Abstract
The tissue factor pathway inhibitor (TFPI) gene encodes a protease inhibitor with a critical role in regulation of blood coagulation. Some genomic variants in TFPI were previously associated with plasma TFPI levels, however, it remains to be further determined whether TFPI variants are associated with other coagulation factors. In this study, we carried out a large population-based study with 2,313 study subjects for blood coagulation data, including fibrinogen levels, prothrombin time (PT), activated partial thromboplastin time (APTT), and thrombin time (TT). We identified significant association of TFPI variant rs10931292 (a functional promoter variant with reduced transactivation) with increased plasma fibrinogen levels (P=0.017 under a recessive model), but not with PT, APTT or TT (P>0.05). Using a large case control association study population with 4,479 CAD patients and 3,628 controls, we identified significant association between rs10931292 and CAD under a recessive model (OR = 1.23, P = 0.005). For the first time, we show that a TFPI variant is found to be significantly associated with fibrinogen levels and risk of CAD. Our finding contributes significantly to the elucidation of the genetic basis and biological pathways responsible for fibrinogen levels and development of CAD.
Keywords: Tissue factor pathway inhibitor (TFPI), Coagulation Factors, Fibrinogen, Coronary Artery Disease (CAD), Association
Introduction
The TFPI gene encodes the tissue factor pathway inhibitor, which is a protease inhibitor regulating the tissue factor (TF)-dependent pathway of blood coagulation, a major factor for thrombosis and risk of cardiovascular disease (Maroney and Mast, 2015; Mast, 2016). The major goal of this study is to determine whether genomic variation in the TFPI gene is associated with the levels of key blood coagulation parameters such as the fibrinogen levels (FIB or Fg) and risk of cardiovascular disease. The TFPI gene is located on chromosome 2q32.1 and spans 101,531 base pairs (http://www.genecards.org/cgi-bin/carddisp.pl?gene=TFPI). TFPI is an anticoagulant protein that is expressed primarily in the vascular endothelium, megakaryocytes, platelets and plasma (Maroney and Mast, 2015; Mast, 2016). There are at least three alternatively spliced isoforms of TFPI in humans, TFPIα, TFPIβ and TFPIγ, which are produced from alternatively spliced mRNAs from a single gene (Maroney and Mast, 2015; Mast, 2016). TFPIα and TFPIβ are the two well-characterized, primary isoforms responsible for inhibiting coagulation (Maroney and Mast, 2015; Mast, 2016). TFPIα is a secreted protein that contains an acidic N- terminal domain, a basic C-terminal domain and 3 tandem Kunitz-type serine protease inhibitor domains, K1, K2 and K3(Maroney and Mast, 2015; Mast, 2016). The K1 domain interacts with FVIIa, the K2 domain with Xa and the K3 domain with protein S (Maroney and Mast, 2015; Mast, 2016). TFPIβ is shorter than TFPIα and contains only two tandem Kunitz-type serine protease inhibitor domains K1 and K2 followed by a GPI-anchor attachment sequence, which anchors TFPIβ to the cell membrane to execute its function (Maroney et al., 2013). The interaction of TFPI with the FVIIa-TF complex potently inhibits the initiation of blood coagulation (Maroney and Mast, 2015; Mast, 2016). The C-terminus of TFPIα interacts with factor V, which activates factor V and inhibits prothrombinase during the initiation of blood coagulation (Maroney and Mast, 2015; Mast, 2016; Wood et al., 2013).
Blood coagulation is initiated with the formation of the FVIIa-TF complex at a site of blood vessel injury (Hoffman and Monroe, 2007). This proteolytically activates FX protease (Hoffman and Monroe, 2007). Activated FX (FXa) mediates the formation of thrombin from prothrombin (FII) together with FVa, calcium and phospholipid (prothrombinase complex) (Hoffman and Monroe, 2007). Thrombin mediates the formation of fibrin (FIa) from fibrinogen (FI) (Hoffman and Monroe, 2007). Cross-linking of the fibrin monomers by FXIIIa results in the formation of a fibrin clot (Hoffman and Monroe, 2007). TFPI inactivates the TF–FVIIa complex in concert with FXa, thereby inhibiting coagulation (Maroney and Mast, 2015; Mast, 2016).
Increased coagulation may increase risk of thrombosis and thromboembolism, whereas decreased coagulation may be associated with bleeding (Hayward et al., 2012). A person’s coagulation activity can be measured by a blood coagulation test, which is usually ordered if there is a problem of bleeding or an unexplained blood clot (thrombosis) (Chee, 2014; Hayward et al., 2012). Blood coagulation panels typically include the fibrinogen level, prothrombin time (PT), activated partial thromboplastin time (APTT)and thrombin time (TT) (Chee, 2014; Hayward et al., 2012).
Genetic factors play an important role in a person’s coagulation activity (Souto et al., 2000). For example, the heritability of plasma fibrinogen levels was estimated to be from 24% to 50% (de Lange et al., 2001; de Lange et al., 2006; Souto et al., 2000; Yang et al., 2003). In 2009, a genome-wide association study (GWAS) with 22,096 study subjects of European ancestry identified 4 genetic loci for circulating fibrinogen levels, including a single nucleotide polymorphism (SNP) in the FGB gene encoding the fibrinogen β-chain and SNPs in IRF1 (interferon regulatory factor 1), PCCB (propionyl coenzyme A carboxylase), and NLRP3 (NLR family pyrin domain containing 3 isoform) (Dehghan et al., 2009). In 2011, a large-scale candidate gene association study with 23,634 European Americans (EA) and 6,657 African Americans (AA) revealed that EA and AA populations share risk SNPs associated with fibrinogen levels, such as FGB rs1800787 and FGG rs2066861 and variants in IL6R, IL1RN, and NLRP3 (Wassel et al., 2011). In 2013, a multiethnic meta-GWAS with >100,000 subjects identified 23 fibrinogen loci and 15 of them are new loci (Sabater-Lleal et al., 2013). The most recent meta-GWAS involved 120,246 study subjects and identified 18 new loci for fibrinogen levels (de Vries et al., 2016). However, all fibrinogen loci identified to date can explain only 3% of the heritability of circulating fibrinogen levels (de Vries et al., 2016). The majority of heritability remains missing, a phenomenon referred to as “missing heritability”. Identification of new SNPs associated with fibrinogen levels will further our understanding of the heritability and genetic architecture of plasma fibrinogen levels.
Due to the important role of TFPI in blood coagulation, we studied association of a functional variant, the T-287C variant in the TFPI promoter/regulatory region (SNP rs10931292) (Amini and Iles, 2008) with coagulation parameters, including fibrinogen levels, PT, APTT, and TT. We identified significant association with fibrinogen levels only. We further studied the association between SNP rs10931292 and coronary artery disease (CAD) because plasma fibrinogen levels were significantly associated with risk of CAD (Danesh et al., 2005). We found that SNP rs10931292 conferred a significant risk of CAD.
