Abstract
In this study, we have characterized a novel set of extracellular enzymes produced by Penicillium chrysogenum strain HKF2. A draft genome data of 31.5 Mbp was generated and annotation suggested a total of 11,243 protein-coding genes out of which 609 were CAZymes, majority of which were found to have homology with Penicillium rubens, Penicillium chrysogenum followed by Penicillium expansum and Penicillium roqueforti. The prominent CAZyme genes identified in the draft genome encoded for enzymes involved in the production of prebiotics such as inulo-oligosaccharides and fructo-oligosaccharides. Corresponding enzyme assay indicated that the isolate possessed the potential to produce 11.8 and 3.8 U/mL of β-fructofuranosidase and inulinase, respectively. This study highlights the significance of Effluent Treatment Plants as novel and under-explored niche for isolation of fungi having the potential for production of prebiotics synthesizing enzymes.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1132-3) contains supplementary material, which is available to authorized users.
Keywords: Prebiotic, Penicillium, CAZymes, Draft genome
Prebiotics are non-digestible food ingredients which selectively promote the growth and activity of beneficial bacteria in the colon, thereby positively improving the health status of the host (Patel and Goyal 2012; Gujar et al. 2018). Popular prebiotics which include inulo-oligosaccharides (IOS), fructo-oligosaccharides (FOS), chito-oligosaccharides (COS), isomalto-oligosaccharides (IMO), and galacto-oligosaccharides (GOS) have gained much attention in pharmaceutical and food sector (Patel and Goyal 2011). Penicillium sp. is known to produce range of various carbohydrate active enzymes (CAZymes), e.g., exoinulinase (EC 3.2.1.80), β-fructofuranosidase (EC 3.2.1.26), endochitinase (EC 3.2.1.14), α-glucosidase (EC 3.2.1.20) and β-galactosidase (EC 3.2.1.23) which are used for industrial production of IOS, FOS, COS, IMO and GOS, respectively (Gyo et al. 2009; Park and Oh 2010; Prata et al. 2010; Patel and Goyal 2011; Singh and Shukla 2012; Schafhauser et al. 2015; Saqib et al. 2017).
Penicillium chrysogenum strain HKF2 was isolated from activated sludge samples from effluent treatment plant (ETP) located at Ankaleshwar in Gujarat State of India (Deshmukh et al. 2014). The details of the isolation have been provided in Table S1. In the present study, the strain was screened for production of extracellular β-fructofuranosidase and inulinase enzymes. Submerged fermentation was carried out for production of the two enzymes (Prata et al. 2010; Trivedi et al. 2012). Samples taken at a regular interval of 24 h were used as the source of crude enzyme, while the enzyme activities were determined as per the protocols developed by Sangeetha et al. (2004) and Trivedi et al. (2012). One unit of FFase activity was defined as the amount of enzyme required to produce 1 µmol of glucose per minute at 55 °C with 55% sucrose at pH 5.5 (Sangeetha et al. 2004). One unit of inulinase activity was defined as the amount of enzyme necessary to release one micromole of fructose per minute at 60 °C with 1% inulin at pH 5 (Trivedi et al. 2012). Production of FFase and inulinase by the fungal isolate is shown in Fig. S1. Increase in FFase activity was observed which reached a peak in 72 h (11.8 U/mL) with further incubation resulting in decrease in enzyme activity. The corresponding inulinase activity was also found to increase gradually to 1 U/mL in 48 h followed by a rapid increase to maximum level of 3.8 U/mL in 96 h.
For understanding the genetic potential of P. chrysogenum strain HKF2, whole-genome sequencing approach was used for mining of genes of various prebiotics synthesizing enzymes present in the genome. Genomic DNA was isolated with FastDNA SPIN Kit 116540600 (MP Biomedicals, USA) followed by preparation of paired-end and mate-pair sequencing libraries with mean sizes 639 and 679 bp, respectively. The libraries were sequenced (2 × 150 bp) on Illumina HiSeq 2500 platform. High-quality reads generated were used for de novo assembly using Soapdenovo2 assembler (Luo et al. 2012; He et al. 2017) resulting in a draft genome of 31.5 Mbp containing 296 contigs and 94 scaffolds (Table 1). Genome–genome comparison between P. chrysogenum strain HKF2 and closest matching P. chrysogenum strain P2niaD18 performed using MAUVE v.2.4.0 software (Darling et al. 2004) is shown in Fig. S2.
Table 1.
