Abstract
Vertebrates harbor both symbiotic and pathogenic bacteria on the body and various mucosal surfaces. Of these surfaces, the intestine has the most diverse composition. This composition is dependent upon various environmental and genetic factors, with diet exerting the maximum influence. Significant roles of the intestinal bacteria are to stimulate the development of a competent mucosal immune system and to maintain tolerance within the intestine. One manner in which this is achieved is by the establishment of epithelial integrity by microbiota found in healthy individuals (healthy microbiota); however, in the case of a disrupted intestinal microbiome (dysbiosis), which can be caused by various conditions, the epithelial integrity is compromised. This decreased epithelial integrity can then lead to luminal products crossing the barrier, generating a systemic proinflammatory response. In addition to epithelial integrity, healthy intestinal commensals metabolize indigestible dietary substrates and produce short-chain fatty acids, which are bacterial metabolites that are essential for colonic health and regulating the function of the intestinal immune system. Intestinal commensals are also capable of producing neuroactive molecules and neurotransmitters that can affect the function of the vagus nerve. The observations that intestinal dysbiosis is associated with different diseases of the nervous system, suggests that cross-talk occurs amongst the gut, the nervous system, and the immune system.
Electronic supplementary material
The online version of this article (10.1007/s13311-017-0601-4) contains supplementary material, which is available to authorized users.
Keywords: Immunomodulation, Neuronal, Dysbiosis, Intestinal, Microbiome
Introduction
The human intestine is colonized by a large number of microorganisms, including around 1014 types of bacteria that are involved in various functions. The colonization of the gut begins at birth and continues throughout the teenage years, forming a unique intestinal microbiome signature for each individual. The composition of an individual’s gut microbiota is influenced by many factors, including diet, geographical location, genetics, age, and sex [1]. Recent studies have provided evidence that the intestinal microbiome has a mutualistic relationship with the host and helps regulate the host’s innate and adaptive immune systems.
In this mutualistic relationship, the human body acts as a host providing gut microbes with the nutrients that they need. In return, the microbes help with the catabolism of indigestible carbohydrates and fatty-acid chains leading to the production of short-chain fatty acids (SCFAs) [2]. The SCFAs produced by gut microbes are essential for many physiological pathways, as well as for regulating immune responses. For example, butyrate, a SCFA, is required for colonic health and epithelial integrity. Other benefits that the gut microbiota provide to the host include the production of vitamins, such as vitamin B, as well as the secretion of hormones that signal the body to store fat [3]. Along with aiding in nutrient uptake, hormone signaling, and vitamin production, some of these bacteria also help to shape and to regulate the immune system.
However, the above-described beneficial effects come from beneficial commensals. In order to remove deleterious and pathogenic bacteria, the host has a significant amount of lymphoid tissue called mucosa-associated lymphoid tissue (MALT), which consists of 70% of the body’s lymphoid tissue. Immunologically active T and B lymphocytes are found in the MALT, which routinely samples the microbes that enter the gut in order to detect and eliminate pathogenic bacteria. This regulation of the gut microbiota can be achieved in many ways, as various bacterial species express different cell surface receptors, as well as different metabolites. Intestinal epithelial and immune cells will recognize conserved microbial sequences called microbe-associated molecular patterns, using receptors called pattern recognition receptors [4]. Detection of microbe-associated molecular patterns by the MALT will lead to epithelial cells producing regenerating Protein-III gamma (Reg III gamma), which is an antibacterial lectin that targets Gram-positive bacteria [5]. This is, therefore, one example of how intestinal microbiota, through the release of natural antigens, can stimulate and modulate our mucosal immune system. In fact, analysis of the transcriptomes of healthy adult C3H/HeN mice raised in germ-free conditions (no fecal microbiota), conventional housing, and mice colonized with fecal microbiota showed that 45% of genes induced by the fecal microbiota were assigned to immune response pathways [6].
Pathogenic bacteria and in some cases, symbiotic-turned-pathogenic bacteria, will certainly stimulate the immune response. Another way that intestinal microbiota can affect the immune response is through the alteration of the overall composition of the intestinal microbiota. Altered gut microbial composition has been associated with obesity and inflammatory bowel disease [6], and as well as with systemic diseases like multiple sclerosis (MS) and rheumatoid arthritis [1, 7]. Examples of environmental factors that may alter a patient’s microbiome include noncommensal bacteria, antibiotics, diet, lifestyle, and exercise. Such shifts in the intestinal microbiome would affect health by: changing nutrient uptake by the host, altering the host’s ability to mount appropriate immune responses, and disrupting the host’s ability to tolerate their own intestinal commensal bacteria.
