Abstract
The M protein is the major surface-associated virulence factor of group A Streptococcus (GAS) and an antigenically variable target of host immunity. How selection pressures to escape immune recognition, maintain indispensable functions, and mask vulnerabilities have shaped the sequences of the >220 M protein types is unclear. Recent experiments have shed light on this question by showing that hidden within the antigenic variability of many M protein types are sequence patterns conserved for recruiting human C4b-binding protein (C4BP). Other host factors may be recruited in a similar manner by conserved but hidden sequence patterns in the M protein. The identification of such patterns may be applicable to the development of a GAS vaccine.
Keywords: M protein, Group A Streptococcus, Antigenic Variation
Antigenic Variation, Functional Indispensability, and Vulnerability Masking
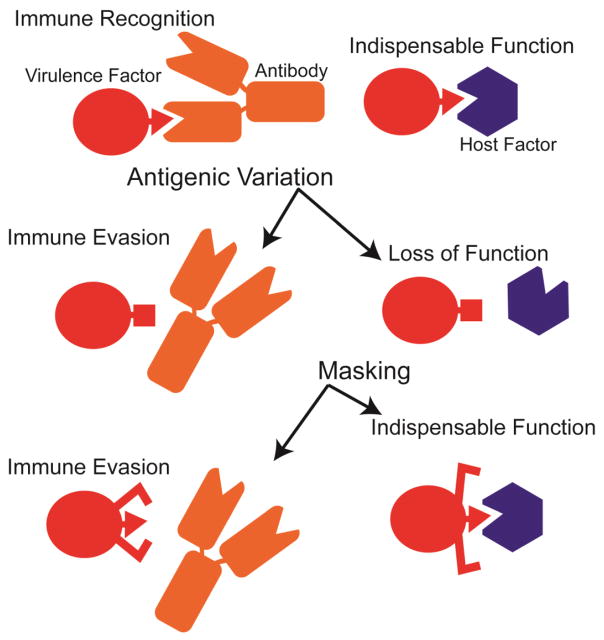
Antigenic variation is a favored means for escape from host immune surveillance and neutralization by microbial pathogens. Small changes in the sequence of microbial virulence factors are usually sufficient to attenuate recognition by neutralizing antibodies (Fig. 1). At the same time, such changes have the potential to limit or destroy the essential function of virulence factors. These functions are generally quite precise and specified by sequences and structures that are poorly tolerant to sequence variation. Such functionally indispensable sequences and structures represent vulnerabilities for the pathogen. In turn, pathogens have evolved strategies for masking these vulnerabilities. Examples from HIV and influenza virus include host glycosylation, which is seen as “self,” and functional structures that exist only upon induced conformational changes. Recent work on HIV [1], influenza virus [2], and dengue virus [3] has revealed that it is possible for host immunity to thwart these masking strategies in the form of broadly neutralizing antibodies.
Figure 1. Selection Pressures on Virulence Factors.
The sequences and structures of virulence factors are shaped by antigenic variation to escape immune recognition and sequence conservation to maintain indispensable functions. Masking strategies enable virulence factors to both evade host immunity and maintain indispensable functions. A masking strategy involving an induced conformational change is depicted.
The Gram-positive bacterial pathogen Streptococcus pyogenes (or group A Streptococcus, GAS) has been long known to evade immunity through antigenic variation of the M protein [4, 5], its major surface-associated virulence factor. However, how the M protein has been shaped by antigenic variation, functional indispensability, and masking strategies is less clear. This review is focused on sequence variable regions of the M protein, and recent results showing that certain variable regions recruit common host factors despite their sequence diversity. In particular, this review deals with the emerging theme of sequence conservation hidden within M protein variability.
Antigenic Variation in the M protein
GAS is a leading cause of global morbidity and mortality [6]. With an estimated >500,000 deaths annually, GAS ranks among the top 10 causes of mortality from infectious disease [6]. GAS is responsible for mucosal infections (e.g. pharyngitis and impetigo), acute invasive diseases (e.g., necrotizing fasciitis and streptococcal toxic shock syndrome), and autoimmune sequelae (e.g., rheumatic heart disease and glomerulonephritis) [7, 8].
The M protein has a significant role in the entire range of conditions caused by GAS. Covalently attached to the GAS cell wall, the M protein forms a dense fibrillar coat that extends ~500 Å into the extrabacterial space [9]. The M protein can also be found in released, soluble form during routine growth [10] or infection [11]. Throughout nearly its entire length, the M protein is composed of a dimeric, parallel α-helical coiled coil [9, 12–14], explaining its fibrillar appearance. However, portions of the coiled coil are quite far from canonical and contain key irregularities [13, 15, 16], as detailed below.
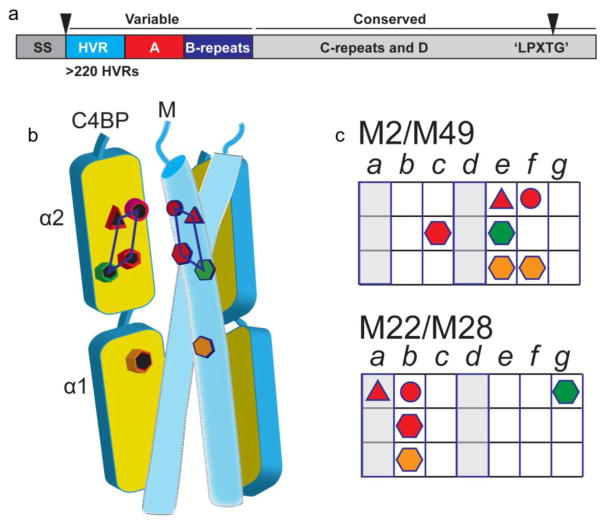
Sequence variation is localized to the N-terminal portion of the mature M protein, whose length on average is ~320–440 amino acids [17] (Fig. 2a). The N-terminal 50 amino acids of the mature M protein are denoted the hypervariable region (HVR) and distinguish one M type from another [18]. Extending a further 100–150 amino acids beyond the HVR are sequence variable regions denoted the A region or A repeats and B-repeats, depending on the M protein type; certain M protein types lack one or the other [19]. The A and B designations are based on relative position rather than sequence identity. Following these regions are the conserved C-repeats and D region, with the latter containing an ‘LPXTG’ motif for cell wall anchoring (Fig. 2a).
