Abstract
Specific, chemically modified aptamers (X-Aptamers) were identified against two immune checkpoint proteins, recombinant Programmed Death 1 (PD-1) and Programmed Death Ligand 1 (PD-L1). Selections were performed using a bead-based X-Aptamer (XA) library containing several different amio acid functional groups attached to dU at the 5-position. The binding affinity and specificity of the selected XA-PD1 and XA-PDL1 were validated by hPD-1 and hPD-L1 expression cells, as well as by binding to human pancreatic ductal adenocarcinoma tissue. The selected PD1 and PDL1 XAs can mimic antibody functions in in vitro assays.
1. Introduction
Immune checkpoint antibodies (1–4) are emerging as highly promising therapeutic agents. Recent studies have demonstrated that immune checkpoints contribute not only maintaining physiologic self-tolerance but are also implicated in the down-regulation of anti-tumor immunity. To restore latent anti-tumor immunity, many efforts have focused on antibody-based interventions targeting CTL antigen 4 (CTLA-4) (1,5,6) and programmed cell death protein 1 (PD-1) (7,8) on T lymphocytes, and its principal ligand (PD-L1) (9) on tumor cells. Antibodies targeting regulatory molecules on T cells such as CTLA-4 and PD-1 have demonstrated clinical activity across various tumor types (1,10). Ipilimumab (11,12), an antibody targeting CTLA-4, appears to restore tumor immunity at the priming phase, whereas anti-PD-1/PD-L1 antibodies restore immune function in the tumor microenvironment. Anti-tumor activity was achieved in patients with melanoma (12) and renal cancer (13), as well as those with non-small-cell lung, bladder and head and neck cancers, even tumors not to be sensitive to immunotherapy (14).
However, antibodies are expensive to produce, highly immunogenic, and relatively unstable to environmental stress. The major adverse events with immunomodulatory antibodies are inflammatory pathologies. Repeated or long-term use of antibodies can stimulate humoral immune responses and overproduction of cytokines (11,14). Aptamers generally avoid these adverse events while also offering significant advantages such as lower cost, non-immunogenicity (15), and easy modification for various applications (16,17). They are also easier to produce with no batch-to-batch variation. At least one aptamer (Macugen) was approved by FDA for therapeutic use (18) and several others are in various phases of clinical development (19).
Unfortunately, many traditional aptamers developed using SELEX do not possess the binding affinity and specificity required for therapeutic use (20,21). More functionally diverse aptamers that interact more robustly with targeted proteins hold significant promise for improving the performance of traditional aptamers (16,22,23). XAs (24) are a unique alternative to traditional SELEX aptamers. XAs are selected in one round using bead-based libraries synthesized by solid phase synthesis (24–26). In such libraries, the variety and combination of modifications are far superior to what can be accomplished via enzymatic methods (22–24,27–29). Modifications included in the library used for this selection were the 5-position modified deoxyurdines: 5-amino-, 5-phenol-, and 5-indole- (www.am-biotech.com). Here we report the use of such a library in a single round selection to PD-1 and PD-L1. The XAs discovered were characterized using experiments involving stably transfected mouse L929 fibroblasts expressing hPD-1 or hPD-L1 protein.
2. Material and methods
Standard solvents and reagents were purchased from either Sigma-Aldrich, Chemgenes or Alfa Aesar. The XA library was obtained from AM Biotechnologies, Houston, TX, USA. A detailed selection protocol is available from their website www.am-biotech.com (24). EZ-Link NHS-PEG4-Biotin was purchased from Thermo Pierce.
2.1 Fusion proteins and monoclonal antibodies
Recombinant human PD-1 Fc chimera protein and recombinant human PD-L1/B7-H1 Fc chimera protein were purchased from R&D Systems (Minneapolis, MN) and used for aptamer identification. Recombinant human IgG1 Fc was used as a control. Anti-hPD-1 antibody (clone NAT105) and isotype control mouse IgG1, κ; Anti-hPD-1 antibody (clone 28–8) and isotype control Rabbit IgG, (clone EPR25A) were purchased from Abcam (Cambridge, FL). Goat anti-mouse IgG H&L (Alexa Fluor® 488) and goat anti-rabbit IgG H&L (Alexa Fluor® 488) were used as secondary antibodies for all the immune staining.