Materials and Methods
Study subjects
All study subjects for this study were selected from the Gene ID database, which is one of the largest databases with clinical data and DNA samples for cardiovascular diseases in China. Both cardiovascular patients and controls without cardiovascular diseases were enrolled. All subjects belong to the ethnic group of Han by self-description. This study was approved by the Ethics Committee on Human Subject Research at Huazhong University of Science and Technology and local Institutional Review Boards (IRB) on Human Subject Research at participating hospitals. Written informed consent was obtained from the study participants. This study conformed to the guidelines set forth by the Declaration of Helsinki.
For the association study between TFPI SNP rs10931292 and coagulation parameters, clinical diagnostic data on fibrinogen levels, PT, APTT, and TT were extracted from the GeneID database.
For the case control association study between TFPI SNP rs10931292 and CAD, each study subject was carefully evaluated by at least two independent cardiologists using the American College of Cardiology/American Heart Association criteria for a diagnosis of CAD and myocardial infarction (MI) as described previously by us(Chen et al., 2016; Li et al., 2013; Shen et al., 2007; Shen et al., 2008a; Shen et al., 2008b; Shen et al., 2012; Shen et al., 2013; Shen et al., 2014; Tu et al., 2013; Wang et al., 2011; Wang et al., 2004; Xu et al., 2014). We classified individuals with ≥70% luminal stenosis in at least 1 main vessel, percutaneous coronary angioplasty (PCA), coronary artery bypass graft (CABG), and MI as CAD cases. We classified individuals with typical chest pain sustained for at least 30 min, characteristic electrocardiographic patterns of acute MI, and elevation of troponin I or T and cardiac enzymes such as creatine kinase-MB and lactate dehydrogenase as being affected with MI. The control subjects are those individuals without a history CAD or MI or detectable stenosis of a coronary artery by coronary angiography. We excluded study subjects with congenital heart disease, type I diabetes mellitus, myocardial spasm, and myocardial bridge identified by angiography. We extracted demographic and other relevant clinical information, if present, from the medical records from the selected study subjects.
Isolation of human genomic DNA and SNP genotyping
Human genomic DNA was purified from whole blood samples using the Wizard Genomic DNA Purification kit (Promega, USA). Each DNA sample was analyzed for quantity and quality by spectrophotometry (Nanodrop 1000, Thermo Fisher Scientific, Wilmington, DE, USA) and agarose gel electrophoresis.
For association studies, we selected SNP rs10931292 in the TFPI promoter and regulatory region because this variant was shown to be a functional variant that affects the transcriptional activation of the TFPI promoter in a luciferase reporter assay (Amini and Iles, 2008). TFPI SNP rs10931292 was genotyped by the TaqMan assay using a Roche 480 Light Cycler (Roche, Germany) as described previously by us (Chen et al., 2015; Chen et al., 2016; Huang et al., 2015b; Li et al., 2013; Shen et al., 2009; Tu et al., 2013; Wang et al., 2011; Xiong et al., 2013). Twenty-four samples were randomly selected for Sanger sequencing analysis. The sequencing results were compared to the TaqMan genotyping data, and 100% matching was found between the two genotyping platforms.
Statistical analysis
Genotyping data were analyzed for Hardy-Weinberg equilibrium using PLINK version 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/archive.shtml). For analysis of the association between TFPISNP rs10931292 and coagulation parameters (Fg/FIB, PT APPT, and TT), we performed linear modeling by incorporating covariates of age and gender using statistical program SPSS (version 17.0). The linear regression analysis was carried out under three different genetic models, including an additive model, an autosomal dominant model or an autosomal recessive model.
For the association analysis between TFPI SNP rs10931292 and CAD, we used the X2 test. The analysis of allelic association was carried out using 2×2 Pearson χ2 contingence tables, whereas genotypic association analysis was performed using 2×3 Pearson χ2 contingence tables under three different genetic models, including an additive model, an autosomal dominant model or an autosomal recessive model. P values and corresponding odds ratios (ORs) with 95% confidential intervals were also calculated. The statistical analysis was performed using PLINK version 1.07 or SPSS version.17.0. For case control association analysis, multiple logistic regression analysis was performed to adjust significant covariates of age and gender for CAD using SPSS version.17.0.
We used PS software 3.0.12 to calculate the statistical power and sample sizes (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize) as described (Chen et al., 2015; Chen et al., 2016; Huang et al., 2015b; Li et al., 2013; Xu et al., 2014; Yin et al., 2017). The statistical power can be calculated with special parameters, including the minor allele frequency (MAF), OR or effect size, the numbers of cases and controls, and the Type I error of 0.05.
Results
Significant association of TFPI SNP rs10931292 with circulating fibrinogen levels
TFPI plays an important role in regulation of blood coagulation (Maroney and Mast, 2015; Mast, 2016), therefore, we hypothesized that genomic variants of TFPI may be associated with some coagulation parameters. To test the hypothesis, we performed a population-based study to analyze the association between TFPI SNP rs10931292 and quantitative traits of fibrinogen levels, PT, APTT and TT. We searched our GeneID database for all individuals with available data on fibrinogen levels, PT, APTT and TT. A total of 2,313 subjects were found to have data on coagulation parameters (Table 1) and used for association analysis with TFPI SNP rs10931292. The clinical and demographic characteristics of the 2,313 study subjects are shown in Table 1. For the association of TFPI SNP rs10931292 with levels of fibrinogen, PT, APTT and TT, our study population can provide 70%, 74%, 80%, and 78% of statistical power, respectively, with the effect size (β) of 0.5, MAF of 0.31 and a type I error of 0.05.
Table 1.
Characteristics of the study subjects for the association analysis between TFPI SNP rs10931292 and blood coagulation parameters.
| Characteristics | N or % |
|---|---|
| Total number, n | 2,313 |
| Male number, n (%) | 1,429 (61.78%) |
| Age, years (mean±SD) | 64.93±11.97 |
| CAD, n | 1871 (80.9%) |
| Fg/FIB (Fibrinogen level) (mean±SEM)* (g/L) | 4.07±0.08 |
| PT (Prothrombin time) (Mean±SEM) (second) | 12.83±0.13 |
| APTT (Activated partial thromboplastin time) (mean±SEM) (second) | 34.88±1.50 |
| TT (Thrombin time)(mean± SEM) (second) | 16.56±0.19 |
All 2,313 study subjects were genotyped for a functional SNP in the promoter/regulatory region of the TFPI gene, rs10931292. No deviation from the Hardy-Weinberg equilibrium was observed for SNP rs10931292 (P> 0.01). Linear regression analysis revealed significant association between rs10931292 and fibrinogen levels under an additive model (P = 0.026) and an autosomal recessive model (P = 0.017) (Table 2). Individuals with the GG genotype showed a significantly higher fibrinogen levels than those with other genotypes (Fig. 1A). The study population included 1,871 CAD patients and 442 non-CAD individuals (Table 1), however, the association between TFPI SNP rs10931292 and fibrinogen levels remain significant after adjusting for CAD (Table 2).