Figure 2. Conserved Sequence Patterns Hidden within Variability.
a. Variability is localized to the N-terminal portions of the M protein (HVR, A-repeats or A region, and B-repeats), while the C-terminal portions (C-repeats and D region) are conserved. Nascent M protein is processed into mature M protein through the proteolytic removal (denoted by gray triangles) of an N-terminal signal sequence (SS) and proteolytic processing of a C-terminal ‘LPXTG’ motif, the latter resulting in covalent attachment of the M protein to the peptidoglycan.
b. C4BP has five contact sites (depicted by colored symbols) that interact with complementary amino acids of the M protein HVR. The four contact sites on the C4BP α2 domain form a quadrilateral, and the one site on the α1 domain consists of an Arg nook. The Arg sites are depicted as hexagons.
c. Heptad pattern of C4BP-contacting residues (denoted as colored symbols corresponding to panel b) in M2 and M49 (top) and in M22 and M28 (bottom) HVRs. The core heptad a and d positions are in gray.
More than 220 distinct M protein types have been identified through sequencing of the HVR [17]. The variability of this region is ascribable to immune pressure, as the HVR is a target of opsonizing antibodies [20–23]. Such antibodies are generally type-specific, recognizing the immunizing M type but not other M types [23, 24], although there are recently discovered exceptions to this, as discussed below [25, 26]. Type-specificity has been a problem in the development of an M protein-based GAS vaccine. At present, no GAS vaccine exists. The HVR is weakly immunogenic [21, 27, 28], which along with its sequence variability provides an additional means of escape from immune surveillance. This weak immunogenicity is likely related to the fact that the HVR is sensitive to proteolysis and is susceptible to removal from the M protein due to the action of host or bacterial proteases (e.g., elastase or SpeB, respectively) [27]. Proteolytic sensitivity typically correlates with a lack of structural ordering, and consistent with this tendency, the HVR of the M1 protein was observed to be structurally irregular [13].
Functional indispensability appears also to be at play in M protein HVRs. While HVR sequences differ between M types, these sequences are stable within the type, which suggests an indispensable function for each of the HVRs. While a number of host factors have been identified to interact with M protein variable regions [19], they fall far short of the >220 M types and number only in the handful. Recent work examining the interaction of M protein HVRs with human C4b-binding protein (C4BP) offers an answer to this puzzle [29].
Hiding in Plain Sight
C4BP binds the complement protein C4b and acts as a downregulator of the complement system. The binding of C4b by C4BP results in disabling of the C3 convertase of the classical and lectin pathways (i.e., C4bC2a). C4BP also acts as a cofactor for complement factor I [30], a serine protease that cleaves and inactivates C4b. C4BP is recruited to the GAS surface by the M protein [31–34], which effects a marked decrease in the production of the major opsonin C3b (a product of the C3 convertase), thus preventing complement activation and reducing GAS clearance by C3b receptor-bearing phagocytes [35]. C4BP also competes with opsonic antibodies for binding to the M protein, providing an additional means for enhancing GAS survival [35, 36].
Loss of C4BP binding to the GAS surface through mutation of the M protein has been shown to result in a 3- to 13-fold greater susceptibility to killing by whole human blood [35, 36]. Since murine C4BP does not bind the M protein [34], a transgenic mouse line expressing human C4BP was generated to examine the role of C4BP in GAS pathogenesis [37]. In this model, a significant effect was seen between wild-type and the humanized C4BP transgenic mice in response to intravenous inoculation with a C4BP-binding strain of GAS. At a high inoculating dose of this strain, ~80% of humanized C4BP transgenic mice died after 8 days as compared to only ~20% of wild-type mice [37].
Remarkably, the M protein HVR is the C4BP-binding determinant. This has been demonstrated through in vitro binding studies examining more than 10 M protein types (including Protein H, an M-like protein) [29, 31, 32, 38–40]. Equally remarkably, a common site on C4BP is targeted by M protein HVRs, as shown by competition studies [31, 32]. A larger scale study of C4BP binding was carried out with whole bacteria, comprising 100 GAS strains of differing M types [31]. An overwhelming majority of these strains (~90%) bound C4BP. While it is has not been established that the M protein was responsible for C4BP binding in each of these cases, no factor besides an M or M-like protein (e.g., Protein H in certain M1 strains, and Enn18 in M18 strains) [39–41] has been documented to bind C4BP. Additionally, no region in the M protein besides the HVR has been documented to bind C4BP.
Further delineation of C4BP-binding was provided by classification of 175 M protein types into two clades, X (with 85 M proteins) and Y (with 84 M proteins) [38]. In this study, direct binding studies showed that nine of 12 M protein types belonging to clade X bound C4BP, whereas none of 12 M protein types belonging to clade Y did, suggesting that C4BP-binding is restricted to clade X. There was some disagreement between this in vitro binding study and the whole bacteria study, as some of the M proteins identified as C4BP-nonbinders in vitro [38] corresponded to C4BP-binders in the whole bacteria study [31]. C4BP binding in these particular strains may be dependent on M-like proteins, or there may be other C4BP-binding proteins in GAS yet to be identified.
The basis for the broad but specific recognition between HVRs and C4BP was elucidated recently by structural studies [29]. Four M protein HVRs (M2, M22, M28, and M49) were co-crystallized with the α1α2 domains of C4BP, which are sufficient for interaction [34]. The C4BP α1 and α2 domains are each composed of ~60 amino acid complement control protein (CCP) modules, which are found in many complement proteins. Despite the sequence divergence of the HVRs, a common set of contacts to the C4BP α1 and α2 domains was observed [29] (Fig. 2b). This set consisted primarily of electrostatic interactions supplemented by a few hydrophobic ones. The C4BP α2 domain presented a quadrilateral of contacts consisting of a hydrophobic site, a hydrogen bonding site, and two arginines; the α1 domain presented an Arg nook, with both hydrophobic and electrostatic sites (Fig. 2b). The prominence of arginines in C4BP is likely to be consequential, as arginine is especially versatile for interactions. It possesses both a guanidinium head group for electrostatic interactions and a long alkyl body for hydrophobic interactions. Indeed, arginines at antibody combining sites have been noted to promote promiscuity [42]. In effect, the quadrilateral and Arg nook act as a ‘reading head’ that interrogates the HVR coiled-coil structure for a pattern of chemically complementary amino acids.
M2, M22, M28, and M49 all present similar spatial patterns of such complementary amino acids [29]. The coiled-coil heptad locations of these amino acids differ, with M2 and M49 similar to one another and belonging to one subset, and M22 and M28 similar to one another and belonging to a second subset (Fig. 2c). These differences are reflected in the fact that the M2 and M49 coiled coils run roughly parallel to C4BP, while the M22 and M28 coiled coils are positioned crosswise to C4BP.