Interactions Amongst the Intestinal Microbiome and the Neuronal and Immune Systems
Germ-free (GF) mice have been crucial in our understanding of the complex interactions that occur amongst the intestinal microbiome and the neuronal and immune systems. For example, GF mice have a poorly developed mucosal immune system characterized by reduced IgA production, smaller Peyer’s patches, and altered expression of Toll-like receptors (TLRs) [8, 9]. GF mice also have lower levels of serotonin [10]. In addition, GF mice have reduced numbers of enteric neurons, which is associated with decreased gut motility and lower excitability of neurons, suggesting a role of the microbiota in the development of the enteric nervous system [11].
The decreased levels of TLRs in GF mice suggest that the expression of TLRs is influenced by the presence of intestinal microbiota. Two lines of mice, the TLR2–/– and TLR4–/– mice, have also been used to show the interaction between the intestinal microbiome and the nervous system. Both TLR2 and TLR4 are TLRs, which are pattern recognition receptors that can detect antigens from gut microbes. Mice that have TLR2 and TLR4 knocked out (TLR2–/– and TLR4–/–) have reduced gut motility and enteric neurons [12], similar to GF mice. In addition, TLR2–/– mice also have an altered neurochemical profile and reduced expression of glial fibrillary acidic protein (GFAP) in the myenteric plexus. As TLR2 expression on enteric glial cells is enhanced by certain microbes, the microbiota/TLR pathways may be involved in the development of diseases associated with the enteric nervous system.
It should also be noted that pathways between the nervous and immune system also exist, in which stimuli from the nervous system leads to changes in the immune system. For example, neurotransmitters like norepinephrine can bind to the β2 adrenergic receptor expressed by T helper 1 cells. This will then lead to a change in the production of cytokines by the T helper 1 cells, such as a reduction in interleukin (IL)-2 production [13].
The above-described studies in total demonstrate that communication does occur amongst the immune system, the nervous system, and the intestinal microbiome. In order to describe how these communication pathways are disrupted in neuronal diseases, this review will first discuss the communication pathways that are normally present and functioning, such as the microbiota–gut–brain axis, and then a review of those immune-associated neurological disorders that have also been associated with changes in the intestinal microbiome (dysbiosis).
The microbiota–gut–brain axis is essentially an extension of the gut–brain axis, which is a bidirectional communication pathway between the gut and the brain. The communication along the gut–brain pathway is achieved primarily by the vagal nerve, and the hypothalamic–pituitary–adrenal axis [14]. The term microbiota was added (microbiota–gut–brain axis) because studies have shown that the intestinal microbiota can communicate with the gut–brain axis. For example, some bacteria have evolved to produce hormone-like chemicals and monoamine neurotransmitters in the intestine, which can then directly stimulate the intestinal immune and nervous systems [15–17]. The sympathetic nervous system also serves as a conduit between the brain and the immune system, and many immune cells produce neuroactive products. These neuroactive products include: vasoactive intestinal peptide (mast cells and neutrophils), somatostatin (delta cells), adrenocorticotropic hormone, endorphins, and substance P (latter three by macrophages). Specific mechanisms have been identified by which intestinal microbiota can affect the enteric nervous system. For example, intestinal microbiota can induce the production of neurotransmitters. Spore-forming bacteria have been shown to augment the production of serotonin by enterochromaffin cells of the intestinal epithelium via the upregulation of the enzyme tryptophan-hydroxylase-1. Serotonin in the gut is regulated by serotonin-selective reuptake transporter, which is expressed by the intestinal epithelium and controlled by TLRs [10]. G protein-coupled receptors, GPR41 and GPR43, are expressed by epithelial cells and enteric neurons. Both can be activated by SCFAs [18]. SCFA-induced activation of GPR43 promotes the secretion of hormone glucagon-like peptide-1, which is involved in gastrointestinal transit [19]. Another mechanism by which SCFAs affect enteric neurogenesis is by inhibition of histone deacetylase activity. Treatment of cultured enteric neurons with butyrate has been shown to enhance the expression of excitatory choline acetyltransferase and enhance the acetylation of H3K9 [20]. SCFAs in conjunction with other dietary factors and bile acids can also regulate gut motility [12]. Thus, there are ample pathways in which each system, nervous, immune, and intestinal microbiome, can all communicate with each other.