Features of draft genome sequence of Penicillium chrysogenum strain HKF2
| Features | Penicillium chrysogenum strain HKF2 draft genome |
|---|---|
| No. of reads | 42,170,766 |
| Pair end read length | 150 bp |
| Genome assembly size | 31,484,772 |
| Number of contigs | 6194 |
| Number of scaffolds | 94 |
| Contig N50 | 10,874 |
| Gaps between scaffolds | 0 |
| Scaffold N50 | 3,449,782 |
| No. of gene model | 11,243 |
| No. of exon per gene | 3.15 |
| Mean protein length | 476.36 |
| GC content | 53.21% |
| No. of tRNA | 188 |
| NR annotated | 11,172 |
| SwissProt annotated | 7580 |
| KEGG annotated | 2128 |
| KOG annotated | 6003 |
| Pfam annotated | 8174 |
| CAZymes | 609 |
The repeat sequences were masked using RepeatMasker and the RepBase library, and a total of 588,673 bp (1.87%) were masked, and the repeats masked assembly was used for coding gene prediction. Coding sequences (CDS) in the genome were predicted by GeneMarkES (Borodovsky and Lomsadze 2011) which resulted in a total of 11,243 protein-coding genes. For the predicted CDS, a similarity search was done against NCBI’s non-redundant (nr) database using the BLASTP algorithm. Top-hit distribution proteins revealed that majority of the hits were found to have homology with P. rubens, P. chrysogenum followed by P. expansum and P. roqueforti. All proteins were also searched for similarity against Swiss-Prot, KOG and Pfam databases using BLASTP, rpsblast and Hmmscan algorithms (via webMGA). Gene Ontology (GO) annotation was obtained for nr database annotated proteins using Blast2GO Pro and those with similar functions were allotted to the same GO functional groups (Götz et al. 2008). Analysis of GO sequence distributions revealed the presence of all three GO domains, i.e., Biological Processes (3980), Molecular Function (3349) and Cellular Components (3946). KEGG automatic annotation server (KAAS) was used for assigning orthologs and mapping of proteins to the biological pathways which were then compared against the KEGG database using BLASTP. CAZymes analysis was carried out through dbCAN and all data generated in dbCAN were based on the family classification from CAZy database (Yin et al. 2012).
