Abstract
Multiple sclerosis (MS) is the prototypic complex disease, in which both genes and the environment contribute to its pathogenesis. To date, > 200 independent loci across the genome have been associated with MS risk. However, these only explain a fraction of the total phenotypic variance, suggesting the possible presence of additional genetic factors, and, most likely, also environmental factors. New DNA sequencing technologies have enabled the sequencing of all kinds of microorganisms, including those living in and around humans (i.e., microbiomes). The study of bacterial populations inhabiting the gut is of particular interest in autoimmune diseases owing to their key role in shaping immune responses. In this review, we address the potential crosstalk between B cells and the gut microbiota, a relevant scenario in light of recently approved anti-B-cell therapies for MS. In addition, we review recent efforts to characterize the gut microbiome in patients with MS and discuss potential challenges and future opportunities. Finally, we describe the international MS microbiome study, a multicenter effort to study a large population of patients with MS and their healthy household partners to define the core MS microbiome, how it is shaped by disease-modifying therapies, and to explore potential therapeutic interventions.
Key Words: Gut microbiome, multiple sclerosis, dysbiosis, progression, genetics, B cells, consortia
Introduction
Multiple sclerosis (MS) is an autoimmune disease of the central nervous system characterized by demyelination and axonal loss resulting in characteristic lesions in the brain and spinal cord [1]. MS is thought to result from the loss of tolerance of pre-existing myelin autoreactive T cells, although no triggering factor has yet been identified [2]. While more evidence exists to implicate T cells in MS pathogenesis, the role of B cells and that of the innate immune system (i.e., monocyte/macrophages, microglia, reactive astrocytes, etc.) has recently been explored [3–5]. Indeed, most experimental models point to the necessary role of effector T cells and/or a deficient proportion of regulatory T cells and the first highly effective MS therapies targeted the extravasation of activated T cells into the brain or the exit of activated T cells form lymphoid organs [6, 7]. However, successful new MS therapies point to a somewhat unexpected and prominent role of B lymphocytes, as full depletion of cells expressing the CD20 surface marker results in an almost complete shutdown of disease activity [8, 9].
Genetics
Heritable contributions to MS risk are unquestionable. For several years it has now been clear that the main MS susceptibility association signal maps genome wide to the HLA-DRB1 locus in the major histocompatibility complex. This region is located on the short arm of chromosome 6 at p21.3, spans almost 4000 kb of DNA, and contains ~165 closely linked genes, about half having pivotal roles in the immune system [10]. In the last 10 years several genome-wide association studies have been conducted mostly by the International Multiple Sclerosis Genetics Consortium (IMSGC), and today > 200 independent loci have been reported to contribute to disease pathogenesis [11].
As with many other complex diseases, risk of MS is driven by multiple common variants whose biological effects are not immediately clear. In addition, MS shares a substantial proportion of risk alleles with several other autoimmune disorders, including type 1 diabetes mellitus, rheumatoid arthritis, systemic lupus erythematosus, and Crohn’s disease. This remarkable sharing is the highest among the complex diseases, including cancers, neurological, and metabolic (data not shown). This particular architecture confirms that MS is at its core an autoimmune disease and strongly suggests that susceptibility to autoimmunity is an inherited risk, which, later in life, may be compounded and shaped by additional genetic (and epigenetic) determinants and environmental exposures to ultimately define the effector mechanism (cellular vs humoral) and target organ of the autodestructive process.
Despite this body of evidence, the low concordance between monozygotic twins argues in favor of a larger role for the environment or additional gene:environment interactions. Although environmental exposures are challenging and often impossible to study, MS susceptibility has been associated with smoking, vitamin D levels, and Epstein–Barr virus infection [12–16].
The development of affordable and massively parallel sequencing technologies has enabled the exploration of genomes other than our own, giving birth to the field of metagenomics [17]. It is expected that in the near future large studies will be carried out to characterize the genomes of all microscopic entities that coexist with humans (e.g., bacteria, Archaea, viruses, etc.) and their potential role in triggering or perpetuating the disease [18, 19].