Recent research has shown that some neurological conditions are associated with perturbations of the intestinal microbiota. However, it has been difficult to determine whether or not these changes in the intestinal microbiota are causative of disease, an exacerbating factor for disease, or a consequence of disease. MS is the neurological condition that has been best characterized with regard to the role of dysbiosis. This has been achieved by studying both patients and using an animal model of MS called experimental autoimmune encephalomyelitis (EAE). Intestinal dysbiosis has also been found in other neurological diseases, and the role of the dysbiosis in the pathogenesis of these neurological diseases is even less clear. These diseases include autism spectrum disorder (ASD), amyotrophic lateral sclerosis (ALS), and Parkinson’s disease (PD). That which follows then, will be a series of descriptions and discussions of these neurological diseases and the current understanding of the role of the intestinal microbiome in each disease (Table 1).
Table 1.
Microbial changes in human diseases
| Neuronal disease | Affected neuronal system | Examples of microbiota changes in humans | Mouse models | Mouse models indicate modulatory properties of disease-associated microbiome |
|---|---|---|---|---|
| MS | CNS | Increased Haemophilus, Blautia, and Dorea genera, and decreased Parabacteroides, Adlercreutzia, and Prevotella [21] Increased Methanobrevibacter and Akkermansia, and decreased Butyricimonas [22] |
Yes | Yes [23] |
| ASD | CNS | Decreased in Prevotella [24] Increased Erysipelotrichaceae, Clostridium lituseburense, and Terrisporobacter with low tryptophan; increased Lachnoclostridium bolteae, Lachnoclostridium hathewayi, and Flavonifractor plautii with high serotonin [25] |
Yes | Not tested |
| ALS | CNS, PNS | Reduced Oscillibacter, Anaerostipes, and Lachnospiraceae, and increased Dorea [26] | Yes | Not tested |
| PD | CNS, PNS | Reduction of Prevotellaceae and Lactobacillaceae [27, 28] | Yes | Yes [29] |
MS = multiple sclerosis; CNS = central nervous system; ASD = autism spectrum disorder; ALS = amyotrophic lateral sclerosis; PNS = peripheral nervous system; PD = Parkinson’s disease
Microbiome and MS
The role of the intestinal microbiome in the development of MS has recently become an area of intense research. One recent paper showed that the species richness (alpha diversity) was lower in patients with active MS than in healthy controls [21]. In addition, a Bray–Curtis distance-based community analysis (beta diversity) demonstrated that the microbiomes of patients with relapsing-remitting MS (RRMS) were significantly different than healthy controls. Taxonomic analysis found that Parabacteroides, Prevotella (Bacteroidetes), Adlercreutzia, Collinsella (Actinobacteria), and Erysipelotrichaceae (Firmicutes) were decreased in the fecal microbiota of patients with RRMS as compared with healthy controls [21]. Another publication found MS associated with increases in Methanobrevibacter and Akkermansia and decreases in Butyricimonas [22].
Studies with animal models of MS, EAE, have provided insight into the role of intestinal microbiota in the development of MS. In one study, the authors induced EAE in GF mice and found that the clinical severity was decreased versus EAE induced in specific-pathogen-free conditions [30]. This would indicate that the presence of intestinal microbiota makes the symptoms of EAE more severe. When the GF mice were colonized with segmented filamentous bacteria, mice developed much more severe EAE after induction [30]. How MS-associated gut microbiome influences disease was studied by transferring the microbiomes of patients with MS into GF mice [23]. In this study, the recipient mice retained the signature of the microbiome of the patient with MS, which was characterized by significant increases in Akkermansia and Acinetobacter, with a significant decrease in Parabacteroides. The GF mice induced for EAE that received the MS stool samples had more severe disease than the mice that received stool samples from healthy controls and GF mice that did not receive human stool samples. This increased severity was associated with a decreased number of regulatory T cells in the GF MS mice. Thus, this model proves that the MS microbiome can modulate the immune response and lead to severe disease.