Forty-one further M protein HVRs (including that of Protein H) were identified as having amino acid patterns similar to the M2/M49 or M22/M28 patterns, and were therefore predicted to bind C4BP in like manner [29]. These 41 HVRs comprise nearly half of the M strains identified to bind C4BP in the whole GAS bacteria study [31]. The great majority of these M2/M49- or M22/M28-like predicted C4BP-binders (>90%) belong to clade X [38], consistent with C4BP-binding residing in clade X. However, in vitro binding data indicate that several of the predicted M2/M49-like binders (M97, M102, and M106) do not in fact bind C4BP [38]. It is possible that these M protein HVRs, while having M2/M49-like patterns, also have amino acids that interfere with C4BP binding. In line with this possibility, it was observed that several amino acids of the M2 HVR antagonize C4BP binding [29]. Slightly more than half of the M strains identified to bind C4BP using whole GAS bacteria [31] cannot be ascribed to either the M2/M49 or M22/M28 pattern. A large number of these unascribable M protein HVRs (85%) belong to clade X [38]. This suggests that there are likely to be further and as yet undiscovered M protein HVR patterns involved in binding C4BP.
These structural studies show that hidden within M protein variability are conserved sequence patterns that confer C4BP binding. How does GAS mask these functionally conserved patterns? Part of the answer seems to be that C4BP is tolerant to variation whereas antibodies are much less so. Only one of nine alanine-substitutions in the M2 HVR, at amino acids that contact C4BP, resulted in loss of C4BP binding [29]. Likewise, C4BP was tolerant to a number of single amino acid substitutions in the M22 HVR (except those that disrupted the coiled-coil structure) [31]. In contrast, these same single amino acid substitutions in the M22 HVR disrupted antibody binding [31]. Despite this overall tolerance, the interaction with C4BP is indeed specific and there is a limit to sequence variation. C4BP binding was lost upon several dual alanine-substitutions in the M2 HVR [29]. Another part of the masking strategy appears to be diversion. Surrounding the conserved C4BP-binding pattern are a large number of variable amino acids. The attention of the antibody response appears to be drawn to the variable residues rather than to the conserved C4BP-binding pattern, resulting in a type-specific rather than a type-promiscuous response (Fig. 3a).
Figure 3. Type-specificity and Type-promiscuity of Antibodies.
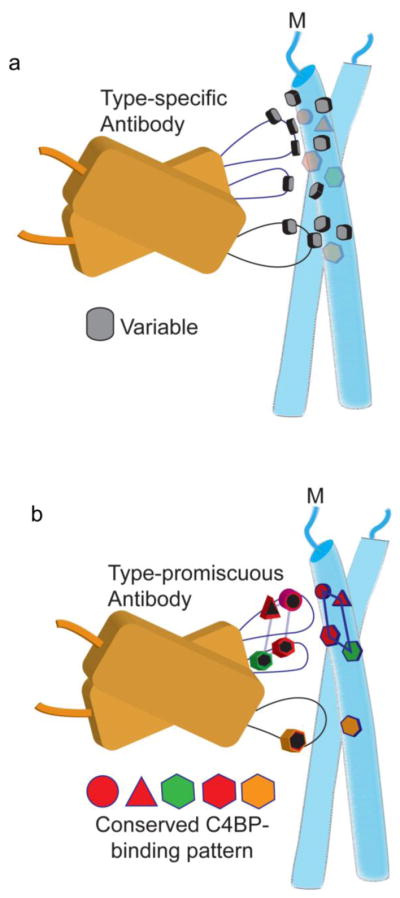
a. Type-specific antibodies contact variable residues of the M protein. Conserved C4BP-binding amino acids are in ghosted symbols. Just the antibody variable domains are shown.
b. An antibody that contacted conserved C4BP-binding amino acids in the M protein would be predicted to be able to recognize multiple M types (i.e., type-promiscuous response).
Can antibodies be made to mimic the type-promiscuous binding mode of C4BP (Fig. 3b)? There is suggestive evidence that this is possible. The HVR alone, in the absence of other portions of the M protein, has been shown to evoke protective immunity [25]. This is in agreement with the observation that non-immunodominant streptococcal epitopes, such as the HVR [21, 27, 28], can serve as protective immunizing antigens [28]. Type-promiscuity was noted in a vaccine composed of 30 M protein HVRs, which elicited responses not only against the 30 immunizing M proteins but also a number of M protein types not included in the vaccine [25, 26]. Intriguingly, 14 of the 30 immunizing M protein HVRs can be assigned to either the M2/M49 or M22/M28 patterns, and 20 of the crossreactive M protein HVRs also can be assigned to either of these two patterns [25, 26]. Thus, it is possible that these crossreactive antibodies adopted a C4BP-like binding mode.
A different approach has been taken to generating crossreactive antibodies that combines information from phylogenetic clustering of M proteins (into clades X and Y) [38] with computational structure-based peptide modeling [43]. In this study, five HVRs from a cluster of 17 HVRs within clade X were used as vaccine antigens. Responses against the immunizing M types were found, as well as responses against the remaining 12 M types in the cluster not included in the vaccine. This cluster included M2, M22, and M28, but not M49. Interestingly, in one of the immunizations, crossreactivity was seen to M49, suggesting that again, in certain cases, antibody crossreactivity may resemble C4BP type-promiscuity [29]. It may be possible to combine the physico-chemical approach [43] with the structure-based approach to identify functionally indispensable sequence patterns [29], for example, those responsible for recruiting C4BP. Antigens possessing such sequence patterns would be expected to elicit a neutralizing response against a broad variety of GAS strains of differing M types. Masking strategies would need to be dealt with, but surrounding conserved sequence patterns with low-information-content sequences may achieve this. An added benefit of an HVR-based vaccine is that the HVR does not elicit autoreactive antibodies [44], which have been a concern for M protein-based vaccines, as the M protein is associated with autoimmune sequelae arising from prolonged streptococcal infection [45]. Further studies on structure- and phylogenetic-based clustering of M protein HVRs may lead to the rational design of an M protein-based GAS vaccine.