A Window Into the Environment
Despite impressive advances in MS genetics, common DNA polymorphisms can only explain, at most, 40% of the phenotypic variance, suggesting the presence of nonclassic genetic factors (epigenetics, gene interactions, etc.), environmental variables, or a combination of both [11]. Interestingly, changes in the microbial composition of the intestinal compartments have been shown to alter the balance between inflammatory and regulatory host responses, and to modulate the phenotype, proliferation, and functional capacity of inflammatory/regulatory cells. Specifically, gut microbiota regulate host immune responses through interactions with lymphocytes and dendritic cells in the lamina propria of the gastrointestinal (GI) tract (particularly the cecum and ileum) [20]. The hypothesis that the triggering factor(s) surrounding the onset and perpetuation of MS can be traced to the gut is now getting more traction [21], and several lines of evidence point towards this being a plausible scenario. While this is still an emerging field, it is becoming clear that different commensal microorganisms have the ability to elicit (mostly low-grade) proinflammatory and anti-inflammatory responses in the host [22]. For example, certain bacteria can promote a T helper (Th)17 response [23], whereas others induce regulatory T cells (Treg) [24]. An excess of Th17-promoting bacteria can lead to a loss of tolerance of autoreactive clones and result in autoimmunity. Similarly, a deficiency in Treg-inducing bacteria can result in a relative loss of this important population and also lead to autoimmunity. An adequate balance of both these kinds of bacterial populations would render a healthy, effective immune response, one that not only promotes the necessary Th17 stimulation for T cells to be able to react to potential pathogens, but that can also be counterbalanced when needed by a functional Treg response.
A different role of the gut microbiome is related to the sheer number of genes they encode. It is estimated that the collective gut microbiome encodes upward of a million genes, which is several times the size of the human genome [25, 26]. This creates the possibility of another mechanism by which bacteria could trigger disease: via the production of mimics. It could be speculated that bacterial gene products (either in the sense or antisense orientation) with sufficient homology to human proteins could activate T cells to cross-react with myelin proteins. This has already been shown in Guillain–Barré syndrome [27] and, more recently, in neuromyelitis optica [28], a human demyelinating disease with features common with MS. Moreover, gut bacteria can directly communicate through a complex web of metabolites, some of which can reach the brain through the bloodstream [29]. In addition, bacteria can produce neurotransmitter-like molecules that signal directly to the brain via the vagus nerve [30]. Another way of communication is through bacterial translocation to the blood stream leading to activation of the innate immune system, which could lead to chronic immune stimulation and disease perpetuation. A prerequisite for the latter would be an increased intestinal permeability, which has been suggested to be present in a subgroup of patients with MS in a recent study [31].
B cells, Antibodies and Their Crosstalk to the Gut Microbiota
While T cells have for a long time been considered key players in the immune pathogenesis of MS, the success of several clinical trials with B-cell depleting therapies both in relapsing-remitting and primary progressive MS has highlighted the essential role of B cells in MS [32]. In addition to being the precursor to antibody-producing plasma cells, B cells modulate the immune response through cytokine production and are highly efficient antigen-presenting cells [33].
In contrast to the significant advancement in our understanding of B-cell immunology in the periphery and the inflamed nervous system, the role of B cells, and the humoral immune response in the context of the gut microbiome in MS remains largely unexplored. There is increasing evidence that the gut microbiota–immune interaction at the mucosal interface is essential for microbial and immune homeostasis [20]. Conversely, the increasing evidence for perturbation of the “healthy” microbial composition in various autoimmune diseases (including MS) impacts both the immunologic, as well as neurohumoral, communication between the gut and the brain—the so-called gut–brain axis [34].
Several lines of evidence show that gut microbiota and microbial products can directly regulate B-cell development, activation, and differentiation [35]. As an example, the prevailing dogma that B cells develop in the bone marrow has been challenged by the striking finding that B-cell development also occurs in the gut, where it is stimulated by resident microbes [36]. Moreover, the induction of regulatory B cells in an autoimmune context depends on a healthy microbiota, as has been shown recently [37]. In a complementary fashion, the immune system (and, in particular, the humoral response), mediates homeostasis of the microbial composition via production and secretion of IgA [38]. IgA is the most abundantly produced immunoglobulin in mammals and plays an essential role in controlling the composition of the microbiota in mice and humans. Recently, IgA coating of bacteria has been shown to identify resident pathosymbionts in inflammatory bowel disease, kwashiorkor, and spondylarthritis [39–41]. Remarkably, oral treatment of mice with experimental colitis with a high-affinity IgA antibody against known pathosymbionts has been used to restore eubiosis [42]. Future studies will be needed to show whether this mechanism also relates to other conditions in which dysbiosis is a feature.
Another line of evidence relates to the presence of antibodies against specific bacteria. Antibodies in the serum of patients with MS but not controls have been shown to target bacterial peptides derived from Acinetobacter species and Pseudomonas aeruginosa, which share sequence homology to myelin basic protein and myelin oligodendrocyte glycoprotein (MOG), suggesting molecular mimicry [43]. Interestingly, recent data from our laboratory revealed that Acinetobacter calcoaceticus is more abundant in microbiome samples from untreated patients with relapsing-remitting MS than in healthy controls [44]. Another study found higher serum reactivity to the epsilon toxin of Clostridium perfringens type B in patients with MS when compared with controls, suggesting prior exposure of patients to this bacterium [45].