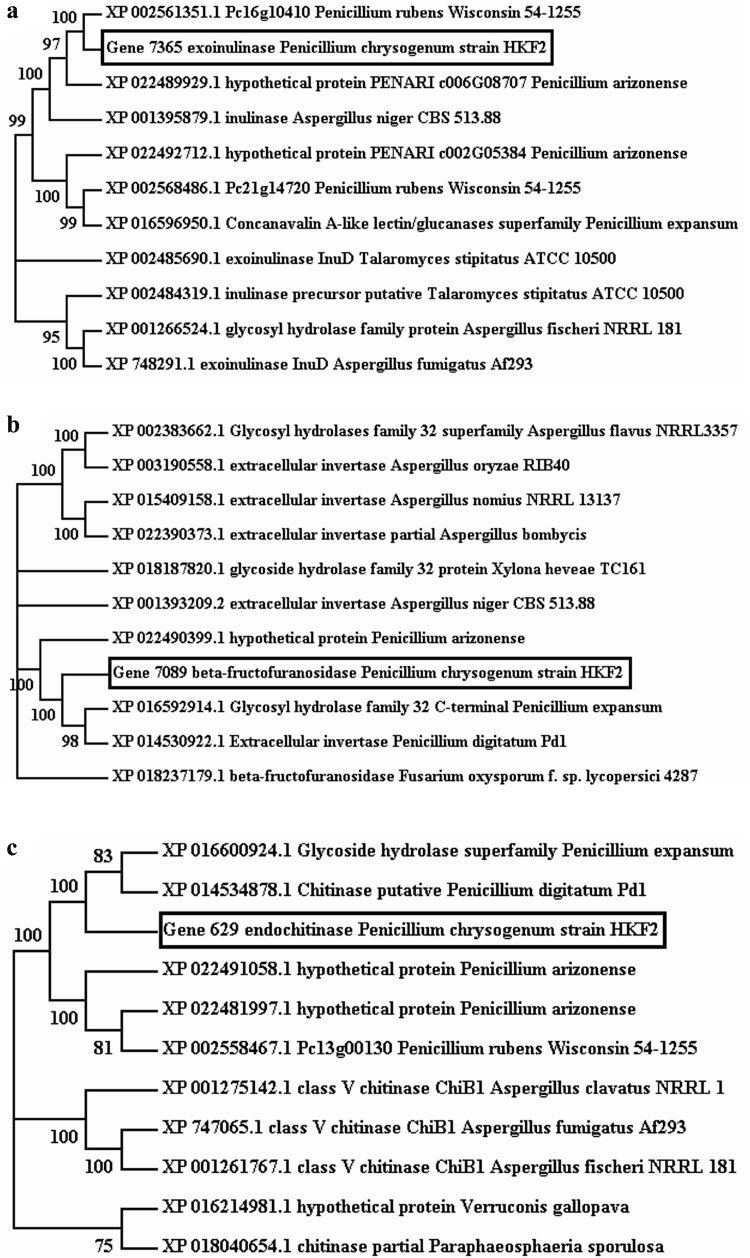
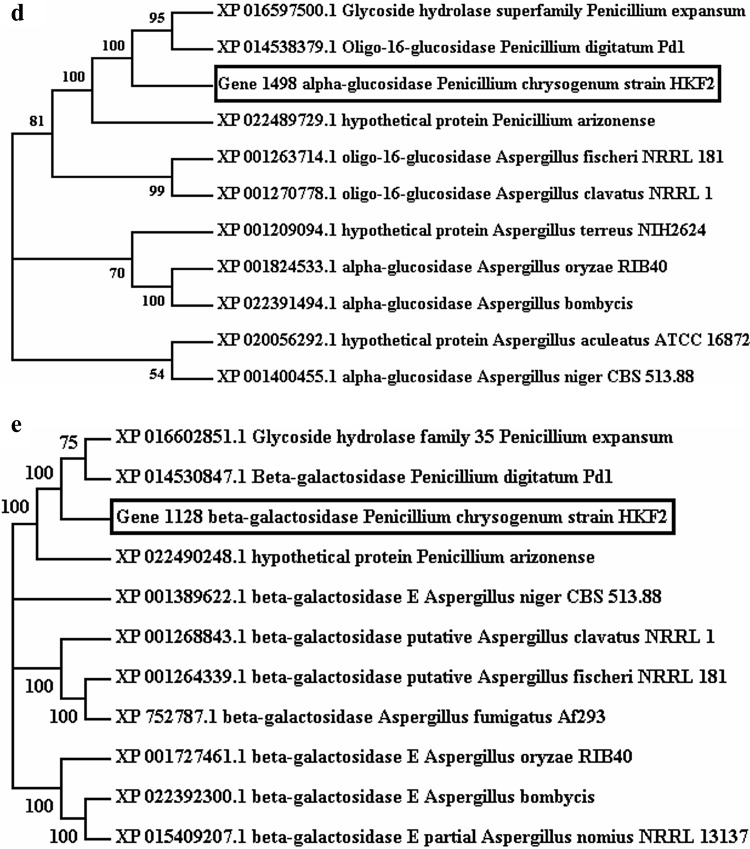
Gene annotation revealed the presence of genes coding for various prebiotic synthesizing enzymes in P. chrysogenum strain HKF2 genome, which were labeled with locus id/tag numbers viz., gene 7365 (exoinulinase), gene 7089 (β-fructofuranosidase), gene 629 (endochitinase), gene 1498 (α-glucosidase) and gene 1128 (β-galactosidase). The amino acid sequence of predicted genes was searched for similarity against reference proteins (refseq_proteins) database using BLASTP (version 2.7.1), and top 10 hits were chosen to study phylogeny of respective enzymes. Sequences were aligned using ClustalW interface in MEGA 7 based on the neighbor joining method, and bootstrap values were based on 1000 replicates (Fig. 1). The nearest neighbor analysis of exoinulinase, β-fructofuranosidase, endochitinase, α-glucosidase and β-galactosidase showed the diversification associated with strain which allowed the survival in such extreme environments as existing in ETPs. Exoinulinase (gene 7365) exhibited 95% similarity with P. rubens strain wisconsin 54-1255 exoinulinase. β-fructofuranosidase (gene 7089) displayed 86% similarity with β-fructofuranosidase of P. expansum isolated from infected apple (Accession Number XP_016592914.1) (Ballester et al. 2015). The genes, gene 629 (endochitinase), gene 1498 (α-glucosidase), and gene 1128 (β-galactosidase) showed 87, 93 and 86% identities, respectively, with corresponding enzymes of P. digitatum Pd1 which was isolated from infected grapefruit (Accession Numbers XP_014534878.1, XP_014538379.1, XP_014530847.1) (Marcet-Houben et al. 2012). Comparative genome analysis was carried out to determine the novelty of extracellular CAZymes secreted by this strain needs further exploration for identifying their role in utilization of complex polysaccharides.
Fig. 1.