Factor H
Although not as well characterized as type-promiscuity in M HVR-C4BP interactions, type-promiscuity also appears to be the mode of factor H (FH) recruitment by M protein HVRs to the GAS surface. Like C4BP, FH is a downregulator of the complement system and composed of CCP domains. FH is a cofactor for the factor I-mediated cleavage of C3b, and promotes the dissociation of the alternative complement pathway C3 convertase (i.e., C3bBb). The conserved C-repeats of M5 and M6 proteins, rather than their variable regions, had been reported to bind FH [46, 47]. However, a thorough re-examination of M protein-FH interactions indicates that FH is bound by HVRs and not the C-repeats [48, 49]. In particular, the M5, M6, and M18 HVRs were found to be sufficient to bind FH [48]. In addition, constructs of M5 and M6 lacking portions of the HVR did not bind FH, whereas those missing the C-repeats continued to bind FH [48]. Furthermore, binding of FH by HVRs is in agreement with the observation that the M5 and M6 HVRs bind FH-like protein 1 (FHL-1) [20, 49], a natural splice variant of FH. This agreement stems from the fact that CCP domain 7 of FH is responsible for binding M proteins [48, 50], and FHL-1 consists of the first seven CCP domains of FH. The FH-binding M5, M6, and M18 proteins belong to clade Y [38], suggesting that C4BP-binding and FH-binding assort to non-overlapping sets of HVRs. The exception to this is the M-like Protein H, which binds both C4BP and FH [39]. C4BP is bound by the Protein H HVR [32], while the portion of Protein H that binds FH has not been determined. Whether FH-binding by HVRs is as widespread as C4BP-binding is not yet clear. Unlike C4BP, there are GAS proteins unrelated to the M protein that have been documented to bind FH, such as Fba [51] and Scl1 [52].
The role of FH in GAS pathogenesis has also been less clear than that of C4BP. Several experiments have suggested little or no role for FH. For example, a GAS strain expressing a truncated M5 that does not bind FH [48] retains resistance to phagocytosis, albeit at a somewhat lower level than wild-type M5 GAS [20]. Since murine FH does not bind the M protein [48], a transgenic mouse line expressing a human-mouse FH chimera that binds the M protein was constructed [48]. In an invasive infection model using an FH-binding GAS strain (i.e., M5), no difference in infection was seen between wild-type and the human-mouse FH transgenic mice [48]. However, countervailing evidence demonstrating a significant role for FH has come from a different mouse transgenic line, in which intact human FH rather than a human-mouse FH chimera was expressed [37]. After intravenous introduction of an FH-binding GAS strain (AP1, which expresses the FH- and C4BP-binding Protein H), 50% of humanized FH transgenic mice died after 8 days as compared to no wild-type mice. Transgenic mice expressing both human FH and human C4BP were even more severely affected by this same GAS strain, which also binds C4BP, with no mice surviving after 8 days.
These recent data from humanized FH transgenic mice [37] support the conclusion that FH-binding is an indispensable function in pathogenesis for some GAS strains, a necessary condition for the evolution of a conserved FH-binding pattern in the M protein HVRs of these strains. However, whether such a pattern exists in M protein HVRs remains an open question.
LL-37 and Histones
A globally disseminated GAS strain of the M1 type has been a leading cause of severe GAS infections during the last several decades [53]. M1 is also the most prevalent M type in the industrialized world [54]. However, the selective pressure maintaining the sequence of the M1 HVR, which binds neither C4BP nor FH, is unclear.
A strong possibility for functional indispensability in the M1 HVR is interaction with the cathelicidin peptide LL-37 [55, 56]. Cathelicidins are cationic antimicrobial peptides that provide a critical first line of innate immune defense against microbial infection [57–59], including by GAS [60]. Cathelicidins are produced by neutrophils, macrophages, mast cells, and keratinocytes along with other epithelial cell types [58–60]. They are synthesized as ~18 kDa precursor proteins which lack antimicrobial activity [61], and are processed by neutrophil proteinase-3 or keratinocyte kallikreins to release mature C-terminal antimicrobial peptides [62–64]. Only one human cathelicidin exists, LL-37 (~4.5 kDa), which is produced from the precursor hCAP-18. LL-37 is α-helical in solution [65] and lipid micelles [66], and functions, like other amphipathic α-helical cationic antimicrobial peptides, by inserting into bacterial plasma membranes and causing bacterial lysis. LL-37 also acts as an immune signaling molecule [67–69], promoting chemotactic responses as well as other activities. LL-37 is also sensed by GAS to elicit changes in virulence factor expression [70].
The GAS M1 strain is highly resistant to killing by LL-37 and neutrophil extracellular traps (NETs) [55], in which LL-37 is abundant. This resistance was shown to be due to the specific interaction between LL-37 and the M1 protein [55, 56]. The M1 HVR along with the downstream, sequence variable A region and B-repeats (Fig. 2a) were found to confer LL-37-binding [55]. The structural details of this interaction are not yet known. Detoxification of LL-37 by the M1 protein is achieved by sequestration of LL-37 into an M1 ‘protein trap’ [56]. This occurs either on the GAS surface by cell wall-attached M1 protein, or in the extracellular space by the released, soluble form of the M1 protein. Sequestration by M1 protein prevents LL-37 from interacting with its target of action, the bacterial plasma membrane [56]. M1 protein also neutralizes the chemotactic properties of LL-37, impairing recruitment of neutrophils to the infection site [56]. In addition, M1 protein binds hCAP-18 and inhibits its maturation to LL-37 [56]. In a murine subcutaneous infection model, the contribution to virulence of the M1 protein was almost entirely attributable to its ability to neutralize the murine ortholog of LL-37, CRAMP [56], which has properties essentially identical to those of LL-37.
LL-37 interaction with M1 GAS was also seen by electron microscopy to lead to release of vesicle-like structures [71]. These vesicle-like structures had LL-37 on their surface, and had proinflammatory effects, causing release of resistin and myeloperoxidase from neutrophils. Concentrated supernatants enriched in the vesicle-like structures contained the M protein, suggesting that the M protein was part of these vesicles.
In a process that appears to be similar to detoxification of LL-37, the N-terminal, sequence variable region of the M1 protein (i.e., HVR, A region, and B-repeats) has also been identified to detoxify histones [72], which are major components of NETs and have antibacterial activity [73]. As with LL-37, the mechanism of histone detoxification appears to be the trapping of histones by the M1 protein, and preventing histones from reaching the bacterial membrane and entering the bacterium.
To date, M1 is the only M protein known to bind and detoxify cathelicidins and histones, but a number of other GAS strains display high levels of resistance against LL-37 [55] and against histones [72]. It is not clear yet whether LL-37-resistance, histone-resistance, or both are conferred by the M protein in these strains, and if so, whether variable regions of the M protein are responsible.