Other studies have administered bacterial lines or probiotics to EAE mice and found that severity of disease is significantly altered. A recently published study by Mangalam et al. [31], administered a probiotic line of Prevotella histicola to a mouse model of EAE and showed that this significantly decreased the severity of the disease and also altered the composition of the microbiome. In a similar study, Lactobacillus plantarum and Bifidobacterium animalis were shown to have a similar inhibitive effect upon the clinical severity of EAE [32]. In this study, CD4+ T cells were shown to be significantly decreased with the co-administration of both probiotic-like strains. A third study by Kwon et al. [33] demonstrated that providing a probiotic mix (IRT5) also decreased the clinical score of the EAE mice, as well as the incidence. Furthermore, the administration of the IRT5 mix resulted in an increase in the number of regulatory T cells in the spinal cord and the reduction of IL-17 producing CD4+ T cells and interferon-γ-positive spinal cord CD4+ T cells.
The above studies on MS and its model demonstrate that changes in the microbiome can alter immune responses and thereby modulate the severity of disease; however, these studies do not prove that changes in the microbiome alone can cause MS. Other factors such as environmental factors will contribute the most to the development of MS.
ASD
A recent study of the microbiome and mycobiome of patients with ASD revealed no differences in the alpha diversity but significant differences in the beta diversity [24]. This analysis of 40 autistic subjects found a stark decrease in Prevotella in the patients with ASD versus neurotypical patients. This study also found that Clostridium cluster XVIII and Escherichia/Shigella positively correlated with constipation in the patients with ASD. Although this study had a small number of patients, it indicates that intestinal dysbiosis may be present in ASD. As such, further research is required to determine the level and degree to which the intestinal microbiome can affect the development of ASD beyond the genetic predisposing factors and environmental factors.
Of great interest are the reports in which certain bacterial groups in ASD are correlated with the production of neurotransmitters. In a study by Luna et al. [25], certain bacterial groups correlated with the change in the levels of tryptophan, serotonin, and a serotonin metabolite (5-hydroxyindoleacetic acid) in the supernatant of colon biopsies from patients with ASD [25], suggesting that the presence of these bacterial groups led to the altered production of these molecules of the serotonin pathway. Increased levels of Erysipelotrichaceae, Clostridium lituseburense, and Terrisporobacter were associated with decreased levels of tryptophan in the supernatant of the colon biopsy cultures. In contrast, higher levels of serotonin in the biopsy itself were correlated with the abundance of Lachnoclostridium bolteae, Lachnoclostridium hathewayi, and Flavonifractor plautii.
In addition, significant correlations between the production of cytokines and abundance of certain bacteria in the intestinal microbiota of patients with ASD was determined [25]. Specifically, a high abundance of C. lituseburense was correlated with increased levels of IL-12p70, IL-17A, IL-1α, IL-5, IL-6, interferon gamma-inducible protein-10, macrophage inflammatory protein-1α, macrophage inflammatory protein-1β, and vascular endothelial growth factor in the sera and IL-1RA in the supernatant of intestinal biopsies. Also, higher levels of IL-15, IL-9, IL-1β, and IL-7 were correlated with higher abundance of L. hathewayi. With sera, monocyte chemoattractant protein-1 was found to be increased in the patients with ASD with gastrointestinal disorders versus neurotypical controls with or without gastrointestinal disorders. This suggests that a high production of inflammatory cytokines and chemokines in the sera and intestine are associated with the different bacterial groups found in the microbiomes of patients with ASD.
ALS
Recent studies have revealed the important role that microglia play in the development of ALS. While a strong connection between the immune system and the neurological system in ALS has been demonstrated, how the dysbiosis in ALS affects (or is affected by) either the immune system or the neuronal system is not as clear. Few studies have been done on the intestinal microbiome of ALS; however, one study of the microbiome in ALS did come from the hSOD1G93A transgenic mouse model of ALS [34]. hSOD1G93A is a mutation of a human Cu, Zn superoxide dismutase that was associated with a familial case of ALS [35]. In that study, the ALS G93A mouse model developed a dysbiosis characterized by significant changes in beta diversity and not alpha diversity versus wild-type controls. The most significant alterations were reduced abundance in the butyrate-producing bacteria Butyrivibrio fibrisolvens, as well as Firmicutes, Peptostreptococcus, and Escherichia coli. Of interest is that the altered intestinal microbiome occurred before the onset of symptoms (2 months of age). The authors also detected an increase in intestinal permeability in the ALS G93 mice.
