Abstract
Studies of human skin microbiome suggest that Propionibacterium acnes strains may contribute differently to skin health and disease. However, the immune phenotype and functions of Th17 cells induced by healthy (PH) vs. acne (PA) skin-associated P. acnes strains are currently unknown. We stimulated PBMCs from healthy donors and observed that PA strains induce higher IL-17 levels than PH strains. We next generated PH and PA strain-specific Th17 clones and show that P. acnes strains induce Th17 cells of varied phenotype and function that are stable in the presence of IL-2 and IL-23. Although PH and PA-specific clones expressed similar levels of LL-37 and DEFB4, only PH-specific clones secreted molecules sufficient to kill P. acnes. Furthermore, electron microscopic studies revealed that supernatants derived from activated PH and not PA-specific clones exhibited robust bactericidal activity against P. acnes, and complete breaches in the bacterial cell envelope were observed. This antimicrobial activity was independent of IL-26, as both natural IL-26 released by Th17 clones and rhIL-26 lacked antimicrobial potency against P. acnes. Overall, our data suggest that P. acnes strains may differentially modulate the CD4+ T cell responses, leading to the generation of Th17 cells that may contribute to either homeostasis or acne pathogenesis.
Keywords: P. acnes, Protective Th17 cells, Ribotypes, Pathogenic Th17 cells, Interleukin 17
Introduction
Acne vulgaris, is the most common inflammatory skin disease of the pilosebaceous unit (PSU) in humans. This disease affects millions of people worldwide, predominantly the pre-, adolescent and post-adolescent populations. (James, 2005; Williams et al., 2012). Propionibacterium acnes is generally believed to play a major role in the pathogenesis of acne, in part by stimulating an inflammatory response. Other factors involved include, increased production of sebum, keratinocyte hyperproliferation, and altered P. acnes bacterial colonization (Leyden et al., 1998; Zouboulis et al., 2005). While most humans harbor P. acnes on their skin, not everyone suffers from acne. P. acnes is the dominant species within the microfollicles and the PSU in both healthy individuals and acne patients (Degitz et al., 2007; Fitz-Gibbon et al., 2013). However, at the strain level, P. acnes distribution has been shown to be significantly different in healthy and acne patients, suggesting that different P. acnes strains may play different roles in acne vulgaris (Fitz-Gibbon et al., 2013).
Like other bacterial species, P. acnes shows phenotypic and genotypic diversity. This diversity at the strain level and its association with both human health and disease is poorly understood (Fitz-Gibbon et al., 2013). Recently, typing of P. acnes has revealed associations of particular strains with different diseases (McDowell et al., 2013a; Tomida et al., 2013), and that various P. acnes strains may be associated with acne (referred to as PA), while others may be associated with healthy skin (referred to as PH) (Fitz-Gibbon et al., 2013; Lomholt and Kilian, 2010; McDowell et al., 2011). Strains of P. acnes have also been shown to have differences in pathogenic potential and secretome profiles (Holland et al., 2010; Nakatsuji et al., 2008; Suh and Kwon, 2015), as they differ in their ability to induce human β-defensin 2, influence cell growth, differentiation, and viability of skin resident cells and activation of both innate and adaptive arms of the immune response (Akaza et al., 2009; Beylot et al., 2014; Bojar and Holland, 2004; Nagy et al., 2006; Nagy et al., 2005).
A role for the adaptive immune response has also been suggested based on the detection of CD4+ T cells in the inflammatory infiltrate from early acne lesions (Norris and Cunliffe, 1988). We and others recently demonstrated that P. acnes is a potent inducer of IL-17 and IFN-γ from CD4+ T cells, and that IL-17+ cells were present in perifollicular infiltrates in biopsies of inflammatory acne lesions, suggesting that acne may be a Th17-mediated disease (Agak et al., 2014; Kistowska et al., 2015; Thiboutot et al., 2014). Moreover, Th17 cells not only characteristically induce the recruitment of neutrophils, which contribute to antibacterial activity, but also cause tissue injury. Reactive oxygen species and lysosomal enzymes are also released by neutrophils, and levels may correlate with severity (Abdel Fattah et al., 2008; Chiu et al., 2003; Levell et al., 1989). Therefore, understanding the mechanisms by which PA and PH associated strains modulate the adaptive responses is essential to understanding the phenotypic differences of the strains and their roles in acne vulgaris.
In this study, we tested the hypothesis that P. acnes strains PA and PH induce Th17 cells with varied phenotypes and functions. The P. acnes strains PA and PH used in this study are representative of the ribotypes found to be strongly associated with healthy and acne-associated skin and are shown in supplementary table 1 (Fitz-Gibbon et al., 2013). We generated PA and PH -specific Th17 clones and evaluated the cytokine profiles and functional activity of Th17-derived molecules against P. acnes in vitro.
Results
P. acnes strains associated with acne disease (PA) induce higher IL-17 levels than strains associated with healthy skin (PH)
Microcomedones from healthy vs. diseased skin have been shown to harbor P. acnes strains from distinct lineages and possess distinct nucleopeptide signatures of 16S ribosomal DNA (rDNA) sequences (Fitz-Gibbon et al., 2013). Although some P. acnes strains are found on healthy skin ribotype 6 (RT6), others have been associated with acne disease (RT4, RT5 and RT8) (McDowell et al., 2013b). We sought to determine the cytokine secretion levels when PBMCs from healthy human donors were stimulated with PH vs. PA associated strains. We observed that PA strains (HL110PA1, HL043PA1 and, HL096 PA1) induced significantly higher levels of IL-17 ranging from 1500-1700pg/ml in comparison to PH associated strains (HLA042PA3, HL110PA3, HL110PA4), which induced IL-17 at a range of 750-1000pg/ml (Fig. 1a) (P<0.001). This PA vs. PH pattern of IL-17 induction was similar in all the donors that we tested.