Intricacy in Protein Dynamics
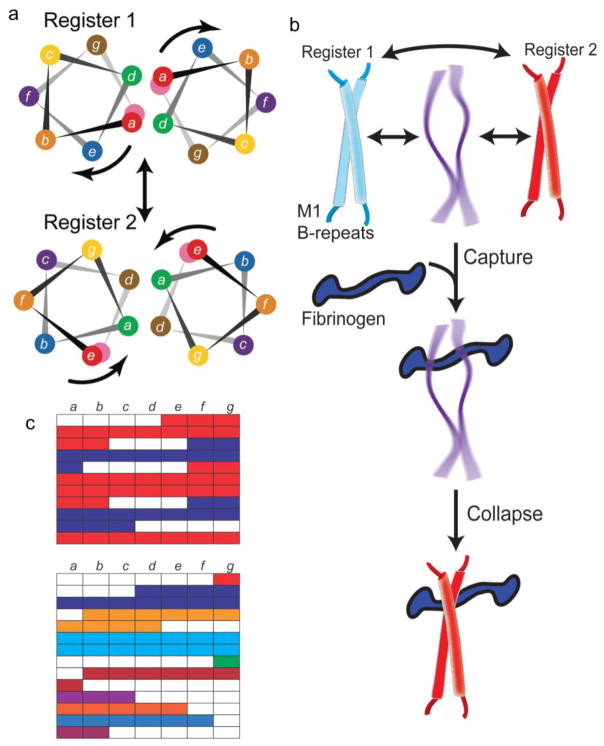
An intricacy in the deceptively simple α-helical, coiled-coil structure of the M protein has been uncovered in the sequence variable B-repeats of the M1 protein [16], which confer binding to fibrinogen (Fg) [74]. Interaction between the M protein and Fg in general protects against phagocytosis [20, 74–76], and in the case of M1, also leads to the formation of a proinflammatory, supramolecular M1-Fg network [15] that is likely involved in toxic shock [11].
The M1 B-repeats are conformationally dynamic and interconvert between two alternate coiled-coil registers, which occupy adjacent helical faces [16] (Fig. 4a). One of the registers, called register 1, is incompatible with Fg-binding [13], while the other, called register 2, corresponds to the conformation observed in the Fg-bound state of the M1 protein [15]. The M1 protein in its free state has been documented to sample both registers 1 and 2 [16]. The B-repeats, in addition, have a dissociated and most likely natively unfolded state [16] (Fig. 4b). The alternate registers and multiple conformations of the B-repeats are due to its highly noncanonical coiled coil sequence [77]. Indeed, the sequence of the M1 B-repeats is a mosaic of short blocks of amino acids that prefer register 1 alternating with short blocks of amino acids that prefer register 2 [15, 77] (Fig. 4c). The M1 B-repeats were mutated such that they contained amino acids ideal for forming a coiled coil entirely in either register 1 or register 2, creating two versions of the M1 protein [13, 16]. Both register 1- and 2-idealized versions of the M1 protein were more stable than the wild-type M1 protein. Unsurprisingly, stabilization of the M1 protein B-repeats in the Fg-nonbinding register 1 resulted in a marked loss of Fg binding [13, 16]. Again unsurprisingly, expression of the register 1-idealized M1 protein on the surface of GAS resulted in a significant decrease in bacterial survival in either a whole blood or neutrophil killing assay [78].
Figure 4. Protein Dynamics in the B-repeats.
a. Helical wheel representation of the M1 B-repeats in register 1 (top) and 2 (bottom). The color coding of residues is preserved in the two registers, and the arrows indicate the rotation of the helical face required to transition from one register to the other. Adapted from Stewart, C.M. et al. (2016) Proc Natl Acad Sci USA 113 (34), 9515–20 [16].
b. Interconversion between multiple conformations in the M1 B-repeats, with the central conformation representing a dissociated and natively unfolded state. The ‘capture-and-collapse’ model of Fg-binding is shown, with the natively unfolded state of the B-repeats capturing Fg, leading to collapse of the B-repeats into register 2.
c. Top, Heptad register predicted for the M1 B-repeats, with register 1 in blue and register 2 in red. Gaps are in white. Bottom, Heptad register predicted for the M5 B-repeats. Contiguous amino acids are colored similarly.
However, surprisingly, stabilization of the M protein B-repeats in the Fg-binding register 2 also resulted in a marked loss of Fg binding and concomitant Fg-dependent proinflammatory activity in whole blood [16]. A key observation that explains this counterintuitive result is that Fg-binding was restored to the register 1- and 2-idealized M1 proteins by chaotropic destabilization of these proteins [16]. This indicates that instability and dynamics in the M1 protein are essential to Fg binding. A ‘capture-and-collapse’ mechanism was suggested to account for this behavior (Fig. 4b). In this mechanism, a dynamic and intrinsically disordered conformation of the M1 protein B-repeats is responsible for the initial capture of Fg. This dynamic conformation in the B-repeats then collapses into a register 2 coiled coil due to stabilization provided by Fg-binding energy [16]. Thus, the amino acids that destabilize the coiled coil but do not directly contact Fg are as important for Fg-binding as those that directly contact Fg.
Fibrinogen has been shown to interact with several other M proteins that have B-repeat regions, including M3, M6, M12, M14, M18, M19, M23, M54, and M57 [38, 79], which all belong to clade Y (as does M1) [38]. As noted above, the B-repeats are variable in sequence and do not share considerable sequence identity. Like the HVR, the B-repeats are weakly immunogenic [27], but unlike the HVR, they do not elicit opsonic antibodies [20]. Thus, the selective pressure driving their variation is unresolved.
The M5 B-repeats are the only B-repeats besides the M1 B-repeats shown to bind Fg [74]. The M5 B-repeats (2.4 repeats, 25 amino acids) are only 25% identical in sequence to the M1 B-repeats (2.2 repeats, 28 amino acids). While the M1 and M5 B-repeats bind different sites on Fg [74], the M5 B-repeats share with the M1 B-repeats a highly noncanonical coiled-coil sequence. The M5 B-repeats also appear to be composed of a mosaic of multiple registers (Fig. 4c). However, the M5 B-repeats appear to be even more complex than the M1 B-repeats. Rather than the alternating three or four amino acid gaps diagnostic of two competing registers in the M1 B-repeats, the M5 B-repeats have no clear pattern of alternating gaps. The noncanonical nature of the M5 B-repeats suggest that the ‘capture-and-collapse’ mechanism of Fg-binding may hold true for the M5 B-repeats as well. Less detailed information is known about other Fg-binding M proteins. To date, all Fg-binding M proteins have bound fibrinogen fragment D (FgD) [38, 74, 79], and the interaction between various M proteins and FgD has been noted to have complex binding and dissociation kinetics [79], which is consistent with a ‘capture-and-collapse’ mechanism. Further experiments are needed to determine the generality of this mechanism for M protein-Fg interactions.