A later study evaluated the intestinal (fecal) microbiomes of 6 patients with ALS and compared them with those of 5 healthy controls [26]. In that study, changes in beta diversity were detected. Specifically, the genera Oscillibacter, Anaerostipes, and Lachnospiraceae were significantly reduced in the patients with ALS compared with the healthy controls, and the genus Dorea were increased. It is interesting to note that both Anaerostipes and Lachnospiraceae are butyrate producers, and as a consequence of their reduced levels in ALS, we can surmise that butyrate production was decreased in these patients with ALS versus healthy controls. To determine if the microbiomes of patients with ALS truly are producing significantly lower levels of butyrate as the mouse model and microbiome analysis of patients with ALS would suggest, more studies need to be done on larger numbers of patients with ALS.
In support of the theory that it is the decreased levels of butyrate that contribute to the pathogenesis of ALS, a study utilizing the G93A ALS mice demonstrated that the administration of sodium phenylbutyrate (PBA) significantly ameliorated gross spinal cord atrophy in the ALS mice [36]. In addition, reactive astrogliosis was also significantly reduced in the PBA-treated mice. However, it should be mentioned that animal models such as this one use high doses of PBA (100–900 mg/kg mouse) [37]. This assessment is based on the fact that the concentration of butyrate is approximately 1 to 32.6 g/kg of stool [37, 38]. If the concentration of butyrate in the stool is used as a guide for biologically relevant concentrations, then the typical amount of butyrate (PBA) injected into mice is much higher than we would expect to be in the gut. More studies need be done to determine what exactly the concentrations of butyrate within the intestine are and whether or not this concentration is variable within a day.
Sodium butyrate has also been shown to act as an anti-inflammatory agent for LPS-induced responses in primary microglia but not for proliferating microglial cells isolated from rats [39]. Butyrate can signal through GPR109a, which is not only expressed in T cells, but has also been found in microglia [40]. While the beneficial effects of butyrate have been shown in the colon, very few studies have probed its beneficial neurological effects. Restoration of the blood–brain barrier in GF mice was achieved when colonized with Clostridium tyrobutyricum. This restoration was due to an increase in the expression of the tight junction proteins, occludin, and claudin-5, suggesting a crosstalk amongst the intestinal metabolites, the brain, and the immune system [41]. Thus, observations from mouse models and human studies suggest that the decreased levels of butyrate-producing bacteria contribute to or are a factor of disease pathogenesis in ALS.
PD
A reduced abundance of Prevotellaceae and Lactobacillaceae was observed in patients with PD versus healthy controls [27, 28]. This reduction of Prevotellaceae and Lactobacillaceae, both anti-inflammatory in nature, could result in inflammatory conditions in PD. In another study, 19 patients with PD and 14 age-matched healthy controls were evaluated for colonic inflammation [42]. Transcript levels of proinflammatory cytokines, tumor necrosis factor-α, interferon-γ, and IL-6, measured from colonic biopsies were significantly increased in the colons of patients with PD. A correlated increase in glial markers, GFAP and Sox-10 mRNA, was also observed in the colon biopsies of patients with PD. Additionally, the proinflammatory cytokine mRNA levels and GFAP and Sox-10 mRNA levels were correlated with disease duration.
In addition to colonic inflammation, patients with PD have higher permeability of their gut than controls [15], and this permeability may arise as a result of a dysbiosis of the intestinal microbiota and/or other factors. Patients with PD also had significantly decreased levels of the SCFAs acetate, propionate, and butyrate in their feces [28]. These particular SCFAs are recognized by the intestinal immune system and promote the production of IgA, much of which is specific for microbiota [43]. The importance of the intestinal microbiota in stimulating the production of intestinal IgA is demonstrated in GF mice, as they produce almost no intestinal IgA [44]. Because patients with PD develop colitis and have a decreased production of SCFAs, it is possible that their production of intestinal IgA may also be decreased.