Fig. 1. P. acnes strains associated with acne disease induce significant IL-17 responses.
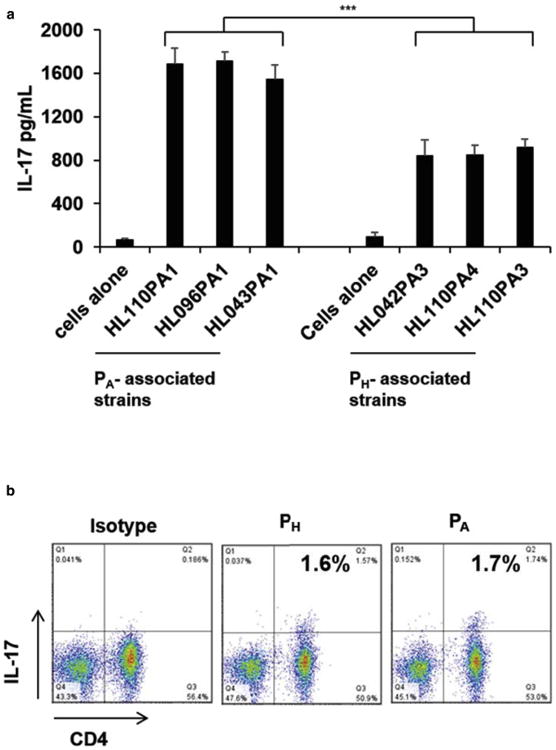
(a) PBMCs were cultured (2-5 × 106/well) in the presence of live PA and PH strains (1MOI). Levels of IL-17 accumulated in the culture supernatants were measured using ELISA. Results from three donors were combined and the variation demonstrated as mean ± SD. (***p ≤ 0.001 compared to PH associated strains). (b) For Sterile cell sorting, PBMCs were stimulated with PH and PA strains (1MOI) for 16 h and IL-17 secretion determined using a cytokine secretion capture assay. The cells were further stained with α-CD4 antibodies, and the CD4+IL-17+ cells were sorted under sterile conditions and cloned at 0.3cells/well. Each panel is representative of four independent experiments.
Cloning efficiency and specificity of P. acnes-responsive CD4+ IL-17+ T cell clones
The observation that PA strains induced significantly higher levels of IL-17 in comparison to PH associated strains suggested that P. acnes strains might have the ability to modulate the immune response at the T cell level. In order to explore this possibility, we generated PA and PH-specific clones by stimulating PBMCs of four healthy donors with PA and PH associated strains as previously described (Agak et al., 2014). Both PA and PH strains induced IL-17 secretion from PBMCs ranging from 1.5% -2% (Fig. 1b). We next sterile sorted and cloned the CD4+ IL-17+ population. We achieved a cloning efficiency ranging from 26 to 34% (Supplementary Table 2). We next selected antigen-specific T cell clones using T cell proliferation assays (Fig. 2a-b). Upon stimulation with α-CD3/α-CD28, both PA and PH-specific clones produced IL-17 ranging from 29-42%, and expressed Th17 associated genes (Fig. 2c-e). PA and PH-specific clones expressed functional markers of Th17 cells. In addition, treatment with α-HLA-DR antibody, but not with α-MHC I antibody, led to a strong reduction of T cell proliferation (Fig. 2f-g), indicating that proliferation and cytokine secretion by PA and PH-specific clones is dependent on MHC II. Thus, our T-cell cloning strategy can successfully generate CD4+IL-17+T-cells with desired specificities.
Fig. 2. PH and PA Th17 clones are specific and express Th17 associated genes.
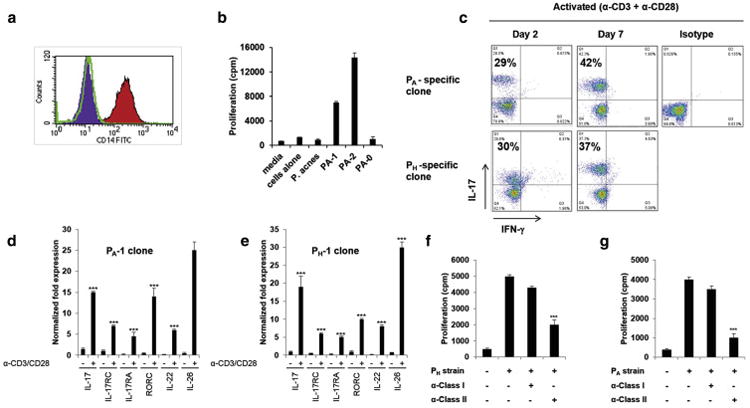
(a) Autologous CD14+ monocytes were isolated by positive selection and used as APCs. (b) Specificity of PA and PH specific clones as determined by 3[H] thymidine incorporation in T cell proliferation assays. (c) FACS on clones stimulated with a-CD3/a-CD28 (0.75μg/ml) and stained with antibodies to IL-17 and IFN-γ. (d-e) Real-time PCR of Th17 associated genes analyzed 6 h following (-) no or α-CD3/α-CD28 stimulation. Gene expression was normalized to the housekeeping gene GAPDH and quantified by the comparative method 2 -ΔΔCT, (n=4). (f-g) Effects of blocking MHC class I and II (10μg/ml) in autologous monocytes as measured in T cell proliferation assays. Data represent mean ± SD (***p ≤ 0.001).