However, a recent study reported decreased levels of antibodies against α1,3-galactose (Gal) in patients with MS and clinically isolated syndrome versus controls [46]. Gal is a highly immunogenic sugar moiety produced by gut microbiota. Anti-Gal antibodies appear in almost all individuals within the first months of life following immunization by Gal produced by the microbiota and can represent a percentage of circulating immunogloblins [47]. This suggests specific mechanisms for an anti-Gal decrease, such as a modified gut microbiota in patients with MS, more specifically affecting microorganisms producing Gal.
While changes in microbial composition associated with diseases are only beginning to be understood, there is a lot to be discovered with regard to the reciprocal interaction between the altered gut microbiome and B cells.
Dysbiosis in MS?
A simple strategy to explore the involvement of intestinal microbiota in MS relies on comparing microbiota between MS patients and healthy donor cohorts as controls. The most established method for studying the presence of microbes in stool samples is by sequencing one or more variable regions of the 16S ribosomal RNA gene. Since 16S rRNA is conserved across most bacteria and Archaea and the variable regions comprise a small fragment of DNA, this is highly convenient as a screening method. In addition, it is relatively inexpensive and the analysis does not entail much complexity.
Several studies suggesting alteration of relative proportions of gut bacteria in MS have been reported (Table 1) [48–52]. While several individual taxa were reported as present at different proportions between cases and controls, none of the studies was able to identify and independently replicate a significant dysbiosis in MS. However, 2 recently published studies by 2 independent groups have shown an increased abundance of Akkermansia muciniphila in patients with MS versus household controls or their monozygotic twin pair without MS [44, 53]. Nonetheless, even at the lower taxa level, results are highly variable across different studies, highlighting challenges including sample size, heterogeneity, disease-modifying treatment, sample collection, sequencing technique, and analysis strategy. Most notably, at least half of the patients in any given study were receiving some form of disease-modifying therapy at the time of sample collection. Recent studies have highlighted the strong effect of cancer and immunomodulatory therapies on the composition of the gut microbiota [54, 55]. Interestingly, at least in mice this effect seems to be bi-directional, as indicated by reports on the differential ability of gut microbes to metabolize and ultimately influence the bioavailability of therapeutic drugs [56, 57]. In MS, to date, the effect of disease-modifying treatment on the microbiota and vice versa has been poorly understood. To answer these questions and overcome current heterogeneity in patient populations, larger studies will have to assess the gut microbiota composition in patients before and on treatment. Moreover, technical challenges related to differences in sampling and use of standardized experimental and analytical tools currently pose problems on study comparability [58, 59].
Table 1.
Overview of the current literature on the microbiome as determined by 16S rRNA gene analysis in multiple sclerosis
| Reference | No. of patients (disease course) |
Treatment | Controls (household?) | Ethnicity | OTUs or genera increased in MS |
|---|---|---|---|---|---|
| [52]* | 7 (RRMS) | 5 treated (GA), 2 untreated |
8 (no) | White |
Akkermansia
Faecalibacterium Coprococcus |
| [51] | 20 (RRMS) | 13 treated (IFN-β and/or PSL), 7 untreated |
40+10 (no) | Asian |
Eggerthella lenta
Streptococcus thermophiles/salivarius Clostridia cluster XIVa and IV (including Faecalibacterium prausnitzi, Coprococcus comes) Anaerostipes hadrus Eubacterium rectale |
| [50] | 31 (RRMS) | 20 treated (IFN-β, NTZ, GA), 11 untreated |
36 (no) |
Pseudomonas,
Pedobacter Mycoplana Blautia |
|
| [48] | 60 (RRMS) | 28 untreated | 43 (no) | White, black (n = 2) |
Akkermansia
Methanobrevibacter Butyricimonas Paraprevotella Haemophilus Slackia |
| Tremlett et al. [44] | 18 (RRMS) | 9 untreated, 9 treated (IFN-β, NTZ, GA) |
17 (no) | White (50%), nonwhite (50%) |
Bilophila
Bifidobacterium Desulfovibrio Christensenellaceae |
| [44, 49] | 71 (RRMS) | 71 untreated | 71 (yes) | White |
Acinetobacter calcoaceticus
Akkermansia muciniphila Eggerthella lenta |
| [49, 53] | 34 (22 RRMS, 7 SPMS, 3 CIS, 2 PPMS) |
15 untreated,
19 treated (13 IFN-β, 4 NTZ, 1 GA, 1 AZT) |
34 (monozygotic twins) | White | Akkermansia muciniphila |
*Subanalysis of a pre-/postvitamin D supplementation study
OTU = operational taxonomic unit; RRMS = relapsing-remitting multiple sclerosis; GA = glatirameracetate; IFN = interferon; PSL = prednisolone; NTZ = natalizumab; AZT = azathioprine; SPMS = secondary progressive multiple sclerosis; CIS = clinically isolated syndrome
Another major potential confounder for studies on the gut microbiome is diet, whose influence on the gut bacterial composition is arguably stronger than that of genetics [60]. While perfectly controlling for the effect of diet is admittedly difficult for any study, recruiting matched household controls is a valid strategy that helps mitigate the heterogeneity of otherwise unrelated controls. At the very minimum, a detailed recording of food intake and other environmental variables (e.g., smoking, exercise, etc.) is highly advisable.