Table 2.
Analysis of association of TFPI SNP rs10931292 with coagulation parameters.
| Coagulation Indicator | Sample Size (AA/AG/GG) | Padj | Effect [B (95% CI)] | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Additive | Dominant | Recessive | Additive | Dominant | Recessive | ||
| Fibrinogen | 980/790/305 | a0.026 | 0.131 | 0.017 | 0.53 (0.06–1.00) | 0.51 (−0.15–1.17) | 1.14 (0.21–2.08) |
| b0.027 | 0.136 | 0.018 | 0.52 (0.06–0.98) | 0.51 (−0.16–1.17) | 1.13 (0.20–2.07) | ||
| Prothrombin time | 1,084/886/335 | 0.289 | 0.492 | 0.239 | −0.19 (−0.55–0.17) | −0.18 (−0.69–0.33) | −0.44 (−1.17–0.29) |
| 0.283 | 0.487 | 0.232 | −0.20 (−0.56–0.16) | −0.18 (−0.69–0.33) | −0.45 (−1.17–0.29) | ||
| Activated partial thromboplastin time | 1,079/866/330 | 0.308 | 0.274 | 0.606 | −2.16 (−6.32–2.00) | −3.32 (−9.27–2.63) | −2.23 (−10.68–6.23) |
| 0.353 | 0.300 | 0.679 | −1.97 (−6.13–2.19) | −3.15 (−9.10–2.80) | −1.78 (−10.24–6.68) | ||
| Thrombin time | 992/786/296 | 0.595 | 0.915 | 0.216 | 0.14 (−0.38–0.67) | −0.04 (−0.79–0.71) | 0.67 (−0.40–1.74) |
| 0.595 | 0.915 | 0.216 | 0.14 (−0.38–0.67) | −0.04 (−0.79–0.71) | 0.68 (−0.40–1.75) | ||
Padj, P value obtained from multiple linear modeling after adjustment for age and gendera or for age, gender and CADb.
Additive model = GG/AG/AA; Dominant model = GG+AG/AA; Recessive model = GG/AG+AA.
Figure 1. Analysis of association of TFPI SNP rs10931292 and coagulation indicators.
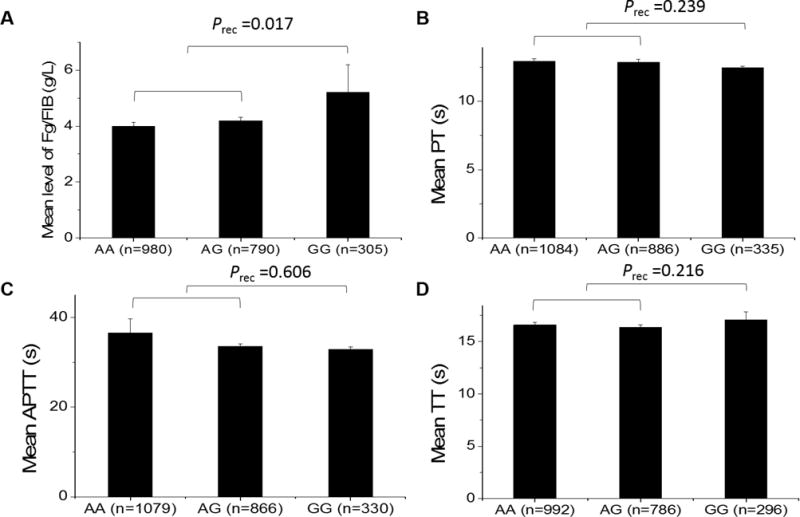
(A) Fibrinogen levels (Fg/FIB). Mean fibrinogen levels for different genotypes: NAA=980 subjects, 3.99 ± 0.15 g/L; NAG=790 subjects, 4.18 ± 0.14 g/L; NGG=305 subjects, 5.21 ± 0.97 g/L.(B) PPT. Mean PPT for different genotypes: NAA=1084 subjects, 12.92 ± 0.20 s; NAG=886 subjects, 12.86 ± 0.23 s; NGG=335 subjects, 12.46 ± 0.10 s.(C) APTT. Mean APTT for different genotypes: NAA=1079 subjects, 36.52 ± 3.14 s; NAG=866 subjects, 33.59 ± 0.46 s; NGG=330 subjects, 32.89 ± 0.49 s. (D) TT. Mean TT for different genotypes: NAA=992 subjects, 16.58 ± 0.24 s; NAG=786 subjects, 16.34 ± 0.28 s; NGG=296 subjects, 17.08 ± 0.76 s. Prec, P value after adjustment with age and gender under a recessive model.
No significant association was detected between TFPI SNP rs10931292 and PT, APTT or TT under any genetic model (Table 2 and Fig. 1A–D).
Significant association between TFPI SNP rs10931292 and CAD
Fibrinogen levels play an important role in coagulation and hemostasis as well as inflammation and are a well-established risk factor for cardiovascular disease (Danesh et al., 2005; Lowe and Rumley, 2014). Moreover, the fibrinogen level was shown to be increased in atherosclerosis and CAD (Koenig, 1999). As shown above, there is a significant association between TFPI SNP rs10931292 and fibrinogen levels (Fig. 1A and Table 2). Therefore, we hypothesized that TFPI SNP rs10931292 is associated with risk of CAD. To test the hypothesis, we performed a large-scale case control association study to analyze the association between TFPI SNP rs10931292 and the binary trait of CAD. Our study involved a case population with 4,479 CAD patients and a control population with 3,628 non-CAD controls (Table 3). The clinical and demographical features of the case group and the control group are shown in Table 3. Under the population parameter setting of OR of 1.2 for CAD, and the MAF of 0.31 for SNP rs10931292 (HapMap CHB Dataset), our samples (4,479 CAD cases and 3,628 controls) can provide a statistical power of 100% to detect an association between rs10931292 and CAD with a type I error of 0.05.
Table 3.
Clinical and demographical characteristics of the case-control study population for testing association between TFPI SNP rs10931292 and CAD.
| Characteristics | CAD Group | Control Group | P |
|---|---|---|---|
| Number (n) | 4,479 | 3,628 | – |
| MI, n (%) | 1,083 (24.18%) | 0 | <0.001 |
| Male, n (%) | 2707 (60.44%) | 2242 (61.80%) | 0.22 |
| Age, years (mean± SD)* | 65.27±12.55 | 58.79±11.28 | <0.001 |
| Hypertension, n (%) | 1982 (44.25%) | N/A | – |
| DM, n (%) | 631 (14.09%) | N/A | – |
| Smoker, n (%) | 1115 (24.89%) | N/A | – |
| Drinker, n (%) | 773 (17.26%) | N/A | – |
| Total cholesterol (mmol/L)* | 5.97 ± 1.33 | N/A | – |
| LDL cholesterol (mmol/L)* | 2.76 ±0.15 | N/A | – |
| HDL cholesterol (mmol/L)* | 1.55 ± 0.28 | N/A | – |
| Triglyceride (mmol/L)* | 1.74 ± 0.12 | N/A | – |
Data were presented as mean ± SEM; N/A. data not available or not complete.