IL-23 is a stabilizing factor for in-vitro developed Th17 lineages
IL-23 has been shown to be important in the maintenance of Th17 cells (Veldhoen et al., 2006). To specifically address the role of IL-23 as a stabilizing factor for the in vitro developed PH and PA specific clones, we tested the ability of the clones to survive in vitro both in the presence and absence of IL-23. In the absence of IL-23, the proportion of cells secreting IL-17 was sharply reduced on day 12 from 84% to 72%, and by day 40 all the clones lost the ability to secrete IL-17 (Fig. 3). Furthermore, upon stimulation with α-CD3/α-CD28 or APCs incubated/pulsed with microbe, these “exTh17” cells mainly secreted IL-10 and IL-26 (Supplementary Fig. 1a). We next tried to rescue the IL-17 phenotype in the day 28 and 40 “exTh17” clones expressing 19% and 0% IL-17, respectively, which had been cultured without IL-23. Surprisingly, the loss of the Th17 phenotype was irreversible, as “exTh17” clones could not regain IL-17 protein secretion, even though both IL-23 treated and untreated clones demonstrated IL-23R expression as measured by qPCR (Supplementary Fig 1b-c). However, we noted no loss of IL-17 secretion in PH and PA - specific clones that were maintained in the presence of IL-23 (Supplementary Fig. 2).
Fig. 3. IL-23 is a stabilizing factor for in-vitro developed PA and PH- specific Th17 clones.
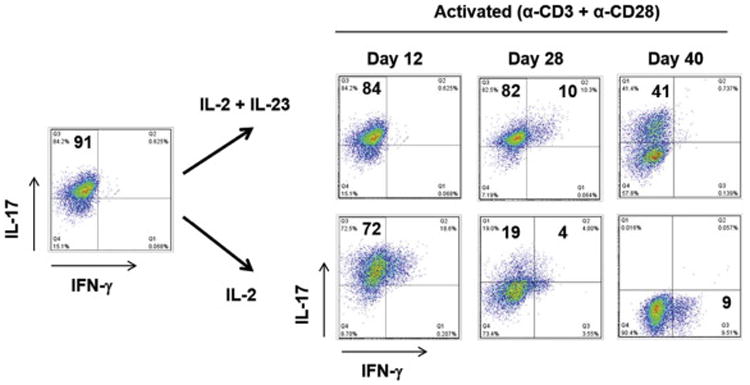
PA and PH- specific clones cultured with IL-2 and with/without IL-23 were stimulated with α-CD3/α-CD28 (0.75μg/ml) on days 12, 28, and 40 and stained with antibodies to IL-17 and IFN-γ. Data is representative of more than five independent experiments using clones derived from four different donors.
PH and PA strains induce Th17 clones of varied phenotype
We developed a cloning system that uses the whole PH and PA microbes and autologous monocytes as APCs. This approach takes advantage of the complexity of the P. acnes microbes, which provides, at the same time, a large number of antigens and a variety of stimuli to innate receptors to elicit polarizing cytokines. Therefore, to investigate whether PA and PH -specific clones express and secrete functional molecules associated with Th17 cells, clones underwent costimulation by α-CD3/α-CD28 antibodies. After 24-hour stimulation, we observed that PA and PH strains induced similar levels of antimicrobial encoding genes, cathelicidin and DEFB4 expression (Fig. 4a and supplementary Fig. 2a). Interestingly, PA- specific clones expressed both Th17 and Th1 associated transcriptional factors RORC and tbet, in contrast to PH- specific clones that only expressed IL-17 (Fig. 4b). In addition, ELISA cytokine profiles indicate that PA -specific clones secreted IFN-γ, IL-17, IL-22, and IL-26 whereas the PH- specific clones secreted IL-10, IL-17, IL-22, and IL-26, and maintained this capacity at later time points (Fig. 4c-f and supplementary Fig. 2b-e). Non-stimulated clones did not secrete IFN-γ, IL-10, or any Th17 associated cytokine (data not shown). These findings suggest that PH and PA strains can modulate the T cell responses in vitro leading to induction of an IL-17/IFN-γ (IFN-γ+IL-17+IL-22+IL-26+) and/or the IL-17/IL-10 (IL-10+IL-17+IL-22+IL-26+) producing phenotypes.
Fig. 4. Gene expression and cytokine profiles of PA and PH-strain specific clones.
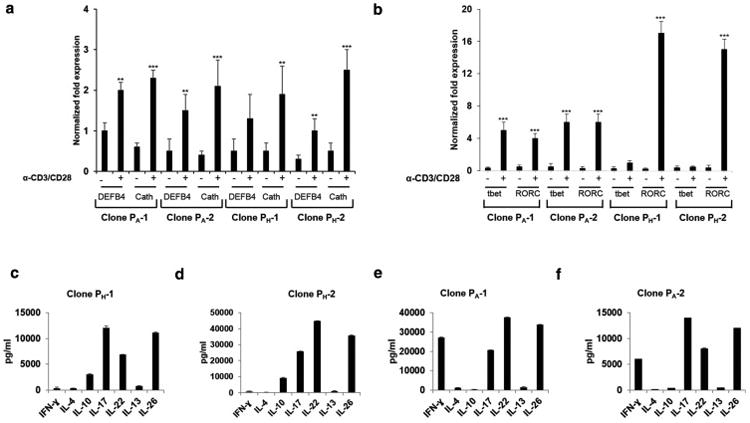
(a-b) DEFB4, Cathelicidin (Cath), tbet, and RORC analyzed 24 h following (-) no or α-CD3/α-CD28 (0.75μg/ml) stimulation representative prTh17 clones (PH-1 and PH-2) and paTh17 (PA-1 and PA-2) generated from two different donors (D1 and D2) are shown. The results were also consistent for prTh17 and paTh17 clones generated from donors D3 and D4 (Supp. Fig 2). Gene expression was normalized to GAPDH. Data are representative of three independent experiments (n=3). Data represent mean ± SD (**p ≤ 0.05; ***p ≤ 0.001). (c-f) PA and PH-strain specific clones were stimulated as above, and cytokine levels accumulated in the culture supernatants were measured using ELISA.