A potentially interesting finding in 4 of the studies is the elevated proportion of the genus Akkermansia in MS versus controls [44, 48, 52, 53]. Akkermansia belongs to the mucin-degrading bacteria that convert mucin to short-chain fatty acids and has been reported to have both regulatory and proinflammatory properties [61]. Moreover, it has been correlated to proinflammatory pathways and activation of the complement cascade [62]. The proinflammatory feature most likely results from its mucus-degrading ability leading to an exposure of immune cells to microbial antigens as has been shown in an intestinal inflammation model of Salmonella typhimurium where Akkermansia lead to disease exacerbation [63]. Interestingly, we recently showed that microbiome samples from patients with MS with detected A. muciniphila have been shown to exert a proinflammatory phenotype on peripheral blood T lymphocytes in vitro, which was replicated by exposure to isolated A. muciniphila [44].
While 16S rRNA gene sequencing is a popular screening method, it is unable to consistently detect species-level (even less strain-level) taxonomies, thus limiting the interpretability of the analyzed sequences to just describing the presence or absence of certain (mostly higher) taxa. However, whole-genome shotgun (metagenomic) sequencing investigates every gene in the organism’s genome and thus could be more informative. For example, it can lead to identification of genes (or gene fragments) with homology to human sequences, thus prompting more focused searches for mimics that could trigger an autoimmune response in humans. Also, detailed metagenomic sequencing can offer a global view on the aggregate metabolic capability of a given microbial population. Reports on the importance of bacterial polymorphisms have recently described a discovery only made possible by performing high-depth metagenomic sequencing [64].
Most commensal bacteria are human symbionts and are needed for proper development of our immune system and to regulate multiple physiological processes. These include nutrient breakdown and absorption, promote integrity of the gut, and develop and regulate brain physiology. Microbiota also regulate metabolic pathways, including fatty-acid synthesis, and hormonal and antioxidant metabolisms. Thus, a detailed understanding of metabolite production by gut bacteria and how they interact and modulate our own cellular and physiological machinery is of outmost importance. Therefore, combining shotgun-sequencing data with metabolomics analysis (typically by mass spectrometry) can be a powerful strategy to explore the underlying biochemical processes driven or regulated by bacteria.
One limitation of sequencing DNA from stool material is that one might be sampling the colonic bacterial population, when that of the small intestine might be more relevant [65]. In addition to the variation in microbial composition along the GI tract, there are also significant differences in the localization of the microbiota to the mucosa. It is their proximity to the epithelial cells that determines the interaction with the host cells and immune system. However, at present there is no alternative to sampling the upper GI tract in a noninvasive manner.
From Association to Causation
Although potentially informative, the genomic investigation of microbiome populations is inherently an associative approach, and does not establish a causal involvement of eventually detected differences (see [66] for an elaborated discussion). Specifically, the finding that certain organisms are present at the same time as the disease or phenotype of interest could either mean that they are causal or reactive (i.e., reverse causality) to the process under study. In the case of MS, no studies have been yet reported to unequivocally prove that the observed differences are the cause, and not a consequence, of the disease process.
While proving this experimentally poses a considerable challenge, perhaps the most convincing strategy is to transfer human microbiota into germ-free (GF) mice and evaluate their effect after colonization. This approach has been successfully employed in other settings (obesity, Parkinson’s disease, etc.) and offers a more mechanistic view of any identified differences [60, 67–72]. In groundbreaking work, Berer et al. [21] derived a GF version of a transgenic mouse that spontaneously develops relapsing-remitting disease and closely resembles human MS [21]. In this model a large proportion of CD4+ T cells carries a T-cell antigen receptor recognizing MOG peptide 92–106 in the context of major histocompatibility complex class II. They were able to show that the initiation of disease is dependent on the presence of the healthy gut microbiota leading to the activation and expansion of autoreactive T cells. Subsequently, these T cells recruit autoantibody-producing MOG-specific B cells from the endogenous repertoire, which cooperatively trigger autoimmune demyelinating encephalomyelitis with a relapsing disease course, which is similar to MS. Two recent independent studies demonstrated that colonization of GF mice with MS donor microbiota inhibits Treg differentiation and exacerbates disease severity in 2 different experimental mouse models of MS [44, 53]. These 2 studies are the first to show a proinflammatory effect of the different microbiota composition in human MS compared with control in vitro and in vivo. This raises the question whether dysbiosis in MS perpetuates (or even triggers) chronic inflammation.