SNP rs10931292 was genotyped in the case control population. Hardy-Weinberg disequilibrium tests showed that the genotyping data for SNP rs10931292 did not deviate from a randomly mated population (P> 0.01). The allelic association analysis did not identify significant association between rs10931292 and CAD (P = 0.08) (Table 4). However, significant association was identified between rs10931292 and CAD under a recessive genetic model (P = 0.001 before adjustment; P = 0.005 after adjustment for gender and age) (Table 5). This may be due to the possibility that the effect of two copies of the risk allele under a recessive model may be stronger than that of a single copy under a study of allelic association. When the study population was divided into different groups, the association between rs10931292 and CAD remained significant for early onset CAD and male CAD under a recessive model, with adjusted P values of 0.049 and 0.016, respectively (Table 5).
Table 4.
Analysis of allelic association between TFPI SNP rs10931292 and CAD.
| Population (n, case/control) | Phwe | Risk Allele | Frequency (Case/Control) |
Before adjustment | After adjustment | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Pobsa | OR(95%CI) | Padjb | OR(95%CI) | ||||
| Overall CAD (4,479/3,628) | 0.08 | G | 0.34/0.33 | 0.08 | 1.06 (0.99–1.14) | 0.08 | 1.07 (0.99–1.15) |
| Early-onset CAD (1,541/3,628) | 0.08 | G | 0.33/0.33 | 0.94 | 1.00 (0.89–1.14) | 0.64 | 1.04 (0.89–1.20) |
| Female CAD (1,698/1,329) | 0.33 | G | 0.35/0.33 | 0.51 | 1.04 (0.93–1.16) | 0.62 | 1.03 (0.91–1.16) |
| Male CAD (2,707/2,242) | 0.20 | G | 0.33/0.32 | 0.05 | 1.09 (1.00–1.19) | 0.07 | 1.09 (0.99–1.19) |
Phwe, P value from Hardy-Weinberg disequilibrium tests; Pobs, observed P value; Padj, P value after adjustment for covariates of age and gender; OR, odds ratio; 95% CI, 95% confidence interval; Early-onset CAD, males age 50 years or younger; females age 55 years or younger at the first diagnosis of the disease.
Table 5.
Analysis of genotypic association between TFPI SNP rs10931292 and CAD under three different genetic models.
| Population (n, case/control) | Genetic model | Before adjustment | After adjustment | ||
|---|---|---|---|---|---|
|
| |||||
| Pobsa | OR (95%CI) | Padjb | OR (95%CI) | ||
| Overall CAD (4479/3628) | Additive | 0.089 | 1.06 (0.99–1.13) | 0.088 | 1.06 (0.99–1.14) |
| Dominant | 0.826 | 1.01 (0.92–1.11) | 0.618 | 1.03 (0.93–1.13) | |
| Recessive | 0.001 | 1.25 (1.09–1.44) | 0.005 | 1.23 (1.06–1.42) | |
| Early-onset CAD (700/3628) | Additive | 0.944 | 1.00 (0.89–1.14) | 0.644 | 1.04 (0.90–1.20) |
| Dominant | 0.428 | 1.07 (0.91–1.26) | 0.061 | 1.21 (0.99–1.46) | |
| Recessive | 0.167 | 1.19 (0.93–1.53) | 0.049 | 1.35 (1.00–1.81) | |
| Female CAD (1698/1329) | Additive | 0.524 | 1.04 (0.93–1.15) | 0.634 | 1.03 (0.92–1.16) |
| Dominant | 0.625 | 1.04 (0.89–1.21) | 0.804 | 1.02 (0.87–1.20) | |
| Recessive | 0.038 | 1.27 (1.01–1.60) | 0.169 | 1.19 (0.93–1.52) | |
| Male CAD (2707/2242) | Additive | 0.058 | 1.09 (1.00–1.18) | 0.079 | 1.08 (0.99–1.18) |
| Dominant | 0.424 | 1.05 (0.93–1.18) | 0.409 | 1.05 (0.93–1.19) | |
| Recessive | 0.006 | 1.28 (1.07–1.53) | 0.016 | 1.25 (1.04–1.50) | |
Pobs, observed P value (X2 test); Padj, P value after adjustment for covariates (logistic regression analysis by adjusting for gender and age); OR, odds ratio; 95% CI, 95% confidence interval; Early-onset CAD, males age 50 years or younger; females age 55 years or younger at the first diagnosis of the disease; Additive model = GG/AG/AA; Dominant model = GG+AG/AA; Recessive model = GG/AG+AA.
Discussion
In this large-scale population study with 2,313 study subjects, we assessed potential genetic association between the TFPI gene and coagulation, specifically focusing on the data from a coagulation test panel (fibrinogen levels, PT, APTT and TT). We identified significant association between TFPI SNP rs10931292 and plasma fibrinogen levels (Table 2). The minor allele G/C of SNP rs10931292 was associated with a significant increase of fibrinogen levels (Fig. 1). The association of TFPI SNP rs10931292 with coagulation parameters was found to be specific to fibrinogen levels because no significant association was detected for PT, APTT or TT (Table 2). To the best of our knowledge, this is the first time that a TFPI variant is found to be significantly associated with fibrinogen levels. Because all genetic variants identified to date account for only 3% of heritability of fibrinogen levels (de Vries et al., 2016; Sabater-Lleal et al., 2013), our finding of a novel fibrinogen-associated variant in TFPI contributes to the elucidation of the genetic basis and biological pathways of fibrinogen levels.
An increased plasma fibrinogen level is a well-established risk factor for cardiovascular disease (Danesh et al., 2005; Lowe and Rumley, 2014). Therefore, we assessed whether TFPI SNP rs10931292 was associated with risk of CAD. Using a large case control association study population with 4,479 CAD patients and 3,628 controls, we identified significant association between TFPI SNP rs10931292 and CAD under a recessive genetic model (Table 5). Individuals with the GG/CC genotype had a significantly increased risk of CAD (OR = 1.23, P = 0.005) (Table 5). The minor allele G/C is the risk allele. To the best of our knowledge, this is the first report showing significant association between a TFPI variant and CAD. Together, our data suggest that TFPI SNP rs10931292 is associated with increased fibrinogen levels and increased risk of CAD (Tables 2). Specifically, the GG genotype was associated with an increased fibrinogen levels, thereby conferring a significant risk of CAD.