Not all Th17 clones secrete molecule(s) that are sufficient to kill P. acnes
Given the importance of IL-26 as a cationic amphipathic protein that is able to kill extracellular bacteria via membrane pore formation (Meller et al., 2015), we next sought to determine whether Th17-derived molecules had microbicidal activity against P. acnes. We conducted CFU assays as previously described (McInturff et al., 2005; Schmidt et al., 2015) using supernatants derived from activated PH and PA-specific clones. All Th17 clones derived from PH-associated strains exhibited robust bactericidal activity against P. acnes strains that we tested (Fig. 5a and supplementary Fig. 3a-b). Against P. acnes strains HL096PA1 (Fig. 5a), greater than 5-log reductions in CFU were observed at a dilution of 1:4. We used the antimicrobial peptide LL-37 as a positive control. LL-37 (0.1-100 μM) was also effective against all strains of P. acnes. (Fig. 5b). In contrast, no single clone derived from PA-associated strains exhibited microbicidal activity against P. acnes. Even though we observed a 1-log reduction in CFU using neat supernatants for some PA-specific clones, including PA-1, this was not significant (Fig. 5c). We therefore classified the IL-10+ IL-17+ IL-22+ IL-26+-secreting clones as protective Th17 (prTh17) and the IFN-γ+ IL-17+ IL-22+ IL-26+-secreting clones as pathogenic Th17 (paTh17) as our data corroborate recent evidence that broadly categorize ROR+Th17 cells into two groups: first, host protective cells Th17 that express both IL-17 and IL-10 (Esplugues et al., 2011; Hirota et al., 2013; McGeachy et al., 2007) and, second, a highly inflammatory population that express IL-17, IL-22 and IFN-γ (Langrish et al., 2005; Zheng et al., 2007).
Fig. 5. Supernatants derived from activated prTh17 clones are bactericidal against P. acnes.
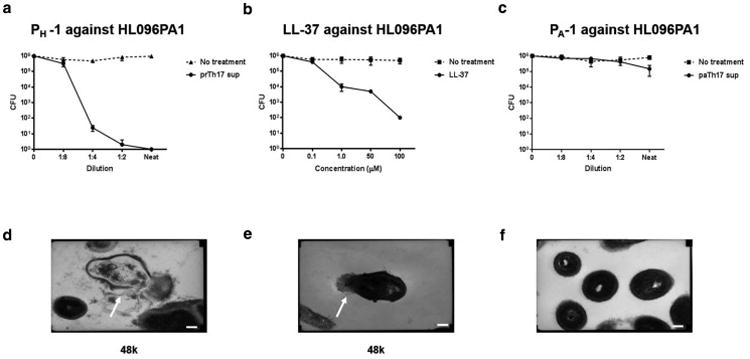
CFU assay results for P. acnes strain HL096PA1 after 3 h incubation with (a) prTh17 clone PH-1 supernatants, (b) LL-37 (0.1-100μm), and (c) paTh17 clone PA-1 supernatants. Data shows average CFU from three independent experiments (n=3), error bars are mean ± SD. (d-f) Representative transmission electron microscopy (TEM) micrographs of (d) P. acnes strain HL096PA1 after 3 h incubation with activated clone PH-1supernatants, (e) LL-37, and (f) activated clone PA-1supernatants. TEM were imaged at 48K. Compared with the activated PA-1 supernatants (1:2 dilution and neat), the bacteria exposed to activated PH-1 supernatants (1:2 dilution and neat) and LL-37 (50μM and 100μM) exhibit cellular differences indicative of stresses on the membrane. Scale bar = 0.5μm.
We next used electron microscopy to determine whether supernatants from activated prTh17, like other antimicrobial peptides, kill bacteria by pore formation and membrane disruption. Transmission EM micrographs of untreated P. acnes illustrate their normal pleomorphic structure (Supplementary Fig. 4a). After a 3-hour treatment with supernatants derived from activated clone PH-1 (prTh17), we observed complete breaches in the cell envelope, with leakage of cytosol to the extracellular environment (Fig. 5d). The same microbicidal activity was observed for LL-37 (Fig. 5e). We further observed that the killing molecule(s) in these supernatants was heat sensitive, as the antimicrobial activity was completely abrogated by heat inactivation (Supplementary Fig 3d), and that IL-26 neutralizing antibody had little to no effect on the ability of prTh17 clones to kill P. acnes in vitro (Supplementary Fig 3e-f). In contrast, PA-1 (paTh17) derived supernatants had no effect on P. acnes (Fig. 5f). We further observed that both PA and PH strains induce IL-26 secretion in PBMCs (Supplementary Fig. 4b) and that IL-26 was highly expressed in both paTh17 and prTh17 clones (Supplementary Fig. 4c). We therefore assessed whether the phenotypic characteristics of paTh17 and prTh17 would be correlated with IL-26 function as IL-26 has recently been described as an antimicrobial protein that efficiently kills extracellular bacteria (Meller et al., 2015). We validated that the antimicrobial activity against P. acnes by prTh17 cells was not due to the action of IL-26, as both natural IL-26 released by a Th26 clone and rhIL-26 lacked antimicrobial potency against P. acnes in CFU assays (Supplementary Fig. 4d-f). In further experiments, we examined whether this molecule(s) was effective against other Gram positive and negative bacteria. As previously reported (Meller et al., 2015), we observed an inhibitory or killing effect when E. coli and S. aureus were incubated with both natural IL-26 released from a Th26 clone and increasing doses of rhIL-26 (Supplementary Fig. 5).