While experimentation with GF animals is arguably the method of choice to investigate the causal effect of specific microbiota, breeding and keeping experimental animals under GF conditions is a complex undertaking and somehow limits the broad applicability of this method. An alternative solution is pretreatment with an antibiotic cocktail consisting of vancomycin, metronidazole, ampicillin, and neomycin to eliminate the vast majority of gut microbes before the experiment [73]. However, this method also has caveats as it does not completely render the animals GF and introduces further confounders. It is important to note that GF animals are not physiologically normal with particular deficiencies in the gut-associated lymphoid organs and overall immune functions (enlarged bowel, fewer and less cellular Peyer’s patches, fewer CD4+ and CD8+ T cells in lamina propria and mesenteric lymph nodes, reduced production of secretory IgA, among others) and these may also result in potential confounders [22].
Evaluating the consequences of a change in the relative proportions of gut communities in a systematic manner is a nearly impossible task. The metabolism of any given organism can potentially modify that of its neighbors and, in turn, it is itself modified by those in the vicinity. This interdependence, characteristic of all complex systems, is what ultimately determines the emergent behavior of the whole ensemble. A reductionist alternative is to evaluate the effect of select individual bacteria in in vitro co-cultures with host immune cells. For example, extracts of individually grown bacteria (obtained from commercially available sources) can be added to cultures of human or mouse peripheral blood mononuclear cells to evaluate their effect on T-cell polarization or proliferation of specific T- or B-cell populations [44]. One caveat of this approach is that it depends on accurately matching sequencing information from the patient’s sample to available culturable strains.
An emerging (hybrid) alternative is the individual microisolation and colony growing of bacterial organisms from a donor’s sample under specialized conditions (e.g., anaerobic). Since this technique enables the individual growing of most organisms in a given sample, it is then possible to create defined microbial mixes (i.e., formulation) and test them either in vitro or in vivo [74]. This approach has the advantage that it uses the exact same strains directly isolated from the individual and eliminates ambiguities in deducing the most likely match from a given sample to a commercially available, culturable strain.
Consortia
The lack of differences in beta diversity and the small overall effects observed in studies of most diseases so far (including MS) highlight the need to study large populations so as to increase power and to minimize potential confounders (disease-modifying treatment, diet, ethnicity, etc.).
An example on the power of consortia of investigators to increase the pace of discovery is the IMSGC (see above). While the genetic association between the HLA and MS was discovered in 1972, the MS genetics field was unable to discover any non-HLA associations until 2007, when independent research groups formed the IMSGC [75]. Since then, the ever-increasing cohort sizes enabled the discovery of up to 200 independent non-HLA associations [11].
The MS Microbiome Consortium (MSMC) was assembled in 2014 with support from the National MS Society and the US Department of Defense. The MSMC brings together MS investigators from the University of California San Francisco (S.E. Baranzini and B.A.C. Cree) and Icahn School of Medicine at Mount Sinai (P. Casaccia and I. Katz Sand) with microbiologists and microbiome researchers from Caltech (S. Mazmanian) and University of California San Diego (R. Knight). The goal of the MSMC is to investigate the role of the gut microbiota in MS, as well as to evaluate the interrelationship between disease-modifying therapies and gut microbial communities in patients with MS. In addition to recruiting patients for molecular and immunological studies, the MSMC is an avenue for collaboration and cross-discipline training among a diverse group of investigators.
More recently the International MS Microbiome Study was constituted, with support from the Valhalla Charitable Foundation and was modeled after the IMSGC, with the premise that the whole is more than the sum of its parts. This ambitious program brings together all investigators from the MSMC and additional leaders in MS and microbiome research from Harvard (D. Kasper, H. Weiner, T. Chitnis), University of Pittsburgh (Z. Xia), Max Planck Institute (H. Wekerle, R. Hohlfeld), University of Edinburgh (S. Chandran), Biodonostia (D. Otaegui) and FLENI (J. Correale). The main goal of the International MS Microbiome Study is to recruit and analyze the gut microbiome of 2000 patients with MS. All types of MS are allowed and while we strive to obtain samples from individuals while they are naïve to therapeutic drugs, patients taking any of the most commonly used disease-modifying treatments (interferon-β, glatiramer acetate, dimethyl fumarate, fingolimod, and natalizumab) are also allowed. Inclusion of individuals treated with B-cell-depleting therapies is currently being discussed. A unique feature of this collection is that all patients will be recruited with a matching healthy household control. We hypothesize that a shared environment will minimize the confounding effects of factors known to influence the composition of the gut microbiome, such as diet, the presence of children, pets, urban versus rural environment, and others. Stool and blood samples are collected for all qualifying participants. Among the data variables collected are an extensive dietary questionnaire, detailed demographics, and clinical parameters.
We anticipate that the knowledge gained from this study will lead to the design of an entirely new kind of clinical trial that will test our ability to rationally manipulate the gut microbiota in order to alter the course of MS (Fig. 1).