TFPI SNP rs10931292 is a T/A to C/G substitution located 287 bp upstream of the transcriptional start site (T-287C). Luciferase assays with a reporter with the TFPI promoter/regulatory region (−1999/+229) fused to the luciferase gene showed that the C allele had a much lower transactivation activity than the T allele (P = 0.009) in human microvascular endothelial cells (Amini and Iles, 2008). A reduced expression of TFPI may increase coagulation, resulting in increased risk of thrombosis, CAD and MI. We previously reported the first GWAS for CAD in the Chinese population and identified a SNP in the ADTRP gene (rs6903956) that reduces expression of ADTRP and increases risk of CAD and MI (Wang et al., 2011). The finding was replicated by multiple independent studies (Guo et al., 2012; Huang et al., 2015a; Tayebi et al., 2013). Lupu et al (2011) showed that ADTRP regulated the TFPI expression level in endothelial cells. SNP rs6903956 reduces the expression level of ADTRP, which reduces expression of TFPI, and increases risk of coagulation, thrombosis, CAD and MI (Luo et al., 2016). Therefore, TFPI SNP rs10931292 may use the similar mechanism as ADTRP SNP rs6903956 in increasing risk of CAD.
Genomic variants in TFPI were previously studied for their association with TFPI plasma levels, and other important factors important for coagulation. Dennis et al (Dennis et al., 2015) performed a review of genetic risk variants for plasma levels of TFPI and meta-analysis. They showed that SNPs rs5940 and rs7586970 in TFPI were associated with the plasma levels of TFPI, but no association was found for TFPI SNPs rs10153820 and rs10931292 (the variant analyzed in the present study) (Dennis et al., 2015). TFPI variants were also studied for their association with venous thrombosis, but both significant and negative associations were reported (Bezemer and Rosendaal, 2007; Hessner and Luhm, 2000; Kleesiek et al., 1999). No association was reported for TFPI SNP rs10931292 and venous thrombosis yet (Bezemer and Rosendaal, 2007). Opstad et al (2010) previously studied several SNPs in TFPI for their association using 1,001 CAD patients and 204 controls, but did not identify any significant association between TFPI SNPs and CAD. One reason for their failure to identify any significant association between TFPI SNPs and CAD may be due to the small sample size, in particular, the small group of controls. In this study, we performed a large-scale case control association study with 4,479 CAD patients and 3,628 non-CAD controls and identified significant association between TFPI SNP rs10931292 and CAD under a recessive model (Table 5). The same TFPI SNP rs10931292 was also significantly associated with plasma fibrinogen levels (Table 2). However, we acknowledge that these significant associations need to be further validated in other independent Chinese populations and populations from other ethnic background. Moreover, one other limitation of this study is that for logistic regression analysis for CAD, only age and sex were included as covariates because other data such as BMI, lipid levels, hypertension and diabetes were missing for most of the general population control subjects.
In summary, we have identified a significant association between TFPI SNP rs10931292 and increased fibrinogen levels and risk of CAD. Our study, for the first time, implicates the TFPI genetic variation in the regulation of plasma fibrinogen levels and developmental of CAD.
Acknowledgments
We thank the study subjects for their participation and support of this study and all members of the GeneID team for help and assistance.
Funding:
This study was supported by the China National Natural Science Foundation grants (31430047, 81630002 and 31671302), Chinese National Basic Research Programs (973 Programs 2013CB531101 and 2012CB517801), Hubei Province’s Outstanding Medical Academic Leader Program, Hubei Province Natural Science Program (2014CFA074 and 2016CFB224), the China National Natural Science Foundation grant (91439129, NSFC-J1103514), NIH/NHLBI grants R01 HL121358 and R01 HL126729, Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education, and the “Innovative Development of New Drugs” Key Scientific Project (2011ZX09307-001-09). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- TFPI
Tissue factor pathway inhibitor
- CAD
Coronary artery disease
- MI
Myocardial infarction
- FIG or Fg
Fibrinogen levels
- PT
Prothrombin time
- APTT
Activated partial thromboplastin time
- TT
Thrombin time
- IRF1
Interferon regulatory factor 1
- PCCB
Propionyl coenzyme A carboxylase
- NLRP3
NLR family pyrin domain containing 3 isoform
- OR
Odds ratio
- 95%CI
95% confidence interval
Footnotes
Author Contributions:
D. H. N., C.C.T., F.H. and Y.Z. contributed equally to this study. Study concept and design: Q. K. W., C. X. and D.H.N. Acquisition of data: D.H.N., C.C.T., F.H., Y.Z., D.W., J.F., S.L. and S.C. Analysis and interpretation of data: D.H.N., C.C.T., Q.C., C.X. and Q. K. W. Drafting of the manuscript: D.H.N., C.C.T. and Q. K. W. Critical revision of the manuscript for important intellectual content: C.C.T., C.X. and Q.C. Statistical analysis: D.H.N. and C.C.T. Obtained funding: Q. K. W and C.X. Study supervision: Q. K. W., Q.C. and C.X.
Compliance with Ethical Standards
Conflict of Interest
All authors have no conflict of interest.
Ethical approval:
All procedures performed in studies involving human participants were in accordance with the ethical standards of College of Life Science and Technology, Huazhong University of Science and Technology and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent:
Informed consent was obtained from all individual participants included in the study.
References