The M1 B-repeats have also been shown to bind the human A, B, and H blood antigens, which are glycans, with the greatest affinity displayed to the H-antigen [80]. Blood antigen- and Fg-binding are specified by different amino acids in the M1 B-repeats [15]. Intriguingly, the register 1-idealized version of the M1 B-repeats displayed reduced binding to the H-antigen. This suggests that protein dynamics may also be important for blood group antigen interaction, or that register 2 or another conformation in the M1 B-repeats is important. The interaction between the M1 protein and the H-antigen was found to enhance binding to and invasion of H1-antigen-expressing human buccal epithelial cells [80]. This increased M1-dependent adhesion contrasts with a report that found decreased adhesion to human pharyngeal keratinocytes due to the M1 protein, with the decrease in adhesion enhanced by Fg [81]. These keratinocytes presumably lacked the H-antigen.
The M1 B-repeats have recently been shown to be involved in triggering pyroptosis in macrophages [82]. This activity was independent of Fg. The M1 protein was shown to activate the NLRP3 inflammasome, leading to activation of caspase-1 and consequent maturation and release of the proinflammatory cytokine IL-1β. This process required clathrin-mediated endocytosis of the released, soluble form of the M1 protein. Soluble M1 protein was also shown to trigger IL-1β activation and release in vivo, when introduced intraperitoneally into mice. The host factor responsible for recognition of the M1 B-repeats is currently unknown.
The M1 B-repeats have an unusual multiplicity of functions, making them a promising target for intervention. A complex relationship exists between sequence and structure in this region, belying the simplicity of the coiled coil. An ideal coiled coil is structurally regular and thus capable of sampling a very narrow conformational space, as opposed to a dynamic coiled coil which is able to sample a considerably wider conformational space. The multiplicity of functions in the M1 B-repeats would seem to demand a multiplicity of conformations rather than a regular structure. In addition, the multiplicity of conformations may also serve a masking function in creating a moving target for immune recognition.
Concluding Remarks
The antigenic variability in the M protein has been an obstacle to the development of a GAS vaccine. However, recent work has uncovered sequence patterns that are conserved in certain M types for recruiting C4BP to the GAS surface. This conservation is hidden in plain sight by the variability of surrounding sequences. Suggestive evidence exists that these conserved sequence patterns can act as antibody epitopes, resulting in an antibody response against multiple M types. A better understanding of the interactions of other host factors with the variable regions of the M protein (see Outstanding Questions) has the potential to guide the design of a broadly neutralizing GAS vaccine.
Outstanding Questions Box.
Are there additional conserved sequence patterns in M protein HVRs for binding C4BP?
Can the type-promiscuity of C4BP be mimicked in antibodies?
How many M protein HVRs bind FH?
Is there a conserved FH-binding pattern hidden in M protein variability?
Do other M proteins besides M1 bind and detoxify LL-37 in a ‘protein trap’?
Is there a conserved LL-37-binding pattern hidden in M protein variability?
Do other M proteins besides M1 bind and detoxify histones in a ‘protein trap’?
Is there a conserved histone-binding pattern hidden in M protein variability?
Is Fg binding generally conserved in M protein B-repeats?
What drives sequence variation in M protein B-repeats?
Is the ‘capture-and-collapse’ mechanism general to the interaction of M proteins with Fg?
Do the B-repeats of M proteins besides M1 trigger pyroptosis in macrophages?
What is the host factor that recognizes the M1 B-repeats in triggering macrophage phagocytosis?
Trends Box.
Recruitment of C4BP by multiple M types occurs through conserved sequence patterns that are hidden within M protein variability.
Phylogenetic analysis offers a way to group the M protein into two broad clades, and clusters within those clades.
Like C4BP, FH is recruited by M protein variable sequences, but the number of M types that bind FH is unclear as are the sequence patterns underlying the recruitment.
The M1 protein variable region detoxifies the antimicrobial peptide LL-37 through a ‘protein trap.’ The M1 protein variable region similarly detoxifies the antibacterial activity of histones.
Instability and conformational dynamics in the M1 B-repeats are required for recruitment of fibrinogen.
The M1 B-repeats have a multiplicity of functions, including binding glycans belonging to blood group antigens and triggering pyroptosis in macrophages.
Acknowledgments
This works was supported by NIH grant R01 AI096837. I thank the members of the lab for comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16(6):571–6. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lingwood D, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489(7417):566–70. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouvinski A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520(7545):109–13. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 4.Lancefield RC. The Antigenic Complex of Streptococcus haemolyticus: Ii. Chemical and Immunological Properties of the Protein Fractions. J Exp Med. 1928;47(3):469–80. doi: 10.1084/jem.47.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todd EW, Lancefield RC. Variants of Hemolytic Streptococci; Their Relation to Type-Specific Substance, Virulence, and Toxin. J Exp Med. 1928;48(6):751–767. doi: 10.1084/jem.48.6.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carapetis JR, et al. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5(11):685–94. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 7.Cole JN, et al. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol. 2011;9(10):724–36. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 8.Walker MJ, et al. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev. 2014;27(2):264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips GN, Jr, et al. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci U S A. 1981;78(8):4689–93. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akesson P, et al. M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem J. 1994;300(Pt 3):877–86. doi: 10.1042/bj3000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herwald H, et al. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 2004;116(3):367–79. doi: 10.1016/s0092-8674(04)00057-1. [DOI] [PubMed] [Google Scholar]
- 12.Manjula BN, et al. Presence of two distinct regions in the coiled-coil structure of the streptococcal Pep M5 protein: relationship to mammalian coiled-coil proteins and implications to its biological properties. Proc Natl Acad Sci U S A. 1985;82(4):1064–8. doi: 10.1073/pnas.82.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNamara C, et al. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 2008;319(5868):1405–8. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andre I, et al. Streptococcal M protein: structural studies of the hypervariable region, free and bound to human C4BP. Biochemistry. 2006;45(14):4559–68. doi: 10.1021/bi052455c. [DOI] [PubMed] [Google Scholar]