Fig. 1.
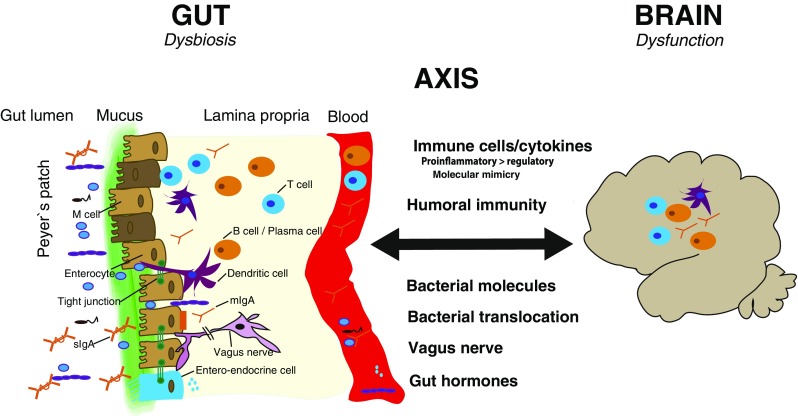
Putative mechanisms of gut–brain communication in the context of dysbiosis and neuroinflammation in multiple sclerosis (MS). The gut mucosal interface is a zone of intensive interaction between the gut microbiota in the luminal space and the immune cells situated in the lamina propria and enriched in lymphoid follicles, the Peyer’s patches. Putative mechanisms by which dysbiosis in the gut can influence central nervous system inflammation in patients with MS are through immune cells through a dysbalance of pro- and anti-inflammatory cytokines (e.g., an excess of T helper 17-promoting bacteria) or molecular mimicry. Other routes of communication include humoral immunity, bacterial molecules (fatty acids, etc.), direct bacterial translocation leading to activation of the innate immune system, and direct communication via the vagus nerve or the release of gut hormones (e.g., 5-hydroxytryptamine). mIgA = monomeric IgA; sIgA = secretory IgA
Concluding Remarks
The role of the gut microbiota in triggering and/or perpetuating MS is only beginning to be understood. The interactions between gut bacteria and cells from the immune system (both innate and adaptive) will likely be characterized in greater detail in the near future. However, a notable body of evidence is already emerging from early studies that put the microbiome at the forefront of immune research in MS. These pioneering studies have explored recruitment strategies, analytical data, and sample collection methods, and have even identified several potential organisms associated with the disease. Perhaps most importantly, this work has paved the road for the next generation of studies that will provide confirmation, functional characterization of key microbes, and, in turn, prepare for interventional strategies that modulate the gut microbiota in a rational and evidence-based manner.
Acknowledgments
This work was supported by grants from the US Department of Defense (W81XWH-15-1-0652), National MS Society (A125014), and the Valhalla Charitable Foundation to S.E.B. S.E.B holds the Heidrich Family and Friends endowed Chair in Neurology. A.-K.P. is supported by a scholarship from the Swiss National Science Foundation (P2SKP3_164938/1).
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372(9648):1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.McFarland HF, Martin R. Multiple sclerosis: a complicated picture of autoimmunity. Nat Immunol. 2007;8(9):913–919. doi: 10.1038/ni1507. [DOI] [PubMed] [Google Scholar]
- 3.Rothhammer V, Quintana FJ. Role of astrocytes and microglia in central nervous system inflammation. Introduction. Semin Immunopathol. 2015;37(6):575–576. doi: 10.1007/s00281-015-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krumbholz M, Derfuss T, Hohlfeld R, et al. B cells and antibodies in multiple sclerosis pathogenesis and therapy. Nat Rev Neurol. 2012;8(11):613–623. doi: 10.1038/nrneurol.2012.203. [DOI] [PubMed] [Google Scholar]
- 5.Prinz M, Priller J. The role of peripheral immune cells in the CNS in steady state and disease. Nat Neurosci. 2017;20(2):136–144. doi: 10.1038/nn.4475. [DOI] [PubMed] [Google Scholar]
- 6.Yednock TA, Cannon C, Fritz LC, et al. Prevention of experimental autoimmune encephalomyelitis by antibodies against alpha 4 beta 1 integrin. Nature. 1992;356(6364):63–66. doi: 10.1038/356063a0. [DOI] [PubMed] [Google Scholar]
- 7.Kappos L, Antel J, Comi G, et al. Oral fingolimod (FTY720) for relapsing multiple sclerosis. N Engl J Med. 2006;355(11):1124–1140. doi: 10.1056/NEJMoa052643. [DOI] [PubMed] [Google Scholar]
- 8.Hauser SL, Bar-Or A, Comi G, et al. Ocrelizumab versus interferon beta-1a in relapsing multiple sclerosis. N Engl J Med. 2017;376(3):221–234. doi: 10.1056/NEJMoa1601277. [DOI] [PubMed] [Google Scholar]
- 9.Montalban X, Hauser SL, Kappos L, et al. Ocrelizumab versus Placebo in primary progressive multiple sclerosis. N Engl J Med. 2017;376(3):209–220. doi: 10.1056/NEJMoa1606468. [DOI] [PubMed] [Google Scholar]
- 10.Campbell RD, Trowsdale J. Map of the human MHC. Immunol Today. 1993;14(7):349–352. doi: 10.1016/0167-5699(93)90234-C. [DOI] [PubMed] [Google Scholar]
- 11.Patsopoulos N, Baranzini SE, Santaniello A, et al. The Multiple Sclerosis Genomic Map: role of peripheral immune cells and resident microglia in susceptibility. Available at: https://www.biorxiv.org/content/early/2017/07/13/143933. Accessed November 6, 2017.