- Amini NA, Iles D. Analysis of a T-287C polymorphism in the tissue factor pathway inhibitor gene and identification of a repressor element in the promoter. Thromb Res. 2008;121:813–819. doi: 10.1016/j.thromres.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Bezemer ID, Rosendaal FR. Predictive genetic variants for venous thrombosis: what’s new? Semin Hematol. 2007;44:85–92. doi: 10.1053/j.seminhematol.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Chee YL. Coagulation. J R Coll Physicians Edinb. 2014;44:42–45. doi: 10.4997/JRCPE.2014.110. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang C, Wang X, Xu C, Wu M, Wang P, Tu X, Wang QK. Significant Association Between CAV1 Variant rs3807989 on 7p31 and Atrial Fibrillation in a Chinese Han Population. J Am Heart Assoc. 2015;4 doi: 10.1161/JAHA.115.001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang X, Wang J, Zhao Y, Wang D, Tan C, Fa J, Zhang R, Wang F, Xu C, Huang Y, Li S, Yin D, Xiong X, Li X, Chen Q, Tu X, Yang Y, Xia Y, Xu C, Wang QK. Genomic variant in CAV1 increases susceptibility to coronary artery disease and myocardial infarction. Atherosclerosis. 2016;246:148–156. doi: 10.1016/j.atherosclerosis.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh J, Lewington S, Thompson SG, Lowe GD, Collins R, Kostis JB, Wilson AC, Folsom AR, Wu K, Benderly M, Goldbourt U, Willeit J, Kiechl S, Yarnell JW, Sweetnam PM, Elwood PC, Cushman M, Psaty BM, Tracy RP, Tybjaerg-Hansen A, Haverkate F, de Maat MP, Fowkes FG, Lee AJ, Smith FB, Salomaa V, Harald K, Rasi R, Vahtera E, Jousilahti P, Pekkanen J, D’Agostino R, Kannel WB, Wilson PW, Tofler G, Arocha-Pinango CL, Rodriguez-Larralde A, Nagy E, Mijares M, Espinosa R, Rodriquez-Roa E, Ryder E, Diez-Ewald MP, Campos G, Fernandez V, Torres E, Marchioli R, Valagussa F, Rosengren A, Wilhelmsen L, Lappas G, Eriksson H, Cremer P, Nagel D, Curb JD, Rodriguez B, Yano K, Salonen JT, Nyyssonen K, Tuomainen TP, Hedblad B, Lind P, Loewel H, Koenig W, Meade TW, Cooper JA, De SB, Knottenbelt C, Miller GJ, Cooper JA, Bauer KA, Rosenberg RD, Sato S, Kitamura A, Naito Y, Palosuo T, Ducimetiere P, Amouyel P, Arveiler D, Evans AE, Ferrieres J, Juhan-Vague I, Bingham A, Schulte H, Assmann G, Cantin B, Lamarche B, Despres JP, Dagenais GR, Tunstall-Pedoe H, Woodward M, Ben-Shlomo Y, Davey SG, Palmieri V, Yeh JL, Rudnicka A, Ridker P, Rodeghiero F, Tosetto A, Shepherd J, Ford I, Robertson M, Brunner E, Shipley M, Feskens EJ, Kromhout D, Dickinson A, Ireland B, Juzwishin K, Kaptoge S, Lewington S, Memon A, Sarwar N, Walker M, Wheeler J, White I, Wood A. Plasma fibrinogen level and the risk of major cardiovascular diseases and nonvascular mortality: an individual participant meta-analysis. JAMA. 2005;294:1799–1809. doi: 10.1001/jama.294.14.1799. [DOI] [PubMed] [Google Scholar]
- de Vries PS, Chasman DI, Sabater-Lleal M, Chen MH, Huffman JE, Steri M, Tang W, Teumer A, Marioni RE, Grossmann V, Hottenga JJ, Trompet S, Muller-Nurasyid M, Zhao JH, Brody JA, Kleber ME, Guo X, Wang JJ, Auer PL, Attia JR, Yanek LR, Ahluwalia TS, Lahti J, Venturini C, Tanaka T, Bielak LF, Joshi PK, Rocanin-Arjo A, Kolcic I, Navarro P, Rose LM, Oldmeadow C, Riess H, Mazur J, Basu S, Goel A, Yang Q, Ghanbari M, Willemsen G, Rumley A, Fiorillo E, de Craen AJ, Grotevendt A, Scott R, Taylor KD, Delgado GE, Yao J, Kifley A, Kooperberg C, Qayyum R, Lopez LM, Berentzen TL, Raikkonen K, Mangino M, Bandinelli S, Peyser PA, Wild S, Tregouet DA, Wright AF, Marten J, Zemunik T, Morrison AC, Sennblad B, Tofler G, de Maat MP, de Geus EJ, Lowe GD, Zoledziewska M, Sattar N, Binder H, Volker U, Waldenberger M, Khaw KT, Mcknight B, Huang J, Jenny NS, Holliday EG, Qi L, Mcevoy MG, Becker DM, Starr JM, Sarin AP, Hysi PG, Hernandez DG, Jhun MA, Campbell H, Hamsten A, Rivadeneira F, Mcardle WL, Slagboom PE, Zeller T, Koenig W, Psaty BM, Haritunians T, Liu J, Palotie A, Uitterlinden AG, Stott DJ, Hofman A, Franco OH, Polasek O, Rudan I, Morange PE, Wilson JF, Kardia SL, Ferrucci L, Spector TD, Eriksson JG, Hansen T, Deary IJ, Becker LC, Scott RJ, Mitchell P, Marz W, Wareham NJ, Peters A, Greinacher A, Wild PS, Jukema JW, Boomsma DI, Hayward C, Cucca F, Tracy R, Watkins H, Reiner AP, Folsom AR, Ridker PM, O’Donnell CJ, Smith NL, Strachan DP, Dehghan A. A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Hum Mol Genet. 2016;25:358–370. doi: 10.1093/hmg/ddv454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange M, de Geus EJ, Kluft C, Meijer P, van Doornen LJ, Boomsma DI, Snieder H. Genetic influences on fibrinogen, tissue plasminogen activator-antigen and von Willebrand factor in males and females. Thromb Haemost. 2006;95:414–419. doi: 10.1160/TH05-09-0596. [DOI] [PubMed] [Google Scholar]
- de Lange M, Snieder H, Ariens RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001;357:101–105. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- Dehghan A, Yang Q, Peters A, Basu S, Bis JC, Rudnicka AR, Kavousi M, Chen MH, Baumert J, Lowe GD, Mcknight B, Tang W, de MM, Larson MG, Eyhermendy S, Mcardle WL, Lumley T, Pankow JS, Hofman A, Massaro JM, Rivadeneira F, Kolz M, Taylor KD, van Duijn CM, Kathiresan S, Illig T, Aulchenko YS, Volcik KA, Johnson AD, Uitterlinden AG, Tofler GH, Gieger C, Psaty BM, Couper DJ, Boerwinkle E, Koenig W, O’Donnell CJ, Witteman JC, Strachan DP, Smith NL, Folsom AR. Association of novel genetic Loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ Cardiovasc Genet. 2009;2:125–133. doi: 10.1161/CIRCGENETICS.108.825224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis J, Kassam I, Morange PE, Tregouet DA, Gagnon F. Genetic determinants of tissue factor pathway inhibitor plasma levels. Thromb Haemost. 2015;114:245–257. doi: 10.1160/TH14-12-1043. [DOI] [PubMed] [Google Scholar]
- Guo CY, Gu Y, Li L, Jia EZ, Li CJ, Wang LS, Yang ZJ, Cao KJ, Ma WZ. Association of SNP rs6903956 on chromosome 6p24.1 with angiographical characteristics of coronary atherosclerosis in a Chinese population. PLoS One. 2012;7:e43732. doi: 10.1371/journal.pone.0043732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward CP, Moffat KA, Liu Y. Laboratory investigations for bleeding disorders. Semin Thromb Hemost. 2012;38:742–752. doi: 10.1055/s-0032-1326780. [DOI] [PubMed] [Google Scholar]