2.2 Next-generation sequencing (NGS)
XA oligonucleotides that bound to PD-1 and PD-L1 were isolated according to the XA selection protocol and amplified into unmodified sequences by PCR. They were prepared for sequencing using the Ion Plus Fragment Library Kit (Life Technologies) for next-generation sequencing (NGS) (Ion Torrent PGM™, Life Technologies). The sequencing data were processed using Aptaligner© software (30). Sequences with a high frequency of occurrence in selection fractions containing the protein targets when compared to protocol controls were selected for synthesis of the chemically modified oligonucleotides (24).
2.3 Cell lines and cell binding assay
Stably transfected mouse L929 fibroblasts expressing hPD-1 or hPD-L1 protein were a kind gift from Dr. Zhiqiang An (University of Texas Health Science Center at Houston, TX) and were used for XA binding validation. Cells were maintained in RPMI-1640 medium supplemented with 10 % fetal bovine serum (FBS) and 1 % penicillin-streptomycin solution (tissue culture reagents purchased from Life Technologies, NY). All experiments were performed at 70–80 % cell confluence with 5 % CO2 at 37 °C. The stably transfected PD-1 L929 cell line or PD-L1 L929 cell line was seeded in chamber slides (Cole-Parmer, IL) and incubated with the biotinylated XA-PD1 or XA-PD-L1 at different concentrations and cell ratios after blocking with universal blocking buffer (Thermo Fisher Scientific, IL). The binding of the XAs to associated cell lines was measured by adding streptavidin fluorescein isothiocyanate (FITC) and measuring fluorescence. Non-transfected L929 cells were used as negative controls.
2.4 Human pancreatic tumor tissue
Human pancreatic cancer tissue was collected at the time of standard care of interventional radiology treatment at the clinic of University of Texas Health Science Center at Houston (UT Health). All tumor samples for this study were collected prior to initiation of any therapy under IRB approved by the UT Health Committee for the Protection of Human Subjects. Fresh pancreatic tumor tissues were embedded with optimal cutting temperature (OCT) (Thermo Fisher Scientific, Waltham, MA) compound and cut into 5 μm sections. Slides were blocked first with universal blocking buffer in TBS (Cat. #36000; Thermo Fisher Scientific). XA-PDL1 (25 nM) or primary PD-L1 antibody, appropriate isotype control antibody were incubated at 37 °C for 1 hour. Following washing, slides were incubated for one hour at room temperature with appropriate secondary antibody and nuclei counterstaining. The relative extent of XA-PDL1 binding to the tumor tissue was assessed by fluorescence microscopy using a Nikon Eclipse TE2000-E inverted microscope.
2.5 Aptamer flow cytometry
Selected XAs (XA-PD1 and XA-PDL1) that specifically bind to PD-1 or PD-L1 proteins were synthesized with 5′-biotinylation. Targeted cells were blocked with universal blocking buffer before incubation with XA-PD1 or XA-PDL1, or scrambled control aptamer at 37 °C for 1 hour. After washing to remove excess XAs, fluorescein-labeled streptavidin (BD Biosciences, CA) were incubated with cells for 30 min at room temperature. Binding of XA-PD1s or XA-PDL1s to the targeted cells were measured by percentage of positive cells and fluorescence intensity with FACScalibur flow cytometry (BD Biosciences, San Jose, CA). Anti-human PD-1 (Abcam, MA) and PD-L1 (BD Biosciences, CA) antibodies were used as positive controls. The equilibrium dissociation constant (Kd) was obtained by fitting the dependence of mean fluorescence intensity of specific binding on the concentration of the XA to the equation.