- 12.Hedstrom AK, Alfredsson L, Olsson T. Environmental factors and their interactions with risk genotypes in MS susceptibility. Curr Opin Neurol. 2016;29(3):293–298. doi: 10.1097/WCO.0000000000000329. [DOI] [PubMed] [Google Scholar]
- 13.Hedstrom AK, Olsson T, Alfredsson L. Smoking is a major preventable risk factor for multiple sclerosis. Mult Scler. 2016;22(8):1021–1026. doi: 10.1177/1352458515609794. [DOI] [PubMed] [Google Scholar]
- 14.Olsson T, Barcellos LF, Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol. 2017;13(1):25–36. doi: 10.1038/nrneurol.2016.187. [DOI] [PubMed] [Google Scholar]
- 15.Rhead B, Baarnhielm M, Gianfrancesco M, et al. Mendelian randomization shows a causal effect of low vitamin D on multiple sclerosis risk. Neurol Genet. 2016;2(5):e97. doi: 10.1212/NXG.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundqvist E, Bergstrom T, Daialhosein H, et al. Cytomegalovirus seropositivity is negatively associated with multiple sclerosis. Mult Scler. 2014;20(2):165–173. doi: 10.1177/1352458513494489. [DOI] [PubMed] [Google Scholar]
- 17.Nealson KH, Venter JC. Metagenomics and the global ocean survey: what's in it for us, and why should we care? Isme J. 2007;1(3):185–187. doi: 10.1038/ismej.2007.43. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopala SV, Vashee S, Oldfield LM, et al. The human microbiome and cancer. Cancer Prev Res. 2017;10(4):226–234. doi: 10.1158/1940-6207.CAPR-16-0249. [DOI] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 21.Berer K, Mues M, Koutrolos M, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479(7374):538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 22.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 25.Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandoval-Motta S, Aldana M, Martinez-Romero E, et al. The human microbiome and the missing heritability problem. Front Genet. 2017;8:80. doi: 10.3389/fgene.2017.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes RA, Cornblath DR. Guillain-Barré syndrome. Lancet. 2005;366(9497):1653–1666. doi: 10.1016/S0140-6736(05)67665-9. [DOI] [PubMed] [Google Scholar]
- 28.Cree BA, Spencer CM, Varrin-Doyer M, et al. Gut microbiome analysis in neuromyelitis optica reveals overabundance of Clostridium perfringens. Ann Neurol. 2016;80(3):443–447. doi: 10.1002/ana.24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gacias M, Gaspari S, Santos PM, et al. Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. Elife 2016; 5. [DOI] [PMC free article] [PubMed]
- 30.Forsythe P, Kunze WA. Voices from within: gut microbes and the CNS. Cell Mol Life Sci. 2013;70(1):55–69. doi: 10.1007/s00018-012-1028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buscarinu MC, Cerasoli B, Annibali V, et al. Altered intestinal permeability in patients with relapsing-remitting multiple sclerosis: a pilot study. Mult Scler. 2017;23(3):442–446. doi: 10.1177/1352458516652498. [DOI] [PubMed] [Google Scholar]
- 32.Hauser SL. The Charcot Lecture | beating MS: a story of B cells, with twists and turns. Mult Scler. 2015;21(1):8–21. doi: 10.1177/1352458514561911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen P, Roch T, Lampropoulou V, et al. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature. 2014;507(7492):366–370. doi: 10.1038/nature12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colpitts SL, Kasper LH. Influence of the gut microbiome on autoimmunity in the central nervous system. J Immunol. 2017;198(2):596–604. doi: 10.4049/jimmunol.1601438. [DOI] [PubMed] [Google Scholar]
- 35.Kim M, Kim CH. Regulation of humoral immunity by gut microbial products. Gut Microbes 2017: 1-8. [DOI] [PMC free article] [PubMed]
- 36.Wesemann DR, Portuguese AJ, Meyers RM, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501(7465):112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosser EC, Oleinika K, Tonon S, et al. Regulatory B cells are induced by gut microbiota-driven interleukin-1beta and interleukin-6 production. Nat Med. 2014;20(11):1334–1339. doi: 10.1038/nm.3680. [DOI] [PubMed] [Google Scholar]
- 38.Kubinak JL, Round JL. Do antibodies select a healthy microbiota? Nat Rev Immunol. 2016;16(12):767–774. doi: 10.1038/nri.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158(5):1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kau AL, Planer JD, Liu J, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7(276):276ra224. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viladomiu M, Kivolowitz C, Abdulhamid A, et al. IgA-coated E. coli enriched in Crohn's disease spondyloarthritis promote Th17-dependent inflammation. Sci Transl Med 2017; 9(376). [DOI] [PMC free article] [PubMed]
- 42.Okai S, Usui F, Yokota S, et al. High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol. 2016;1(9):16103. doi: 10.1038/nmicrobiol.2016.103. [DOI] [PubMed] [Google Scholar]