- Hessner MJ, Luhm RA. The C536T transition in the tissue factor pathway inhibitor (TFPI) gene does not contribute to risk of venous thrombosis among carriers of factor V Leiden. Thromb Haemost. 2000;84:724–725. [PubMed] [Google Scholar]
- Hoffman M, Monroe DM. Coagulation 2006: a modern view of hemostasis. Hematol Oncol Clin North Am. 2007;21:1–11. doi: 10.1016/j.hoc.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Huang EW, Peng LY, Zheng JX, Wang D, Xu QY, Huang L, Wu QP, Tang SB, Luo B, Liu SP, Liu XS, Li ZH, Quan L, Li Y, Shi H, Lv GL, Zhao J, Cheng JD, Liu C. Common Variants in Promoter of ADTRP Associate with Early-Onset Coronary Artery Disease in a Southern Han Chinese Population. PLoS One. 2015a;10:e0137547. doi: 10.1371/journal.pone.0137547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang C, Yao Y, Zuo X, Chen S, Xu C, Zhang H, Lu Q, Chang L, Wang F, Wang P, Zhang R, Hu Z, Song Q, Yang X, Li C, Li S, Zhao Y, Yang Q, Yin D, Wang X, Si W, Li X, Xiong X, Wang D, Huang Y, Luo C, Li J, Wang J, Chen J, Wang L, Wang L, Han M, Ye J, Chen F, Liu J, Liu Y, Wu G, Yang B, Cheng X, Liao Y, Wu Y, Ke T, Chen Q, Tu X, Elston R, Rao S, Yang Y, Xia Y, Wang QK. Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation. PLoS Genet. 2015b;11:e1005393. doi: 10.1371/journal.pgen.1005393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleesiek K, Schmidt M, Gotting C, Schwenz B, Lange S, Muller-Berghaus G, Brinkmann T, Prohaska W. The 536C—>T transition in the human tissue factor pathway inhibitor (TFPI) gene is statistically associated with a higher risk for venous thrombosis. Thromb Haemost. 1999;82:1–5. [PubMed] [Google Scholar]
- Koenig W. Fibrinogen and coronary risk. Curr Cardiol Rep. 1999;1:112–118. doi: 10.1007/s11886-999-0068-y. [DOI] [PubMed] [Google Scholar]
- Li X, Huang Y, Yin D, Wang D, Xu C, Wang F, Yang Q, Wang X, Li S, Chen S, Xiong X, Huang Y, Zhao Y, Wang L, Zhu X, Su Z, Zhou B, Zhang Y, Wang L, Chang L, Xu C, Li H, Ke T, Ren X, Cheng X, Yang Y, Liao Y, Tu X, Wang QK. Meta-analysis identifies robust association between SNP rs17465637 in MIA3 on chromosome 1q41 and coronary artery disease. Atherosclerosis. 2013;231:136–140. doi: 10.1016/j.atherosclerosis.2013.08.031. [DOI] [PubMed] [Google Scholar]
- Lowe G, Rumley A. The relevance of coagulation in cardiovascular disease: what do the biomarkers tell us? Thromb Haemost. 2014;112:860–867. doi: 10.1160/TH14-03-0199. [DOI] [PubMed] [Google Scholar]
- Luo C, Wang F, Qin S, Chen Q, Wang Q. Coronary artery disease susceptibility gene ADTRP regulates cell cycle progression, proliferation and apoptosis by global gene expression regulation. Physiol Genomics. 2016;48:554–564. doi: 10.1152/physiolgenomics.00028.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu C, Zhu H, Popescu NI, Wren JD, Lupu F. Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood. 2011;118:4463–4471. doi: 10.1182/blood-2011-05-355370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney SA, Ellery PE, Wood JP, Ferrel JP, Martinez ND, Mast AE. Comparison of the inhibitory activities of human tissue factor pathway inhibitor (TFPI)alpha and TFPIbeta. J Thromb Haemost. 2013;11:911–918. doi: 10.1111/jth.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney SA, Mast AE. New insights into the biology of tissue factor pathway inhibitor. J Thromb Haemost. 2015;13(Suppl 1):S200–S207. doi: 10.1111/jth.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast AE. Tissue Factor Pathway Inhibitor: Multiple Anticoagulant Activities for a Single Protein. Arterioscler Thromb Vasc Biol. 2016;36:9–14. doi: 10.1161/ATVBAHA.115.305996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opstad TB, Pettersen AA, Weiss T, Arnesen H, Seljeflot I. Gender differences of polymorphisms in the TF and TFPI genes, as related to phenotypes in patients with coronary heart disease and type-2 diabetes. Thromb J. 2010;8:7. doi: 10.1186/1477-9560-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabater-Lleal M, Huang J, Chasman D, Naitza S, Dehghan A, Johnson AD, Teumer A, Reiner AP, Folkersen L, Basu S, Rudnicka AR, Trompet S, Malarstig A, Baumert J, Bis JC, Guo X, Hottenga JJ, Shin SY, Lopez LM, Lahti J, Tanaka T, Yanek LR, Oudot-Mellakh T, Wilson JF, Navarro P, Huffman JE, Zemunik T, Redline S, Mehra R, Pulanic D, Rudan I, Wright AF, Kolcic I, Polasek O, Wild SH, Campbell H, Curb JD, Wallace R, Liu S, Eaton CB, Becker DM, Becker LC, Bandinelli S, Raikkonen K, Widen E, Palotie A, Fornage M, Green D, Gross M, Davies G, Harris SE, Liewald DC, Starr JM, Williams FM, Grant PJ, Spector TD, Strawbridge RJ, Silveira A, Sennblad B, Rivadeneira F, Uitterlinden AG, Franco OH, Hofman A, van DJ, Willemsen G, Boomsma DI, Yao J, Swords JN, Haritunians T, Mcknight B, Lumley T, Taylor KD, Rotter JI, Psaty BM, Peters A, Gieger C, Illig T, Grotevendt A, Homuth G, Volzke H, Kocher T, Goel A, Franzosi MG, Seedorf U, Clarke R, Steri M, Tarasov KV, Sanna S, Schlessinger D, Stott DJ, Sattar N, Buckley BM, Rumley A, Lowe GD, Mcardle WL, Chen MH, Tofler GH, Song J, Boerwinkle E, Folsom AR, Rose LM, Franco-Cereceda A, Teichert M, Ikram MA, Mosley TH, Bevan S, Dichgans M, Rothwell PM, Sudlow CL, Hopewell JC, Chambers JC, Saleheen D, Kooner JS, Danesh J, Nelson CP, Erdmann J, Reilly MP, Kathiresan S, Schunkert H, Morange PE, Ferrucci L, Eriksson JG, Jacobs D, Deary IJ, Soranzo N, Witteman JC, de Geus EJ, Tracy RP, Hayward C, Koenig W, Cucca F, Jukema JW, Eriksson P, Seshadri S, Markus HS, Watkins H, Samani NJ, Wallaschofski H, Smith NL, Tregouet D, Ridker PM, Tang W, Strachan DP, Hamsten A, O’Donnell CJ. Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. 2013;128:1310–1324. doi: 10.1161/CIRCULATIONAHA.113.002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen GQ, Abdullah KG, Wang QK. The TaqMan method for SNP genotyping. Methods Mol Biol. 2009;578:293–306. doi: 10.1007/978-1-60327-411-1_19. [DOI] [PubMed] [Google Scholar]