3. Results
3.1 Selection of hPD-1 and hPD-L1 X-aptamers
The single-cycle XA selection process was performed in two straightforward selection steps (24). In order to have the appropriate folding of the oligonucleotide structure, the XA bead-based library was first heated at 95 °C for 5 min and cooled at room temperature for 30 min. Recombinant hPD-1, hPD-L1 Fc chimeric proteins, or human IgG1 Fc protein were labeled with biotin (EZ-Link NHS-PEG4-Biotin, Thermo Pierce), respectively. After negative selection with human IgG1 Fc protein, unbound XA library beads were incubated with biotinylated human PD-1 or PD-L1 protein for 90 min at room temperature with rotation. Beads carrying XAs bound to protein were recovered by pull-down using streptavidin-functionalized magnetic particles. The oligonucleotides were cleaved from the library beads by a strong base (50 μl 1N NaOH, at 65 °C for 30 min and the reaction was neutralized using 40 μl 2M Tris-Cl). After pelleting the magnetic particles using a magnetic stand, the solution pool containing XAs were fractionated per the selection protocol and incubated with the biotinylated protein for second step selection. True XAs were enriched after the second binding step and then amplified into unmodified oligonucleotides by PCR. The sequences were prepared for sequencing using the Ion Plus Fragment Library Kit for next-generation sequencing. The sequencing data were processed using Aptaligner© software (30). Sequences with high frequency of occurrence from target fraction pull downs over the starting oligonucleotide pool as well as a magnetic particle only control were selected.
3.2 Validation of selected XA-PD1s and XA-PDL1s
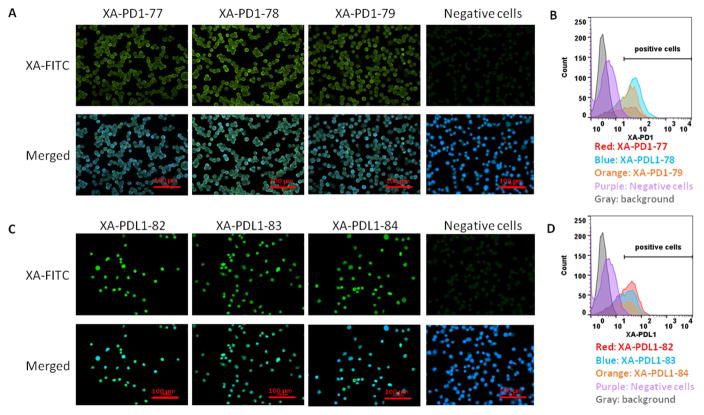
Based on NGS results, the top three XA sequences for each target protein were synthesized and used to determine their binding affinity and specificity for cells. Stably transfected mouse L929 fibroblasts expressing hPD-1 or hPDL-1 protein were used for testing X-Aptamer binding. Flow cytometry analysis indicated all three XA-PD1s bound to PD-1 expressing L929 cells, at 60.1%, 74.2% and 56.5% respectively. Flow cytometry analysis shows that XA-PD1-78 has the highest binding affinity to PD-1 expressing L929 cells (Figure 1 B). Similarly, for XA-PD1s, all three XA-PDL1s bund to PD-L1 expressing L929 cells, at 56.3%, 47.9% and 47.5% respectively. XA-PDL1-82 demonstrated the highest binding affinity to the PD-L1 expressing L929 cells. No binding was observed with PD-1 or PD-L1 negative cells (Figure 1 D). In addition, the binding affinity of the selected XAs to the associated cells was examined by fluorescence microscopy and determined by fluorescence intensity. All the selected XA-PD1s and XA-PD-L1s showed binding to human PD-1 or human PD-L1 expression cells, with some variations in binding intensity (Figure 1 A and C).
Figure 1.
Screening of binding affinity of individual synthesized XA-PD1s and XA-PDL1s. Biotin conjugated XA-PD1s were incubated with human PD-1 expressing L929 cells (A and B) and XA-PDL1s were incubated with human PD-L1 expressing L929 cells (C and D). Non-transfected L929 cells were used as negative control. A and C represent microscope examination of fluorescence intensity of biotinylated XA-PD1s or XA-PDL1s (25 nM) bound to cells in chamber slides. B and D represent flow cytometry analysis of fluorescence intensity of biotinylated XA-PD1s or XA-PDL1s (50 nM) binding to cells in suspension.
