seq Analysis of Human Trigeminal and Dorsal Root Ganglia with a Focus on Chemoreceptors. PloS one. 2015;10(6):e0128951. doi: 10.1371/journal.pone.0128951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Bae KB, Kim MO, Yu DH, Kim HJ, Yuh HS, et al. Overexpression of cathepsin S induces chronic atopic dermatitis in mice. The Journal of investigative dermatology. 2012;132(4):1169–76. doi: 10.1038/jid.2011.404. [DOI] [PubMed] [Google Scholar]
- Kosteletzky F, Namer B, Forster C, Handwerker HO. Impact of scratching on itch and sympathetic reflexes induced by cowhage (Mucuna pruriens) and histamine. Acta dermato-venereologica. 2009;89(3):271–7. doi: 10.2340/00015555-0624. [DOI] [PubMed] [Google Scholar]
- Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, et al. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139(7):1353–65. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil BD, Pundir P, Meeker S, Han L, Undem BJ, Kulka M, et al. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature. 2014 doi: 10.1038/nature14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoiu AD, Tey HL, Coghill RC, Wang H, Yosipovitch G. Cowhage-induced itch as an experimental model for pruritus. A comparative study with histamine-induced itch. PloS one. 2011;6(3):e17786. doi: 10.1371/journal.pone.0017786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VB, Iuga AO, Shimada SG, LaMotte RH, Lerner EA. Cowhage-evoked itch is mediated by a novel cysteine protease: a ligand of protease-activated receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28(17):4331–5. doi: 10.1523/JNEUROSCI.0716-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VB, Lerner EA. Plant cysteine proteases that evoke itch activate protease-activated receptors. The British journal of dermatology. 2010;163(3):532–5. doi: 10.1111/j.1365-2133.2010.09862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VB, Shimada SG, Sikand P, Lamotte RH, Lerner EA. Cathepsin S elicits itch and signals via protease-activated receptors. The Journal of investigative dermatology. 2010;130(5):1468–70. doi: 10.1038/jid.2009.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VB, Sun S, Azimi E, Elmariah SB, Dong X, Lerner EA. Redefining the concept of protease-activated receptors: cathepsin S evokes itch via activation of Mrgprs. Nat Commun. 2015;6:7864. doi: 10.1038/ncomms8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robas N, Mead E, Fidock M. MrgX2 is a high potency cortistatin receptor expressed in dorsal root ganglion. The Journal of biological chemistry. 2003;278(45):44400–4. doi: 10.1074/jbc.M302456200. [DOI] [PubMed] [Google Scholar]
- Schmelz M, Michael K, Weidner C, Schmidt R, Torebjork HE, Handwerker HO. Which nerve fibers mediate the axon reflex flare in human skin? Neuroreport. 2000;11(3):645–8. doi: 10.1097/00001756-200002280-00041. [DOI] [PubMed] [Google Scholar]
- Schonefuss A, Wendt W, Schattling B, Schulten R, Hoffmann K, Stuecker M, et al. Upregulation of cathepsin S in psoriatic keratinocytes. Experimental dermatology. 2010;19(8):e80–8. doi: 10.1111/j.1600-0625.2009.00990.x. [DOI] [PubMed] [Google Scholar]
- Shelley WB, Arthur RP. Mucunain, the active pruritogenic proteinase of cowhage. Science. 1955;122(3167):469–70. doi: 10.1126/science.122.3167.469. [DOI] [PubMed] [Google Scholar]
- Sikand P, Dong X, LaMotte RH. BAM8-22 peptide produces itch and nociceptive sensations in humans independent of histamine release. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(20):7563–7. doi: 10.1523/JNEUROSCI.1192-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikand P, Shimada SG, Green BG, LaMotte RH. Similar itch and nociceptive sensations evoked by punctate cutaneous application of capsaicin, histamine and cowhage. Pain. 2009;144(1–2):66–75. doi: 10.1016/j.pain.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff M, Neisius U, Ikoma A, Fartasch M, Heyer G, Skov PS, et al. Proteinase-activated receptor-2 mediates itch: a novel pathway for pruritus in human skin. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23(15):6176–80. doi: 10.1523/JNEUROSCI.23-15-06176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatemoto K, Nozaki Y, Tsuda R, Konno S, Tomura K, Furuno M, et al. Immunoglobulin E-independent activation of mast cell is mediated by Mrg receptors. Biochemical and biophysical research communications. 2006;349(4):1322–8. doi: 10.1016/j.bbrc.2006.08.177. [DOI] [PubMed] [Google Scholar]
- Tsujii K, Andoh T, Lee JB, Kuraishi Y. Activation of proteinase-activated receptors induces itch-associated response through histamine-dependent and -independent pathways in mice. J Pharmacol Sci. 2008;108(3):385–8. doi: 10.1254/jphs.08200sc. [DOI] [PubMed] [Google Scholar]
- Zhang L, Taylor N, Xie Y, Ford R, Johnson J, Paulsen JE, et al. Cloning and expression of MRG receptors in macaque, mouse, and human. Brain Res Mol Brain Res. 2005;133(2):187–97. doi: 10.1016/j.molbrainres.2004.10.007. [DOI] [PubMed] [Google Scholar]