Neighbor Joining Tree calculated by ClustalW for a Gene 7365 exoinulinase, b Gene 7089 β-fructofuranosidase, c Gene 629 endochitinase, d Gene 1498 α-glucosidase, e Gene 1128 β-galactosidase from Penicillium chrysogenum strain HKF2 constructed by MEGA 7. Numbers displayed on the branches are the bootstrap support obtained through 1000 replications
Nucleotide sequence accession numbers This Whole Genome Shotgun project has been deposited in DDBJ/ENA/GenBank under the accession number MUXA00000000. The version described here is MUXA01000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge the Director, CSIR-NEERI for providing essential resources for the research work [KRC No. CSIR-NEERI/KRC/2017/DEC/EBGD/1]. Vaibhav Gujar is thankful to University Grants Commission (UGC), New Delhi for providing Junior and Senior Research Fellowship for carrying out this research.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1132-3) contains supplementary material, which is available to authorized users.
References
- Ballester A, Marcet-houben M, Levin E, et al. Genome, transcriptome, and functional analyses of Penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol Plant Microbe Interact. 2015;28:232–248. doi: 10.1094/MPMI-09-14-0261-FI. [DOI] [PubMed] [Google Scholar]
- Borodovsky M, Lomsadze A. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinform. 2011 doi: 10.1002/0471250953.bi0406s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R, Mathew A, Purohit HJ. Characterization of antibacterial activity of bikaverin from Fusarium sp. HKF15. J Biosci Bioeng. 2014;117:443–448. doi: 10.1016/j.jbiosc.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Götz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujar VV, Fuke P, Khardenavis AA, Purohit HJ. Annotation and de novo sequence characterization of extracellular β-fructofuranosidase from Penicillium chrysogenum strain HKF42. Indian J Microbiol. 2018 doi: 10.1007/s12088-017-0704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyo Y, Chung K, Gon S, et al. Purification and properties of a chitinase from Penicillium sp. LYG 0704. Protein Expr Purif. 2009;65:244–250. doi: 10.1016/j.pep.2008.12.004. [DOI] [PubMed] [Google Scholar]
- He R, Bai X, Cai P, et al. Genome sequence of Talaromyces piceus 9–3 provides insights into lignocellulose degradation. 3 Biotech. 2017;7:368. doi: 10.1007/s13205-017-1001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Ballester A-R, de la Fuente B, et al. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genom. 2012;13:646. doi: 10.1186/1471-2164-13-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A-R, Oh D-K. Galacto-oligosaccharide production using microbial β-galactosidase: current state and perspectives. Appl Microbiol Biotechnol. 2010;85:1279–1286. doi: 10.1007/s00253-009-2356-2. [DOI] [PubMed] [Google Scholar]
- Patel S, Goyal A. Functional oligosaccharides: production, properties and applications. World J Microbiol Biotechnol. 2011;27:1119–1128. doi: 10.1007/s11274-010-0558-5. [DOI] [Google Scholar]
- Patel S, Goyal A. The current trends and future perspectives of prebiotics research: a review. 3 Biotech. 2012;2:115–125. doi: 10.1007/s13205-012-0044-x. [DOI] [Google Scholar]
- Prata MB, Mussatto SI, Rodrigues LR, Teixeira JA. Fructooligosaccharide production by Penicillium expansum. Biotechnol Lett. 2010;32:837–840. doi: 10.1007/s10529-010-0231-y. [DOI] [PubMed] [Google Scholar]
- Sangeetha PT, Ramesh MN, Prapulla SG. Production of fructo-oligosaccharides by fructosyl transferase from Aspergillus oryzae CFR 202 and Aureobasidium pullulans CFR 77. Process Biochem. 2004;39:755–760. doi: 10.1016/S0032-9592(03)00186-9. [DOI] [Google Scholar]
- Saqib S, Akram A, Halim SA, Tassaduq R. Sources of β-galactosidase and its applications in food industry. 3 Biotech. 2017;7:1–7. doi: 10.1007/s13205-017-0645-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafhauser T, Wibberg D, Rückert C, et al. Draft genome sequence of Talaromyces islandicus (“Penicillium islandicum”) WF-38-12, a neglected mold with significant biotechnological potential. J Biotechnol. 2015;211:101–102. doi: 10.1016/j.jbiotec.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Singh PK, Shukla P. Molecular modeling and docking of microbial inulinases towards perceptive enzyme-substrate interactions. Indian J Microbiol. 2012;52:373–380. doi: 10.1007/s12088-012-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi S, Divecha J, Shah A. Optimization of inulinase production by a newly isolated Aspergillus tubingensis CR16 using low cost substrates. Carbohydr Polym. 2012;90:483–490. doi: 10.1016/j.carbpol.2012.05.068. [DOI] [PubMed] [Google Scholar]
- Yin Y, Mao X, Yang J, et al. DbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:445–451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.