To quantitatively evaluate the binding affinity of selected XA-PD1-78 and XA-PDL1-82, PD-1 or PD-L1 expression cells were incubated separately with biotin-streptavidin conjugates of XAs at various concentrations and analyzed by flow cytometry. Binding affinity of XAs at various dosages with PD-1 or PD-L1 expression cells were determined by mean fluorescence intensity. Saturation curves were fit from these data and the dissociation constant Kd values were determined via nonlinear regression analysis. The dissociation constant Kds of XA-PD1-78 and XA-PDL1-82 for binding with PD-1 or PD-L1 expression cells were determined to be 56.72 nM and 20.18 nM, respectively (Figure 2).
Figure 2.
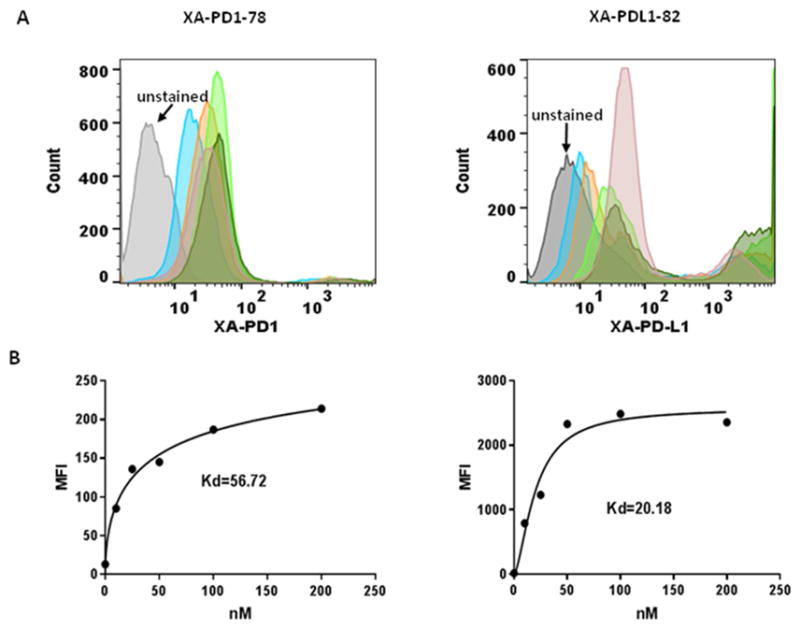
Binding affinity of selected XAs. Biotin-streptavidin conjugated XA-PD1-78 and XA-PDL1-82 were incubated with PD-1 or PD-L1 expression cells at various concentrations and their binding affinity were analyzed by fluorescence intensity with FACScalibur flow cytometry. Histogram of the percent gated fluorescence intensity above background for XA-PD1-78 and XA-PDL1-82 (A). The equilibrium dissociation constant (Kd) was obtained by fitting the dependence of fluorescence intensity of specific binding on the concentration of the aptamers to the equation (B).
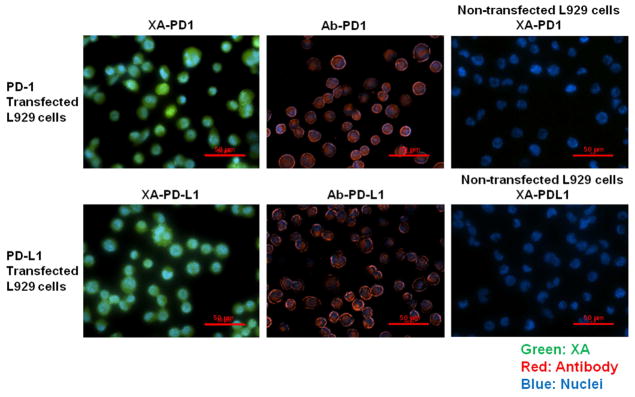
Based on the screening result of binding proficiency, we selected XA-PD1-78 and XA-PDL1-82 for further study. To compare the performance of XA-PD1-78 and XA-PDL1-82 with antibodies, human PD-1 or PD-L1 expressing L929 cells were incubated with either selected XAs or anti-human PD-1 and PD-L1 antibodies. Their binding ability was examined by fluorescence microscopy. Both XA-PD1-78 and XA-PDL1-82 demonstrated similar capacity for binding PD-1 or PD-L1 overexpressing cells, compared to their corresponding antibodies. Specific bindings of selected XA-PD1-78 or XA-PDL1-82 were confirmed with non-transfected L929 cells with no fluorescence detection (Figure 3).