- Shen GQ, Girelli D, Li L, Olivieri O, Martinelli N, Chen Q, Topol EJ, Wang QK. Multi-allelic haplotype association identifies novel information different from single-SNP analysis: a new protective haplotype in the LRP8 gene is against familial and early-onset CAD and MI. Gene. 2013;521:78–81. doi: 10.1016/j.gene.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen GQ, Girelli D, Li L, Rao S, Archacki S, Olivieri O, Martinelli N, Park JE, Chen Q, Topol EJ, Wang QK. A Novel Molecular Diagnostic Marker for Familial and Early-Onset Coronary Artery Disease and Myocardial Infarction in the LRP8 Gene. Circ Cardiovasc Genet. 2014;7:514–520. doi: 10.1161/CIRCGENETICS.113.000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen GQ, Li L, Girelli D, Seidelmann SB, Rao S, Fan C, Park JE, Xi Q, Li J, Hu Y, Olivieri O, Marchant K, Barnard J, Corrocher R, Elston R, Cassano J, Henderson S, Hazen SL, Plow EF, Topol EJ, Wang QK. An LRP8 variant is associated with familial and premature coronary artery disease and myocardial infarction. Am J Hum Genet. 2007;81:780–791. doi: 10.1086/521581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen GQ, Li L, Rao S, Abdullah KG, Ban JM, Lee BS, Park JE, Wang QK. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol. 2008a;28:360–365. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- Shen GQ, Li L, Wang QK. Genetic variant R952Q in LRP8 is associated with increased plasma triglyceride levels in patients with early-onset CAD and MI. Ann Hum Genet. 2012;76:193–199. doi: 10.1111/j.1469-1809.2012.00705.x. [DOI] [PubMed] [Google Scholar]
- Shen GQ, Rao S, Martinelli N, Li L, Olivieri O, Corrocher R, Abdullah KG, Hazen SL, Smith J, Barnard J, Plow EF, Topol EJ, Wang QK. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J Hum Genet. 2008b;53:144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- Souto JC, Almasy L, Borrell M, Gari M, Martinez E, Mateo J, Stone WH, Blangero J, Fontcuberta J. Genetic determinants of hemostasis phenotypes in Spanish families. Circulation. 2000;101:1546–1551. doi: 10.1161/01.cir.101.13.1546. [DOI] [PubMed] [Google Scholar]
- Tayebi N, Ke T, Foo JN, Friedlander Y, Liu J, Heng CK. Association of single nucleotide polymorphism rs6903956 on chromosome 6p24.1 with coronary artery disease and lipid levels in different ethnic groups of the Singaporean population. Clin Biochem. 2013;46:755–759. doi: 10.1016/j.clinbiochem.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Tu X, Nie S, Liao Y, Zhang H, Fan Q, Xu C, Bai Y, Wang F, Ren X, Tang T, Xia N, Li S, Huang Y, Liu J, Yang Q, Zhao Y, Lv Q, Li Q, Li Y, Xia Y, Qian J, Li B, Wu G, Wu Y, Yang Y, Wang QK, Cheng X. The IL-33-ST2L Pathway Is Associated with Coronary Artery Disease in a Chinese Han Population. Am J Hum Genet. 2013;93:652–660. doi: 10.1016/j.ajhg.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, Cheng X, Li C, Yang R, Wang CC, Wu G, Lu QL, Bai Y, Huang YF, Yin D, Yang Q, Wang XJ, Dai DP, Zhang RF, Wan J, Ren JH, Li SS, Zhao YY, Fu FF, Huang Y, Li QX, Shi SW, Lin N, Pan ZW, Li Y, Yu B, Wu YX, Ke YH, Lei J, Wang N, Luo CY, Ji LY, Gao LJ, Li L, Liu H, Huang EW, Cui J, Jia N, Ren X, Li H, Ke T, Zhang XQ, Liu JY, Liu MG, Xia H, Yang B, Shi LS, Xia YL, Tu X, Wang QK. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- Wang Q, Rao S, Shen GQ, Li L, Moliterno DJ, Newby LK, Rogers WJ, Cannata R, Zirzow E, Elston RC, Topol EJ. Premature Myocardial Infarction Novel Susceptibility Locus on Chromosome 1P34-36 Identified by Genomewide Linkage Analysis. Am J Hum Genet. 2004;74:262–271. doi: 10.1086/381560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassel CL, Lange LA, Keating BJ, Taylor KC, Johnson AD, Palmer C, Ho LA, Smith NL, Lange EM, Li Y, Yang Q, Delaney JA, Tang W, Tofler G, Redline S, Taylor HA, Jr, Wilson JG, Tracy RP, Jacobs DR, Jr, Folsom AR, Green D, O’Donnell CJ, Reiner AP. Association of genomic loci from a cardiovascular gene SNP array with fibrinogen levels in European Americans and African-Americans from six cohort studies: the Candidate Gene Association Resource (CARe) Blood. 2011;117:268–275. doi: 10.1182/blood-2010-06-289546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JP, Bunce MW, Maroney SA, Tracy PB, Camire RM, Mast AE. Tissue factor pathway inhibitor-alpha inhibits prothrombinase during the initiation of blood coagulation. Proc Natl Acad Sci U S A. 2013;110:17838–17843. doi: 10.1073/pnas.1310444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Xu C, Li X, Wang B, Wang F, Yang Q, Wang D, Wang X, Li S, Chen S, Zhao Y, Yin D, Huang Y, Zhu X, Wang L, Wang L, Chang L, Xu C, Li H, Ke T, Ren X, Wu Y, Zhang R, Wu T, Xia Y, Yang Y, Ma X, Tu X, Wang QK. BRG1 variant rs1122608 on chromosome 19p13.2 confers protection against stroke and regulates expression of pre-mRNA-splicing factor SFRS3. Hum Genet. 2013;133:499–508. doi: 10.1007/s00439-013-1389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yang Q, Xiong H, Wang L, Cai J, Wang F, Li S, Chen J, Wang C, Wang D, Xiong X, Wang P, Zhao Y, Wang X, Huang Y, Chen S, Yin D, Li X, Liu Y, Liu J, Wang J, Li H, Ke T, Ren X, Wu Y, Wu G, Wan J, Zhang R, Wu T, Wang J, Xia Y, Yang Y, Cheng X, Liao Y, Chen Q, Zhou Y, He Q, Tu X, Wang QK. Candidate pathway-based genome-wide association studies identify novel associations of genomic variants in the complement system associated with coronary artery disease. Circ Cardiovasc Genet. 2014;7:887–894. doi: 10.1161/CIRCGENETICS.114.000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Tofler GH, Cupples LA, Larson MG, Feng D, Lindpaintner K, Levy D, D’Agostino RB, O’Donnell CJ. A genome-wide search for genes affecting circulating fibrinogen levels in the Framingham Heart Study. Thromb Res. 2003;110:57–64. doi: 10.1016/s0049-3848(03)00288-3. [DOI] [PubMed] [Google Scholar]
- Yin D, Naji DH, Xia Y, Li S, Bai Y, Jiang G, Zhao Y, Wang X, Huang Y, Chen S, Fa J, Tan C, Zhou M, Zhou Y, Wang L, Liu Y, Chen F, Liu J, Chen Q, Tu X, Xu C, Wang QK. Genomic Variant in IL-37 Confers A Significant Risk of Coronary Artery Disease. Sci Rep. 2017;7:42175. doi: 10.1038/srep42175. [DOI] [PMC free article] [PubMed] [Google Scholar]