Also of interest are the studies that suggest that dysbiosis in PD could be modulating the nervous system. α-Synuclein is a highly conserved presynaptic protein that was originally associated with PD in 1997 [45], owing to the identification of a mutation in the α-synuclein gene in patients with PD who had a familial history of PD. This mutation leads to abnormal aggregation in both the brain (substantia nigra) and the gut of patients with PD [15, 46], with the level of α-synuclein aggregates in the intestine correlating with intestinal permeability [15]. Also of great interest is another study that was able to demonstrate that the α-synuclein aggregates in the gut appear years (as many as 8) before the development of motor symptoms [47]. This would suggest that the inflammation in the gut, possibly due to aggregation of α-synuclein, precedes the inflammation in the brain. If the inflammation of the gut does precede the inflammation in the brain, it would be of interest to determine if the dysbiosis of the gut precedes the aggregation of the α-synuclein in the gut or vice versa. A recent study showed that fecal microbes from patients with PD, when transplanted into mice that overexpress α-synuclein, can further impair motor function [29], which was associated with an increase in Proteus spp., Bilophila spp., and Roseburia sp., and reduced species of Lachnospiraceae, Rikenellaceae, and Peptostreptococcaceae. Further, the authors showed that gut microbiota is required for microglia activation via α-synuclein. The mouse model of PD upon treatment with a mixture of SCFAs showed microglia morphology similar to specific-pathogen-free mice. It should also be noted that patients with PD have increased levels of constipation and irritable bowel syndrome-like symptoms, both of which are associated with alterations in the intestinal microbiome [48–50]. However, the previously described fecal transfer mouse model would suggest that the PD microbiome enhances motor dysfunction (brain), as well as affects the enteric nervous system [29].
Therapies For Neurological Disorders Based on Targeting Microbiota
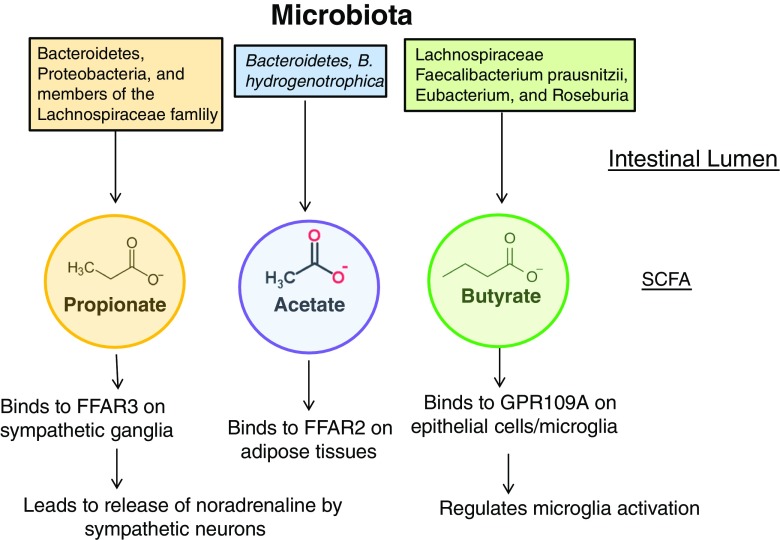
For those neurological disorders in which bacterial species clearly cause or exacerbate the neurological damage, such as Guillain-Barré syndrome and neuromyelitis optica, treatment with antibiotics could be used. However, as the numbers of antibiotic-resistant bacteria increase, this may not be a feasible solution in the future. Hence, research is currently being done on developing bacteriophages that can be used for eradicating pathogenic bacteria [51]. For those neurological disorders in which complex changes of the composition of the intestinal/oral microbiome occur, a more nuanced and complex approach will be necessary. This could be achieved by changing the diet, supplementing the microbiome with commensals of which the individual has low levels, and/or the use of generalized prebiotics and probiotics. Changing the diet of the individual and the use of prebiotics are essentially altering the composition by expanding certain groups of microbiota. Unfortunately, this is not a targeted approach, and there has been limited success with these types of approaches. A much more individualized and targeted approach would be to determine, through next-generation sequencing, which bacterial groups the individual is replete in and then providing a supplement to expand that beneficial bacterial group. Examples of this include the use of P. histicola to treat those patients that have low levels of Prevotella species that are beneficial for the health of patients [52]. Administration of P. histicola to a mouse model of autoimmune disease has demonstrated that this course of treatment can be successful [53]. In addition, the administration of SCFAs has also been successful in mouse models of neurological disorders (Fig. 1). In conclusion, research on how the microbiome interacts with both the brain and the immune system is revealing the complexities of neurological disorders, and research on the use of individualized microbiome targeted therapies for neurological diseases holds great promise.
Fig. 1.