Figure 3.
Comparison of binding performance of selected XA-PD1-78 and XA-PDL1-82 against appropriate antibodies. Human PD-1 or PD-L1 expressing L929 cells were incubated with selected XA-PD1- 78 and XA-PDL1-82 at 25 nM or anti-human PD-1 (5 μg/ml) and PD-L1 (2.4 μg/ml) antibodies. Their binding ability was examined by fluorescence microscope. Non-transfected L929 cells were used as a negative control.
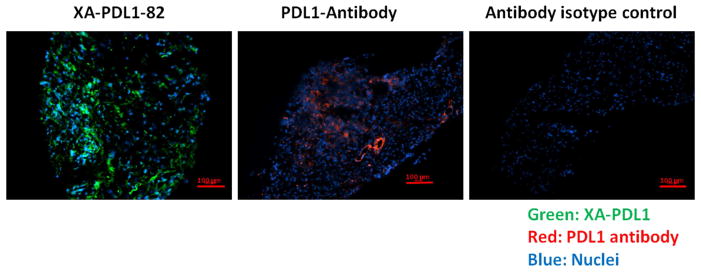
To extend the application of XA-PDL1-82 as a substitute of PD-L1 antibody in clinical immunohistochemistry analysis, XA-PDL1-82 has been used as detection reagent in our preliminary study with PD-L1 antibody in human pancreatic cancers. Sections of fresh frozen pancreatic tumor tissue were incubated with XA-PDL1-82 or PD-L1 antibody. Staining patterns and intensity across slides stained with XA-PDL1-82 or PD-L1 antibody were analyzed. As expected, XA-PDL1-82 could detect PD-L1 expression of pancreatic tumor, either on tumor cells or in the tumor microenvironment on immune-infiltrating cells. XA-PDL1-82 has shown the same capacity to bind PD-L1 expression pancreatic tumor (Figure 4). Based on this preliminary result, our further studies will use XA-PDL1-82 to assess the association between PD-L1 expression on the pancreatic tumor tissue and the likelihood of response to PD-1 blockade treatment.
Figure 4.
Detection of PD-L1 expression on human pancreatic tumor tissue. Sections of human pancreatic ductal adenocarcinoma tissue were incubated with XA-PDL1-82 (25 nM) or PD-L1 antibody (4.8 μg/ml), followed by streptavidin FITC (green) or Goat Anti-Rabbit IgG (Alexa Fluor® 647) secondary antibody at 1/1000 dilution (red). Hoechst 33342 was used to stain the cell nuclei (blue). An IgG isotype was used as negative control. Expression level of PD-L1 was determined by fluorescence intensity of FITC or AF647.
4. Discussion
The beads XA library has ca. 2 × 109 of beads, each bead carrying about 3 × 103 copies of a potential XA consisting of a unique chemically modified strand of DNA. Each XA candidate can include a combination of many natural and modified nucleotides. Specially, AM Biotech’s XA libraries can incorporate modifications that are not compatible with enzymatic incorporation such as phosphorodithioate modification (27,29,31–40) XA libraries can also accommodate multiple different modifications such as multiple different modified versions of dU as described here (24). Any chemical modification can be incorporated into this process at any number of sites, as long as the modification does not prevent determination of the underlying base sequence (26). This extensive chemical diversity enables robust interaction with a target improving specificity.