- 15.Macheboeuf P, et al. Streptococcal M1 protein constructs a pathological host fibrinogen network. Nature. 2011;472(7341):64–8. doi: 10.1038/nature09967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart CM, et al. Coiled-coil destabilizing residues in the group A Streptococcus M1 protein are required for functional interaction. Proc Natl Acad Sci U S A. 2016;113(34):9515–20. doi: 10.1073/pnas.1606160113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMillan DJ, et al. Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study. Clin Microbiol Infect. 2013;19(5):E222–9. doi: 10.1111/1469-0691.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Facklam RF, et al. Extension of the Lancefield classification for group A streptococci by addition of 22 new M protein gene sequence types from clinical isolates: emm103 to emm124. Clin Infect Dis. 2002;34(1):28–38. doi: 10.1086/324621. [DOI] [PubMed] [Google Scholar]
- 19.Smeesters PR, et al. The streptococcal M protein: a highly versatile molecule. Trends Microbiol. 2010;18(6):275–82. doi: 10.1016/j.tim.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Sandin C, et al. Binding of human plasma proteins to Streptococcus pyogenes M protein determines the location of opsonic and non-opsonic epitopes. Mol Microbiol. 2006;59(1):20–30. doi: 10.1111/j.1365-2958.2005.04913.x. [DOI] [PubMed] [Google Scholar]
- 21.Lannergard J, et al. The hypervariable region of Streptococcus pyogenes M protein escapes antibody attack by antigenic variation and weak immunogenicity. Cell Host Microbe. 2011;10(2):147–57. doi: 10.1016/j.chom.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Jones KF, Fischetti VA. The importance of the location of antibody binding on the M6 protein for opsonization and phagocytosis of group A M6 streptococci. J Exp Med. 1988;167(3):1114–23. doi: 10.1084/jem.167.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey M, et al. Streptococcal Immunity Is Constrained by Lack of Immunological Memory following a Single Episode of Pyoderma. PLoS Pathog. 2016;12(12):e1006122. doi: 10.1371/journal.ppat.1006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penfound TA, et al. Protective efficacy of group A streptococcal vaccines containing type-specific and conserved M protein epitopes. Vaccine. 2010;28(31):5017–22. doi: 10.1016/j.vaccine.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dale JB, et al. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine. 2011;29(46):8175–8. doi: 10.1016/j.vaccine.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dale JB, et al. Potential coverage of a multivalent M protein-based group A streptococcal vaccine. Vaccine. 2013;31(12):1576–81. doi: 10.1016/j.vaccine.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lannergard J, et al. Sequence variability is correlated with weak immunogenicity in Streptococcus pyogenes M protein. Microbiologyopen. 2015;4(5):774–89. doi: 10.1002/mbo3.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stalhammar-Carlemalm M, et al. Nonimmunodominant regions are effective as building blocks in a streptococcal fusion protein vaccine. Cell Host Microbe. 2007;2(6):427–34. doi: 10.1016/j.chom.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Buffalo CZ, et al. Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein. Nat Microbiol. 2016;1:16155. doi: 10.1038/nmicrobiol.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gigli I, et al. Modulation of the classical pathway C3 convertase by plasma proteins C4 binding protein and C3b inactivator. Proc Natl Acad Sci U S A. 1979;76(12):6596–600. doi: 10.1073/pnas.76.12.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Persson J, et al. Extreme sequence divergence but conserved ligand-binding specificity in Streptococcus pyogenes M protein. PLoS Pathog. 2006;2(5):e47. doi: 10.1371/journal.ppat.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morfeldt E, et al. Isolated hypervariable regions derived from streptococcal M proteins specifically bind human C4b-binding protein: implications for antigenic variation. J Immunol. 2001;167(7):3870–7. doi: 10.4049/jimmunol.167.7.3870. [DOI] [PubMed] [Google Scholar]
- 33.Blom AM, et al. Human C4b-binding protein has overlapping, but not identical, binding sites for C4b and streptococcal M proteins. J Immunol. 2000;164(10):5328–36. doi: 10.4049/jimmunol.164.10.5328. [DOI] [PubMed] [Google Scholar]
- 34.Accardo P, et al. Binding of human complement component C4b-binding protein (C4BP) to Streptococcus pyogenes involves the C4b-binding site. J Immunol. 1996;157(11):4935–9. [PubMed] [Google Scholar]
- 35.Carlsson F, et al. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J Exp Med. 2003;198(7):1057–68. doi: 10.1084/jem.20030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berggard K, et al. Binding of human C4BP to the hypervariable region of M protein: a molecular mechanism of phagocytosis resistance in Streptococcus pyogenes. Mol Microbiol. 2001;42(2):539–51. doi: 10.1046/j.1365-2958.2001.02664.x. [DOI] [PubMed] [Google Scholar]
- 37.Ermert D, et al. Virulence of group A Streptococci is enhanced by human complement inhibitors. PLoS Pathog. 2015;11(7):e1005043. doi: 10.1371/journal.ppat.1005043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanderson-Smith M, et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis. 2014;210(8):1325–38. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ermert D, et al. Binding of complement inhibitor C4b-binding protein to a highly virulent Streptococcus pyogenes M1 strain is mediated by protein H and enhances adhesion to and invasion of endothelial cells. J Biol Chem. 2013;288(45):32172–83. doi: 10.1074/jbc.M113.502955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnsson E, et al. A highly variable region in members of the streptococcal M protein family binds the human complement regulator C4BP. J Immunol. 1996;157(7):3021–9. [PubMed] [Google Scholar]
- 41.Perez-Caballero D, et al. Interaction between complement regulators and Streptococcus pyogenes: binding of C4b-binding protein and factor H/factor H-like protein 1 to M18 strains involves two different cell surface molecules. J Immunol. 2004;173(11):6899–904. doi: 10.4049/jimmunol.173.11.6899. [DOI] [PubMed] [Google Scholar]
- 42.Birtalan S, et al. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377(5):1518–28. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 43.Dale JB, et al. Structure-based design of broadly protective group a streptococcal M protein-based vaccines. Vaccine. 2017;35(1):19–26. doi: 10.1016/j.vaccine.2016.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNeil SA, et al. Safety and immunogenicity of 26-valent group a Streptococcus vaccine in healthy adult volunteers. Clin Infect Dis. 2005;41(8):1114–22. doi: 10.1086/444458. [DOI] [PubMed] [Google Scholar]
- 45.Carapetis JR, et al. Acute rheumatic fever and rheumatic heart disease. Nat Rev Dis Primers. 2016;2:15084. doi: 10.1038/nrdp.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Casal J, et al. Role of the conserved C-repeat region of the M protein of Streptococcus pyogenes. Mol Microbiol. 1995;15(5):907–16. doi: 10.1111/j.1365-2958.1995.tb02360.x. [DOI] [PubMed] [Google Scholar]