Discussion
The link of Th17 cells to pathologic inflammation is very well established in comparison to our understanding of the role of these cells in host defense. Several studies support their involvement in protection against extracellular bacterial infections such as Klebsiella pneumoniae in mice (Aujla et al., 2008; Happel et al., 2005). However, in humans indirect evidence has recently emerged demonstrating that patients suffering from diseases such as hyper-IgE syndrome, and Mendelian susceptibility to mycobacterial diseases, may have defects in the Th17 cells/pathway (Boniface et al., 2008; Eyerich et al., 2008; Milner et al., 2008; Renner et al., 2008). In addition, both IL-17 and IL-22 induce antimicrobial peptide production from epithelial cells in humans (Liang et al., 2006; Ouyang et al., 2008; Wilson et al., 2007). More recently, Th17-derived IL-26 was shown to have direct antimicrobial activity suggesting their participation in host defense (Meller et al., 2015). Here, we generated PA- and PH-specific clones against six strains of P. acnes and demonstrate that these strains have an ability to differentially modulate the CD4+ T cell responses. Importantly, we show that IL-23 is a stabilizing factor for in vitro developed Th17 lineages, and that prTh17 but not paTh17 secrete molecules that are sufficient to kill P. acnes in vitro. Together, our data suggest that certain subsets of Th17 cells may be involved in providing protection against P. acnes.
Our observations that certain PA strains induced higher IL-17 levels than PH strains suggest that P. acnes strains express different antigenic components/ligands on their surface structure which may be responsible for the differential Th17 responses. These observations corroborate recent results of complete genome studies that compared the genomes of PA associated strain HL096PA1 and that of PH associated strain KPA171202. The studies revealed that, present in HL096PA1 and absent in KPA171202, was a plasmid carrying a tight adhesion locus, which has been shown to enhance colonization and biofilm formation (Kachlany et al., 2000; Tomich et al., 2007). In addition, HL096PA1 also contained genomic islands that were unique to PA-associated strain and may be associated with increased virulence of this lineage in acne (Fitz-Gibbon et al., 2013; Kasimatis et al., 2013). The identification of surface antigens/ligands expressed in PA vs. PH strains that activate PRRs on APCs leading to secretion of inflammatory cytokines, may potentially lead to new diagnostic and therapeutic approaches against acne.
Notably, both paTh17 and prTh17 clones expressed similar levels of antimicrobial proteins cathelicidin and DEFB4. However, despite secreting high levels of IL-26, paTh17 clones demonstrated no antimicrobial activity against P. acnes. This result was surprising as in vitro derived IL-26 was recently shown to be a cationic and amphipathic cytokine that could effectively kill both Gram positive and Gram negative bacteria (Meller et al., 2015). In contrast, prTh17 clones, secreted molecule(s) that were sufficient to kill P. acnes, and we envisage that the high potency of this molecule(s) may be due to synergy with other additional molecules in the culture supernatant.
Th17 cells play a crucial role in controlling different microorganisms in vivo (Khader et al., 2009). We used P. acnes as a model to understand the nature of Th17 responses that may exist in acne vulgaris. Our findings highlight the fact that PH and PA strains differentially modulate the CD4+ T cell responses in vitro leading to induction of IL-17/IFN-γ (paTh17) and IL-17/IL-10 (prTh17) producing cells, and suggest that divergent Th17 responses may exist in acne. Th17 cells have been intensively studied in immune-mediated diseases, but the physiological function of the IL-17/IFN-γ and/or IL-17/IL-10 axis in the context of acne is unknown. Heterogeneous highly inflammatory (Langrish et al., 2005; Zheng et al., 2007) and host protective Th17 responses (Esplugues et al., 2011) have previously been described. In our study, the paTh17 phenotype was mainly induced by PA strains and we associated this phenotype with acne pathogenesis, whereas the PH strains induced the prTh17 phenotype which we associated with protection since activated prTh17 cells demonstrated microbicidal activity against P. acnes in vitro. These data are supported by a previous report that demonstrated that in vitro generated non-pathogenic Th17 cells are able to express IL-10 (McGeachy et al., 2007). Of note, Th17 cells have been shown to respond not only to canonical Th17-dependent pathogens such as Candida albicans, but also to Th1-inducing, intracellular pathogens such as Mycobacterium (Acosta-Rodriguez et al., 2007). Although further studies are required to delineate the mechanisms by which P. acnes strains drive the Th17 to Th1 differentiation program and vice versa, it is likely that poly-functional IL-17/IL-10, but not IL-17/IFN-γ secreting Th17 cells may be beneficial in containing P. acnes. A better mechanistic understanding of how P. acnes strains modulate immune responses can inform future microbiome studies and probiotic designs.
In summary, we demonstrate that P. acnes strains differentially modulate the CD4+ T cell responses, leading to the generation of Th17 cells that may contribute either to homeostasis or pathogenesis in acne vulgaris. It is important to highlight that although RT6 may have only been found on healthy skin to date, the number of subjects have been few, and since current studies indicate that RTs 1, 2 and 3 seem to be evenly distributed in healthy and acne skin, additional studies comparing these P. acnes ribotypes directly isolated from either healthy or acne skin would give a better indication of how acne disease state can be linked to the different functional T cell responses. Our findings and further studies aiming to identify the underlying mechanism(s) by which distinct strains of P. acnes trigger divergent Th17 responses may lead to identification of alternative targets in therapy for acne.