Use the beads XA library developed by Yang and many others (24,26), we have identified DNA aptamers that can specifically bind to human PD-1 and PD-L1 proteins in cell level. The two most highly enriched aptamers, XA-PD1-78 and XA-PDL1-82 were identified and synthesized with proper modifications. Their binding affinity and specificity were verified with hPD-1 or hPD-L1 overexpression cell lines. Compare to PD-1 and PD-L1 antibodies, those selected XA-PD1-78 and XA-PDL1-82 demonstrated similar cell binding intensity. XA-PDL1-82 has shown the same capacity as PD-L1 antibody to bind PD-L1 expression on pancreatic tumor tissue.
5. Conclusions
X-Aptamers XA-PD1-78 and XA-PDL1-82 that specifically bind to human PD-1 and PD-L1 proteins respectively were isolated and their affinity and specificity were verified with hPD-1 or hPD-L1 overexpression cell lines. Compared to PD-1 and PD-L1 antibodies, XA-PD1-78 and XA-PDL1-82 demonstrated similar cell binding intensity. XA-PDL1-82 has shown the same capacity as PD-L1 antibody to bind PD-L1 expression on pancreatic tumor tissue. These XAs provide a synthetic alternative to antibodies for use in research, diagnostic assays, and potentially as a therapeutic. Progress is on the way.
Acknowledgments
Part of the research was supported by National Institutes of Health (NIH) (GM108110).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhou G, Sprengers D, Boor PPC, Doukas M, Schutz H, Mancham S, Pedroza-Gonzalez A, Polak WG, de Jonge J, Gaspersz M, et al. Antibodies Against Immune Checkpoint Molecules Restore Functions of Tumor-infiltrating T cells in Hepatocellular Carcinomas. Gastroenterology. 2017 doi: 10.1053/j.gastro.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Masarova L, Kantarjian H, Garcia-Mannero G, Ravandi F, Sharma P, Daver N. Harnessing the Immune System Against Leukemia: Monoclonal Antibodies and Checkpoint Strategies for AML. Adv Exp Med Biol. 2017;995:73–95. doi: 10.1007/978-3-319-53156-4_4. [DOI] [PubMed] [Google Scholar]
- 3.Ramos RN, Piaggio E, Romano E. Mechanisms of Resistance to Immune Checkpoint Antibodies. Handb Exp Pharmacol. 2017 doi: 10.1007/164_2017_11. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Deppisch N, Ruf P, Eissler N, Lindhofer H, Mocikat R. Potent CD4+ T cell-associated antitumor memory responses induced by trifunctional bispecific antibodies in combination with immune checkpoint inhibition. Oncotarget. 2017;8:4520–4529. doi: 10.18632/oncotarget.13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Via CS, Soloviova K, Puliaiev M, Puliav R, Puliaeva I, Morris SC, Finkelman FD. In vivo IL-4 prevents allo-antigen driven CD8+ CTL development. Clin Immunol. 2017;180:11–24. doi: 10.1016/j.clim.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierce BG, Hellman LM, Hossain M, Singh NK, Vander Kooi CW, Weng Z, Baker BM. Computational design of the affinity and specificity of a therapeutic T cell receptor. PLoS Comput Biol. 2014;10:e1003478. doi: 10.1371/journal.pcbi.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaya CN, Wians FH, Jr, Bryan BA, Torabi A. Enhanced expression of Programmed cell death 1 (PD-1) protein in benign vascular anomalies. Pathology. 2017;49:292–296. doi: 10.1016/j.pathol.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Gutzmer R, Koop A, Meier F, Hassel JC, Terheyden P, Zimmer L, Heinzerling L, Ugurel S, Pfohler C, Gesierich A, et al. Programmed cell death protein-1 (PD-1) inhibitor therapy in patients with advanced melanoma and preexisting autoimmunity or ipilimumab-triggered autoimmunity. Eur J Cancer. 2017;75:24–32. doi: 10.1016/j.ejca.2016.12.038. [DOI] [PubMed] [Google Scholar]