- 47.Fischetti VA, et al. Location of the complement factor H binding site on streptococcal M6 protein. Infect Immun. 1995;63(1):149–53. doi: 10.1128/iai.63.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gustafsson MC, et al. Factor H binds to the hypervariable region of many Streptococcus pyogenes M proteins but does not promote phagocytosis resistance or acute virulence. PLoS Pathog. 2013;9(4):e1003323. doi: 10.1371/journal.ppat.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnsson E, et al. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J Immunol. 1998;161(9):4894–901. [PubMed] [Google Scholar]
- 50.Blackmore TK, et al. M protein of the group A Streptococcus binds to the seventh short consensus repeat of human complement factor H. Infect Immun. 1998;66(4):1427–31. doi: 10.1128/iai.66.4.1427-1431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandiripally V, et al. Acquisition of regulators of complement activation by Streptococcus pyogenes serotype M1. Infect Immun. 2002;70(11):6206–14. doi: 10.1128/IAI.70.11.6206-6214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reuter M, et al. Binding of the human complement regulators CFHR1 and factor H by streptococcal collagen-like protein 1 (Scl1) via their conserved C termini allows control of the complement cascade at multiple levels. J Biol Chem. 2010;285(49):38473–85. doi: 10.1074/jbc.M110.143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu L, et al. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest. 2015;125(9):3545–59. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steer AC, et al. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9(10):611–6. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 55.Lauth X, et al. M1 protein allows Group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1(3):202–14. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LaRock CN, et al. Group A Streptococcal M1 Protein Sequesters Cathelicidin to Evade Innate Immune Killing. Cell Host Microbe. 2015;18(4):471–7. doi: 10.1016/j.chom.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dorschner RA, et al. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Dermatol. 2001;117(1):91–7. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 58.Wong JH, et al. Cathelicidins: peptides with antimicrobial, immunomodulatory, anti-inflammatory, angiogenic, anticancer and procancer activities. Curr Protein Pept Sci. 2013;14(6):504–14. doi: 10.2174/13892037113149990067. [DOI] [PubMed] [Google Scholar]
- 59.LaRock CN, Nizet V. Cationic antimicrobial peptide resistance mechanisms of streptococcal pathogens. Biochim Biophys Acta. 2015;1848(11 Pt B):3047–54. doi: 10.1016/j.bbamem.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nizet V, et al. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414(6862):454–7. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 61.Zaiou M, et al. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J Invest Dermatol. 2003;120(5):810–6. doi: 10.1046/j.1523-1747.2003.12132.x. [DOI] [PubMed] [Google Scholar]
- 62.Murakami M, et al. Postsecretory processing generates multiple cathelicidins for enhanced topical antimicrobial defense. J Immunol. 2004;172(5):3070–7. doi: 10.4049/jimmunol.172.5.3070. [DOI] [PubMed] [Google Scholar]
- 63.Yamasaki K, et al. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 2006;20(12):2068–80. doi: 10.1096/fj.06-6075com. [DOI] [PubMed] [Google Scholar]
- 64.Sorensen O, et al. The human antibacterial cathelicidin, hCAP-18, is bound to lipoproteins in plasma. J Biol Chem. 1999;274(32):22445–51. doi: 10.1074/jbc.274.32.22445. [DOI] [PubMed] [Google Scholar]
- 65.Johansson J, et al. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J Biol Chem. 1998;273(6):3718–24. doi: 10.1074/jbc.273.6.3718. [DOI] [PubMed] [Google Scholar]
- 66.Wang G. Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J Biol Chem. 2008;283(47):32637–43. doi: 10.1074/jbc.M805533200. [DOI] [PubMed] [Google Scholar]
- 67.Yamasaki K, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat Med. 2007;13(8):975–80. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- 68.Alalwani SM, et al. The antimicrobial peptide LL-37 modulates the inflammatory and host defense response of human neutrophils. Eur J Immunol. 2010;40(4):1118–26. doi: 10.1002/eji.200939275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Y, et al. LL-37, the neutrophil granule- and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J Exp Med. 2000;192(7):1069–74. doi: 10.1084/jem.192.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Velarde JJ, et al. The human antimicrobial peptide LL-37 binds directly to CsrS, a sensor histidine kinase of group A Streptococcus, to activate expression of virulence factors. J Biol Chem. 2014;289(52):36315–24. doi: 10.1074/jbc.M114.605394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uhlmann J, et al. LL-37 Triggers Formation of Streptococcus pyogenes Extracellular Vesicle-Like Structures with Immune Stimulatory Properties. J Innate Immun. 2016;8(3):243–57. doi: 10.1159/000441896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dohrmann S, et al. Group A Streptococcal M1 Protein Provides Resistance against the Antimicrobial Activity of Histones. Sci Rep. 2017;7:43039. doi: 10.1038/srep43039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirsch JG. Bactericidal action of histone. J Exp Med. 1958;108(6):925–44. doi: 10.1084/jem.108.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ringdahl U, et al. A role for the fibrinogen-binding regions of streptococcal M proteins in phagocytosis resistance. Mol Microbiol. 2000;37(6):1318–26. doi: 10.1046/j.1365-2958.2000.02062.x. [DOI] [PubMed] [Google Scholar]
- 75.Waldemarsson J, et al. Functional dissection of Streptococcus pyogenes M5 protein: the hypervariable region is essential for virulence. PLoS One. 2009;4(10):e7279. doi: 10.1371/journal.pone.0007279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitnack E, Beachey EH. Antiopsonic activity of fibrinogen bound to M protein on the surface of group A streptococci. J Clin Invest. 1982;69(4):1042–5. doi: 10.1172/JCI110508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lupas A, et al. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 78.Uchiyama S, et al. Coiled-coil irregularities of the M1 protein structure promote M1-fibrinogen interaction and influence group A Streptococcus host cell interactions and virulence. J Mol Med (Berl) 2013;91(7):861–9. doi: 10.1007/s00109-013-1012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Glinton K, et al. Variable region in streptococcal M-proteins provides stable binding with host fibrinogen for plasminogen-mediated bacterial invasion. J Biol Chem. 2017;292(16):6775–6785. doi: 10.1074/jbc.M116.768937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Oliveira DM, et al. Blood Group Antigen Recognition via the Group A Streptococcal M Protein Mediates Host Colonization. MBio. 2017;8(1):e02237–16. doi: 10.1128/mBio.02237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anderson EL, et al. The fibrinogen-binding M1 protein reduces pharyngeal cell adherence and colonization phenotypes of M1T1 group A Streptococcus. J Biol Chem. 2014;289(6):3539–46. doi: 10.1074/jbc.M113.529537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valderrama JA, et al. Group A Streptococcal M Protein Activates the NLRP3 Inflammasome. Nat Microbiol. 2017 doi: 10.1038/s41564-017-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]