Materials and Methods
Bacterial strains
P. acnes strains used in the study are highlighted in (Supplementary Table 1). PA strains (HL110PA1, HL096PA1, HL043PA1), and PH strains (HL042PA3, HL110PA4, HL110PA3) were obtained from Biodefense and Emerging Infections Research Resources Repository (BEI Resources) and cultured as previously described (Agak et al., 2014). The level of endotoxin contaminating the P. acnes was quantified with a Limulus Amoebocyte Lysate assay (BioWhittaker, Radnor, PA) and found to be <0.1 ng/ml-1. Staphylococcus aureus SA113 and Escherichia coli DH5α were grown in Luria broth (LB) overnight at 37°C with agitation. Overnight bacterial cultures were subcultured and incubated until midlog was reached, which was determined to be OD600 = 0.4. Cultures were washed in sterile PBS and renormalized to OD600 = 0.4 in culture media.
PBMC isolation, stimulation and cytokine ELISAs
Peripheral blood mononuclear cells (PBMCs) were obtained from healthy donors with written informed patient consent, as approved by the University of California, Los Angeles Institutional Review Board. PBMCs were then isolated using Ficoll–Paque gradients (GE Healthcare) as previously described (Agak et al., 2014). Briefly, cells were cultured in T cell media (RPMI 1640, 10% heat inactivated human serum (Gemini), 2mM L-glutamine, 10U/ml penicillin and 100 μg/ml streptomycin) and stimulated with different strains of P. acnes at 1 multiplicity of infection (1 MOI). Levels of cytokines accumulated in culture supernatants were measured by ELISA. Samples were assayed in triplicates. Monocyte isolation from PBMC was done using the Monocyte Isolation Kit (Miltenyi Biotec, Auburn, CA) following manufacturer's protocol.
Reagents
IL-4 (BD555194), IL-10 (DY217b), IFN-γ (DY285), IL-13 (DY213), IL-17 (DY317), IL-22 (DY782), IL-26 (CSB-E11716h), rhIL-26 (R&D), rhIL-23 (R&D), rhIL-2 (R&D), Blocking mouse anti human HLA-Class I (W6/32) and anti-HLA-C (Abcam) both used at 10μg/ml, anti-MHC-II (MS163P1ABX) (Fisher), anti-IL-26 mAb1375, clone 197505 (R&D), CD4 (OKT4) (BD), IL-17PE (BD), IFN-γ- FITC (BD), CD14 FITC (BD) LL-37 (Innovagen). Mouse IgG1 and IgG2b isotype matched antibodies were used as controls.
Sterile Cell sorting
PBMCs from healthy donors were stimulated for 16 hours with PH and PA strains, and IL-17 secretion determined using a cytokine secretion capture assay (Cell Enrichment and Detection Kit) following the manufacturer's protocol (Miltenyi). After IL-17 staining, the cells were further stained with α-CD4 antibodies. CD4+ IL-17+ cells were sorted under sterile conditions using Beckton Dickinson FACS Vantage (San Jose, CA). Dead cells were excluded by DAPI staining. Data acquisition and analysis was done using FlowJo software (V7.6).
T cell cloning and proliferation assays
Sorted cells were cloned at 0.3cells/well in Terasaki plates (Nunc Microwell, Sigma-Aldrich) in the presence of unmatched-γ-irradiated (106) PBMCs activated by 2μg/ml phytohaemagglutinin (PHA-Murex) in T cell media supplemented with 100 U/ml rhIL-2. Plates were screened from days 8-12 post cloning and growing clones were transferred to 96 well flat-bottom plates and later expanded in 24 well plates in T cell media supplemented with 100 U/ml IL-2 and 2ng/ml IL-23. Specificity of the clones was confirmed using proliferation assays. Briefly, CD4+IL-17+ T cell clones at 5 × 104 cells/well were seeded into 96-well flat-bottom microplates (BD) containing 2.5 × 104 autologous monocytes in T cell media with/without P. acnes strains (1 MOI) that was originally used as a stimulus to generate the clones. 3[H] thymidine (1 uCi/well) was added 4 h before the culture was terminated. The cells were harvested and assayed by scintillation counting.
T cell stimulation and Flow cytometry
For analysis of cytokine production, T cell clones were stimulated with either control medium or with α-CD3/α-CD28 (BD) in the presence of GolgiPlug (BD). Cells were next stained, and examined by flow cytometry. Samples were acquired on BD Biosciences FacsScan, and analyzed using FlowJo software (V7.6).
RNA isolation, cDNA synthesis and real-time PCR
Total RNA was isolated from T cell clones after polyclonal activation using Trizol reagent (Invitrogen) following manufacturer's protocol and treated with RNase-free DNase. cDNA and real-time PCR was done as previously described (Agak et al., 2014). GAPDH was used as a control. Gene expression level was quantified by the comparative method 2-ΔΔCT. The list of primers used in the study are summarized in Supplementary table 3.