- 9.Guzik K, Zak KM, Grudnik P, Magiera K, Musielak B, Torner R, Skalniak L, Domling A, Dubin G, Holak TA. Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1. J Med Chem. 2017;60:5857–5867. doi: 10.1021/acs.jmedchem.7b00293. [DOI] [PubMed] [Google Scholar]
- 10.Zhuansun Y, Huang F, Du Y, Lin L, Chen R, Li J. Anti-PD-1/PD-L1 antibody versus conventional chemotherapy for previously-treated, advanced non-small-cell lung cancer: a meta-analysis of randomized controlled trials. J Thorac Dis. 2017;9:655–665. doi: 10.21037/jtd.2017.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madden KM, Hoffner B. Ipilimumab-Based Therapy: Consensus Statement From the Faculty of the Melanoma Nursing Initiative on Managing Adverse Events With Ipilimumab Monotherapy and Combination Therapy With Nivolumab. Clin J Oncol Nurs. 2017;21:30–41. doi: 10.1188/17.CJON.S4.30-41. [DOI] [PubMed] [Google Scholar]
- 12.Copur MS, Ramaekers R, Crockett D. Ipilimumab Adjuvant Therapy in Melanoma. N Engl J Med. 2017;376:398–399. doi: 10.1056/NEJMc1615564. [DOI] [PubMed] [Google Scholar]
- 13.Hammers HJ, Plimack ER, Infante JR, Rini BI, McDermott DF, Lewis LD, Voss MH, Sharma P, Pal SK, Razak ARA, et al. Safety and Efficacy of Nivolumab in Combination With Ipilimumab in Metastatic Renal Cell Carcinoma: The CheckMate 016 Study. J Clin Oncol. 2017 doi: 10.1200/JCO.2016.72.1985. JCO2016721985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Liang F, Li W, Wang Q. Risk of treatment-related mortality in cancer patients treated with ipilimumab: A systematic review and meta-analysis. Eur J Cancer. 2017;83:71–79. doi: 10.1016/j.ejca.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 15.Gilboa E, Berezhnoy A, Schrand B. Reducing Toxicity of Immune Therapy Using Aptamer-Targeted Drug Delivery. Cancer Immunol Res. 2015;3:1195–1200. doi: 10.1158/2326-6066.CIR-15-0194. [DOI] [PubMed] [Google Scholar]
- 16.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, Janjic N. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol Ther Nucleic Acids. 2014;3:e201. doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keefe AD, Cload ST. SELEX with modified nucleotides. Curr Opin Chem Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 18.Ng EW, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 19.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Li N, Gorenstein DG. Strategies for the discovery of therapeutic Aptamers. Expert Opin Drug Discov. 2011;6:75–87. doi: 10.1517/17460441.2011.537321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X, Gorenstein DG. Progress in thioaptamer development. Curr Drug Targets. 2004;5:705–715. doi: 10.2174/1389450043345074. [DOI] [PubMed] [Google Scholar]
- 22.Gawande BN, Rohloff JC, Carter JD, von Carlowitz I, Zhang C, Schneider DJ, Janjic N. Selection of DNA aptamers with two modified bases. Proc Natl Acad Sci U S A. 2017;114:2898–2903. doi: 10.1073/pnas.1615475114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelinas AD, Davies DR, Edwards TE, Rohloff JC, Carter JD, Zhang C, Gupta S, Ishikawa Y, Hirota M, Nakaishi Y, et al. Crystal structure of interleukin-6 in complex with a modified nucleic acid ligand. J Bio Chem. 2014;289:8720–8734. doi: 10.1074/jbc.M113.532697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lokesh GL, Wang H, Lam CH, Thiviyanathan V, Ward N, Gorenstein DG, Volk DE. X-Aptamer Selection and Validation. Methods Mol Biol. 2017;1632:151–174. doi: 10.1007/978-1-4939-7138-1_10. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Li X, Prow TW, Reece LM, Bassett SE, Luxon BA, Herzog NK, Aronson J, Shope RE, Leary JF, et al. Immunofluorescence assay and flow-cytometry selection of bead-bound aptamers. Nucleic Acids Res. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Bassett SE, Li X, Luxon BA, Herzog NK, Shope RE, Aronson J, Prow TW, Leary JF, Kirby R, et al. Construction and selection of bead-bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res. 2002;30:e132. doi: 10.1093/nar/gnf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abeydeera ND, Egli M, Cox N, Mercier K, Conde JN, Pallan PS, Mizurini DM, Sierant M, Hibti FE, Hassell T, et al. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 2016;44:8052–8064. doi: 10.1093/nar/gkw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lou X, Egli M, Yang X. Determining Functional Aptamer-Protein Interaction by Biolayer Interferometry. Curr proct Nucleic Acid Chem. 2016;67(7):25.1–15. doi: 10.1002/cpnc.18. [DOI] [PubMed] [Google Scholar]
- 29.Pallan P, Yang X, Sierant M, Abeydeera N, Hassell T, martinez C, Janicka M, Nawrot B, Egli M. Crystal structure, stability and Ago2 affinity of phosphorodithioate-modified RNAs. RSC Adv. 2014;4:64901–64904. [Google Scholar]
- 30.Lu E, Elizondo-Riojas MA, Chang JT, Volk DE. Aptaligner: automated software for aligning pseudorandom DNA X-aptamers from next-generation sequencing data. Biochemistry. 2014;53:3523–3525. doi: 10.1021/bi500443e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X. Solid-phase synthesis of oligodeoxynucleotide analogs containing phosphorodithioate linkages. Curr Protoc Nucleic Acid Chem. 2016;66:4.71.1–4.71.14. doi: 10.1002/cpnc.13. [DOI] [PubMed] [Google Scholar]
- 32.Yang X. Introducing 2′-OMe-thiophosphoramidites. Glen Research. 2015;27:4–5. [Google Scholar]
- 33.Wu SY, Yang X, Gharpure KM, Hatakeyama H, Egli M, McGuire MH, Nagaraja AS, Miyake TM, Rupaimoole R, Pecot CV, et al. 2′-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumour activity. Nature Commun. 2014;5:3459. doi: 10.1038/ncomms4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Sierant M, Janicka M, Peczek L, Martinez C, Hassell T, Li N, Li X, Wang T, Nawrot B. Gene Silencing Activity of siRNA Molecules Containing Phosphorodithioate Substitutions. ACS Chem Biol. 2012;7:1214–1220. doi: 10.1021/cb300078e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Nawrot B, Sierant M, Janicka M, Martinez C, Mierzejewski E, Hassell T. TIDES Research, Technology and Product Development. Boston: 2011. [Google Scholar]
- 36.Yang X. Thiophosphoramidites and their use in synthesizing oligonucleotide phosphorodithioate linkages. Glen Research. 2008;20:4–6. [Google Scholar]
- 37.Fennewald SM, Scott EP, Zhang L, Yang X, Aronson JF, Gorenstein DG, Luxon BA, Shope RE, Beasley DW, Barrett AD, et al. Thioaptamer decoy targeting of AP-1 proteins influences cytokine expression and the outcome of arenavirus infections. J Gen Virol. 2007;88:981–990. doi: 10.1099/vir.0.82499-0. [DOI] [PubMed] [Google Scholar]
- 38.Yang X, Wang H, Beasley DW, Volk DE, Zhao X, Luxon BA, Lomas LO, Herzog NK, Aronson JF, Barrett AD, et al. Selection of thioaptamers for diagnostics and therapeutics. Ann N Y Acad Sci. 2006;1082:116–119. doi: 10.1196/annals.1348.065. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Yang X, Bowick GC, Herzog NK, Luxon BA, Lomas LO, Gorenstein DG. Identification of proteins bound to a thioaptamer probe on a proteomics array. Biochem Biophys Res Commun. 2006;347:586–593. doi: 10.1016/j.bbrc.2006.06.132. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Fennewald S, Luxon BA, Aronson J, Herzog NK, Gorenstein DG. Aptamers containing thymidine 3′-O-phosphorodithioates: synthesis and binding to nuclear factor-kappaB. Bioorg Med Chem Lett. 1999;9:3357–3362. doi: 10.1016/s0960-894x(99)00600-9. [DOI] [PubMed] [Google Scholar]