- 43.Hughes LE, Smith PA, Bonell S, et al. Cross-reactivity between related sequences found in Acinetobacter sp., Pseudomonas aeruginosa, myelin basic protein and myelin oligodendrocyte glycoprotein in multiple sclerosis. J Neuroimmunol. 2003;144(1-2):105–115. doi: 10.1016/S0165-5728(03)00274-1. [DOI] [PubMed] [Google Scholar]
- 44.Cekanaviciute E, Yoo BB, Runia TF, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114:10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rumah KR, Linden J, Fischetti VA, et al. Isolation of Clostridium perfringens type B in an individual at first clinical presentation of multiple sclerosis provides clues for environmental triggers of the disease. PLOS ONE. 2013;8(10):e76359. doi: 10.1371/journal.pone.0076359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Berre L, Rousse J, Gourraud PA, et al. Decrease of blood anti-alpha1,3 Galactose Abs levels in multiple sclerosis (MS) and clinically isolated syndrome (CIS) patients. Clin Immunol. 2017;180:128–135. doi: 10.1016/j.clim.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Galili U. Anti-Gal: an abundant human natural antibody of multiple pathogeneses and clinical benefits. Immunology. 2013;140(1):1–11. doi: 10.1111/imm.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jangi S, Gandhi R, Cox LM, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tremlett H, Fadrosh DW, Faruqi AA, et al. Gut microbiota composition and relapse risk in pediatric MS: a pilot study. J Neurol Sci. 2016;363:153–157. doi: 10.1016/j.jns.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, Chia N, Kalari KR, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyake S, Kim S, Suda W, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLOS ONE. 2015;10(9):e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cantarel BL, Waubant E, Chehoud C, et al. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med. 2015;63(5):729–734. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berer K, Gerdes LA, Cekanaviciute E, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A. 2017;114:10719–10724. doi: 10.1073/pnas.1711233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenhill C. Gut microbiota: Anti-cancer therapies affected by gut microbiota. Nat Rev Gastroenterol Hepatol. 2014;11(1):1. doi: 10.1038/nrgastro.2013.238. [DOI] [PubMed] [Google Scholar]
- 55.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marotz C, Amir A, Humphrey G, et al. DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques. 2017;62(6):290–293. doi: 10.2144/000114559. [DOI] [PubMed] [Google Scholar]
- 59.Kuczynski J, Lauber CL, Walters WA, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2011;13(1):47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Derrien M, Vaughan EE, Plugge CM, et al. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(Pt 5):1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 62.Derrien M, Van Baarlen P, Hooiveld G, et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ganesh BP, Klopfleisch R, Loh G, et al. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLOS ONE. 2013;8(9):e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nayfach S, Rodriguez-Mueller B, Garud N, et al. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26(11):1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerritsen J, Smidt H, Rijkers GT, et al. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6(3):209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wekerle H. Brain autoimmunity and intestinal microbiota: 100 trillion game changers. Trends Immunol. 2017;38(7):483–497. doi: 10.1016/j.it.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Sampson TR, Debelius JW, Thron T, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. 2016;167(6):1469–1480. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goodman AL, Kallstrom G, Faith JJ, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108(15):6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McNulty NP, Yatsunenko T, Hsiao A, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3(106):106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faith JJ, McNulty NP, Rey FE, et al. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333(6038):101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reikvam DH, Erofeev A, Sandvik A, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLOS ONE. 2011;6(3):e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ringeisen BR, Rincon K, Fitzgerald LA, et al. Printing soil: a single-step, high-throughput method to isolate micro-organisms and near-neighbour microbial consortia from a complex environmental sample. Methods Ecol Evol. 2015;6(2):209–217. doi: 10.1111/2041-210X.12303. [DOI] [Google Scholar]
- 75.(IMSGC) IMSGC, Hafler DA, Compston A, et al Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]