CFU assay
P. acnes strains (Supplementary Table 1) were grown under anaerobic conditions in Reinforced Clostridial Medium (Oxoid, Basingstroke, England) for 2 d and collected in mid-log phase. The bacteria were washed three times with the assay buffer (10 mM Tris pH 7.4, supplemented with 0.03% volume trypticase soy broth, Tris-TSB), and enumerated by applying a conversion factor of 7.5 × 107 bacteria per mL=1 OD600. Th17 culture supernatants were diluted in Tris-TSB and the CFU assays performed as previously described (McInturff et al., 2005; Schmidt et al., 2015). For the S. aureus and E. coli CFU assays, bacteria were grown as described above and resuspended in RPMI 1640. Depletion of IL-26 was performed by incubating supernatants with 10ug/ml of neutralizing anti-IL-26 mAb or an isotype mAb for 12 h at 4°C. 100μl reactions (bacteria + Th17 supernatants or rhIL-26 or anti-IL-26) were added to 1.5-ml tubes and incubated at 37°C with shaking for 1, 3, or 24 h after the specified incubation periods, 10-fold serial dilutions were plated on LB plates to quantify surviving CFU.
Electron Microscopy
P. acnes strains at 3×108 CFU/mL were incubated untreated or with supernatants (1:2 dilutions for prTh17 and 1:2 and neat for paTh17) derived from activated clones, LL-37, or rhIL-26 for 3 h, washed twice with PBS, and resuspended in PBS with 2% glutaraldehyde. Samples were processed as previously described (Schmidt et al., 2015).
Statistical analysis
Results are expressed as the means ± SD for the number of separate experiments indicated in each case (n≥3). One-way analysis of variance was used to compare variances within groups and among them. Post hoc two-tailed Student's t-test was used for comparison between two groups. Significant differences were considered for those probabilities ≤ 5% (P≤ 0.05)
Supplementary Material
Acknowledgments
We extend our thanks to Marianne Cillufo for assistance with electron microscopy, and Jeffery Calimlim for assistance with the flow cytometry and cell sorting. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Centers for AIDS Research Flow Cytometry Core Facility supported by NIH Awards CA-16042 and AI-28697, by the JCCC, the UCLA AIDS Institute and the UCLA School of Medicine. This work was supported by a kind donation from the MCJ Amelior foundation and a K01 grant AR071479 (G.A.)
Abbreviations
- PBMCs
Peripheral blood mononuclear cells
- prTh17
Protective Th17 cells
- paTh17
Pathogenic Th17 cells
- PA
P. acnes strains associated with acne
- PH
P. acnes strains associated with healthy skin
- CFU
Colony forming units
Footnotes
Conflict of interest: The authors state no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel Fattah NS, Shaheen MA, Ebrahim AA, El Okda ES. Tissue and blood superoxide dismutase activities and malondialdehyde levels in different clinical severities of acne vulgaris. Br J Dermatol. 2008;159:1086–91. doi: 10.1111/j.1365-2133.2008.08770.x. [DOI] [PubMed] [Google Scholar]
- Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–46. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- Agak GW, Qin M, Nobe J, Kim MH, Krutzik SR, Tristan GR, et al. Propionibacterium acnes Induces an IL-17 Response in Acne Vulgaris that Is Regulated by Vitamin A and Vitamin D. J Invest Dermatol. 2014;134:366–73. doi: 10.1038/jid.2013.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaza N, Akamatsu H, Kishi M, Mizutani H, Ishii I, Nakata S, et al. Effects of Propionibacterium acnes on various mRNA expression levels in normal human epidermal keratinocytes in vitro. J Dermatol. 2009;36:213–23. doi: 10.1111/j.1346-8138.2009.00626.x. [DOI] [PubMed] [Google Scholar]
- Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–81. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beylot C, Auffret N, Poli F, Claudel JP, Leccia MT, Del Giudice P, et al. Propionibacterium acnes: an update on its role in the pathogenesis of acne. J Eur Acad Dermatol Venereol. 2014;28:271–8. doi: 10.1111/jdv.12224. [DOI] [PubMed] [Google Scholar]
- Bojar RA, Holland KT. Acne and Propionibacterium acnes. Clinics in dermatology. 2004;22:375–9. doi: 10.1016/j.clindermatol.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Boniface K, Blom B, Liu YJ, de Waal Malefyt R. From interleukin-23 to T-helper 17 cells: human T-helper cell differentiation revisited. Immunol Rev. 2008;226:132–46. doi: 10.1111/j.1600-065X.2008.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu A, Chon SY, Kimball AB. The response of skin disease to stress: changes in the severity of acne vulgaris as affected by examination stress. Arch Dermatol. 2003;139:897–900. doi: 10.1001/archderm.139.7.897. [DOI] [PubMed] [Google Scholar]
- Degitz K, Placzek M, Borelli C, Plewig G. Pathophysiology of acne. J Dtsch Dermatol Ges. 2007;5:316–23. doi: 10.1111/j.1610-0387.2007.06274.x. [DOI] [PubMed] [Google Scholar]
- Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–8. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyerich K, Foerster S, Rombold S, Seidl HP, Behrendt H, Hofmann H, et al. Patients with chronic mucocutaneous candidiasis exhibit reduced production of Th17-associated cytokines IL-17 and IL-22. J Invest Dermatol. 2008;128:2640–5. doi: 10.1038/jid.2008.139. [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133:2152–60. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel KI, Dubin PJ, Zheng M, Ghilardi N, Lockhart C, Quinton LJ, et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J Exp Med. 2005;202:761–9. doi: 10.1084/jem.20050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, et al. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nature immunology. 2013;14:372–9. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C, Mak TN, Zimny-Arndt U, Schmid M, Meyer TF, Jungblut PR, et al. Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol. 2010;10:230. doi: 10.1186/1471-2180-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James WD. Clinical practice. Acne. N Engl J Med. 2005;352:1463–72. doi: 10.1056/NEJMcp033487. [DOI] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, et al. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol. 2000;182:6169–76. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimatis G, Fitz-Gibbon S, Tomida S, Wong M, Li H. Analysis of complete genomes of Propionibacterium acnes reveals a novel plasmid and increased pseudogenes in an acne associated strain. Biomed Res Int. 2013;2013:918320. doi: 10.1155/2013/918320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2:403–11. doi: 10.1038/mi.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistowska M, Meier B, Proust T, Feldmeyer L, Cozzio A, Kuendig T, et al. Propionibacterium acnes promotes Th17 and Th17/Th1 responses in acne patients. J Invest Dermatol. 2015;135:110–8. doi: 10.1038/jid.2014.290. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. The Journal of experimental medicine. 2005;201:233–40. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levell MJ, Cawood ML, Burke B, Cunliffe WJ. Acne is not associated with abnormal plasma androgens. Br J Dermatol. 1989;120:649–54. doi: 10.1111/j.1365-2133.1989.tb01351.x. [DOI] [PubMed] [Google Scholar]
- Leyden JJ, McGinley KJ, Vowels B. Propionibacterium acnes colonization in acne and nonacne. Dermatology. 1998;196:55–8. doi: 10.1159/000017868. [DOI] [PubMed] [Google Scholar]
- Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. The Journal of experimental medicine. 2006;203:2271–9. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomholt HB, Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One. 2010;5:e12277. doi: 10.1371/journal.pone.0012277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell A, Gao A, Barnard E, Fink C, Murray PI, Dowson CG, et al. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology. 2011;157:1990–2003. doi: 10.1099/mic.0.049676-0. [DOI] [PubMed] [Google Scholar]
- McDowell A, Nagy I, Magyari M, Barnard E, Patrick S. The opportunistic pathogen Propionibacterium acnes: insights into typing, human disease, clonal diversification and CAMP factor evolution. PloS one. 2013a;8:e70897. doi: 10.1371/journal.pone.0070897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell A, Patrick S, Eishi Y, Lambert P, Eady A. Propionibacterium acnes in human health and disease. BioMed research international. 2013b;2013:493564. doi: 10.1155/2013/493564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nature immunology. 2007;8:1390–7. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- McInturff JE, Wang SJ, Machleidt T, Lin TR, Oren A, Hertz CJ, et al. Granulysin-derived peptides demonstrate antimicrobial and anti-inflammatory effects against Propionibacterium acnes. The Journal of investigative dermatology. 2005;125:256–63. doi: 10.1111/j.0022-202X.2005.23805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller S, Di Domizio J, Voo KS, Friedrich HC, Chamilos G, Ganguly D, et al. TH17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat Immunol. 2015;16:970–9. doi: 10.1038/ni.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–6. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I, Pivarcsi A, Kis K, Koreck A, Bodai L, McDowell A, et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect. 2006;8:2195–205. doi: 10.1016/j.micinf.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Nagy I, Pivarcsi A, Koreck A, Szell M, Urban E, Kemeny L. Distinct strains of Propionibacterium acnes induce selective human beta-defensin-2 and interleukin-8 expression in human keratinocytes through toll-like receptors. The Journal of investigative dermatology. 2005;124:931–8. doi: 10.1111/j.0022-202X.2005.23705.x. [DOI] [PubMed] [Google Scholar]
- Nakatsuji T, Liu YT, Huang CP, Zouboulis CC, Gallo RL, Huang CM. Vaccination targeting a surface sialidase of P. acnes: implication for new treatment of acne vulgaris. PLoS One. 2008;3:e1551. doi: 10.1371/journal.pone.0001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JF, Cunliffe WJ. A histological and immunocytochemical study of early acne lesions. The British journal of dermatology. 1988;118:651–9. doi: 10.1111/j.1365-2133.1988.tb02566.x. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–67. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner ED, Rylaarsdam S, Anover-Sombke S, Rack AL, Reichenbach J, Carey JC, et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J Allergy Clin Immunol. 2008;122:181–7. doi: 10.1016/j.jaci.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt NW, Agak GW, Deshayes S, Yu Y, Blacker A, Champer J, et al. Pentobra: A Potent Antibiotic with Multiple Layers of Selective Antimicrobial Mechanisms against Propionibacterium Acnes. J Invest Dermatol. 2015;135:1581–9. doi: 10.1038/jid.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh DH, Kwon HH. What's new in the physiopathology of acne? Br J Dermatol. 2015;172(1):13–9. doi: 10.1111/bjd.13634. [DOI] [PubMed] [Google Scholar]
- Thiboutot DM, Layton AM, Eady EA. IL-17: a key player in the P. acnes inflammatory cascade? J Invest Dermatol. 2014;134:307–10. doi: 10.1038/jid.2013.400. [DOI] [PubMed] [Google Scholar]
- Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–75. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, Weinstock GM, et al. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio. 2013;4:e00003–13. doi: 10.1128/mBio.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Flavell RA, Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nature immunology. 2006;7:1151–6. doi: 10.1038/ni1391. [DOI] [PubMed] [Google Scholar]
- Williams HC, Dellavalle RP, Garner S. Acne vulgaris. The Lancet. 2012;379:361–72. doi: 10.1016/S0140-6736(11)60321-8. [DOI] [PubMed] [Google Scholar]
- Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nature immunology. 2007;8:950–7. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, et al. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–51. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- Zouboulis CC, Eady A, Philpott M, Goldsmith LA, Orfanos C, Cunliffe WC, et al. What is the pathogenesis of acne? Experimental dermatology. 2005;14:143–52. doi: 10.1111/j.0906-6705.2005.0285a.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