Effect of bacterial metabolites upon the host. The intestinal lumen is depicted at the top of the diagram, along with some of the taxa that produce the 3 main bacterial metabolites. These metabolites directly stimulate cells of the host via binding to receptors on epithelial cells and immune cells. Depicted at the bottom are some examples of receptors that the metabolites can bind to, as well as examples of cells/tissue that express these receptors. Dysbiosis causing alterations in the metabolites can impact immune system and neurotransmitters produced by the microbes. SCFA = short-chain fatty acid; FFAR = free fatty acid receptor; GPR = G protein-coupled
Electronic supplementary material
(PDF 498 kb)
Acknowledgements
VT is supported by a grant from Department of Defense and Center of Individualized Medicine, Mayo Clinic.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Taneja V. Arthritis susceptibility and the gut microbiome. FEBS Lett. 2014;588(22):4244–4249. doi: 10.1016/j.febslet.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy CC, Kien CL, Bouthillier L, Levy E. Short-chain fatty acids: ready for prime time? Nutr Clin Pract. 2006;21(4):351–366. doi: 10.1177/0115426506021004351. [DOI] [PubMed] [Google Scholar]
- 3.Vrieze A, Holleman F, Zoetendal EG, de Vos WM, Hoekstra JB, Nieuwdorp M. The environment within: how gut microbiota may influence metabolism and body composition. Diabetologia. 2010;53(4):606–613. doi: 10.1007/s00125-010-1662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 5.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10(2):131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 6.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 7.Rosser EC, Mauri C. A clinical update on the significance of the gut microbiota in systemic autoimmunity. J Autoimmun. 2016;74:85–93. doi: 10.1016/j.jaut.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Luczynski P, McVey Neufeld KA, Oriach CS, Clarke G, Dinan TG, Cryan JF. Growing up in a bubble: using germ-free animals to assess the influence of the gut microbiota on brain and behavior. Int J Neuropsychopharmacol. 2016;19(8). [DOI] [PMC free article] [PubMed]
- 9.Wostmann BS, Pleasants JR, Bealmear P, Kincade PW. Serum proteins and lymphoid tissues in germ-free mice fed a chemically defined, water soluble, low molecular weight diet. Immunology. 1970;19(3):443–448. [PMC free article] [PubMed] [Google Scholar]
- 10.Reigstad CS, Salmonson CE, Rainey JF, 3rd, et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29(4):1395–1403. doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins J, Borojevic R, Verdu EF, Huizinga JD, Ratcliffe EM. Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol Motil. 2014;26(1):98–107. doi: 10.1111/nmo.12236. [DOI] [PubMed] [Google Scholar]
- 12.Obata Y, Pachnis V. The effect of microbiota and the immune system on the development and organization of the enteric nervous system. Gastroenterology. 2016;151(5):836–844. doi: 10.1053/j.gastro.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramer-Quinn DS, Baker RA, Sanders VM. Activated T helper 1 and T helper 2 cells differentially express the beta-2-adrenergic receptor: a mechanism for selective modulation of T helper 1 cell cytokine production. J Immunol. 1997;159(10):4857–4867. [PubMed] [Google Scholar]
- 14.Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front Cell Neurosci. 2015;9:392. doi: 10.3389/fncel.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsyth CB, Shannon KM, Kordower JH, et al. Increased intestinal permeability correlates with sigmoid mucosa alpha-synuclein staining and endotoxin exposure markers in early Parkinson's disease. PLOS ONE. 2011;6(12):e28032. doi: 10.1371/journal.pone.0028032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6(2):111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu WH, Chuang HL, Huang YT, et al. Alteration of behavior and monoamine levels attributable to Lactobacillus plantarum PS128 in germ-free mice. Behav Brain Res. 2016;298(Pt B):202–209. doi: 10.1016/j.bbr.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 18.Nohr MK, Pedersen MH, Gille A, et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154(10):3552–3564. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 19.Marathe CS, Rayner CK, Jones KL, Horowitz M. Effects of GLP-1 and incretin-based therapies on gastrointestinal motor function. Exp Diabetes Res. 2011;2011:279530. doi: 10.1155/2011/279530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soret R, Chevalier J, De Coppet P, et al. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology. 2010;138(5):1772–1782. doi: 10.1053/j.gastro.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Chia N, Kalari KR, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jangi S, Gandhi R, Cox LM, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cekanaviciute E, Yoo BB, Runia TF, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114(40):10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strati F, Cavalieri D, Albanese D, et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5(1):24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luna RA, Oezguen N, Balderas M, et al. Distinct microbiome–neuroimmune signatures correlate with functional abdominal pain in children with autism spectrum disorder. Cell Mol Gastroenterol Hepatol. 2017;3(2):218–230. doi: 10.1016/j.jcmgh.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang X, Wang X, Yang S, et al. Evaluation of the microbial diversity in amyotrophic lateral sclerosis using high-throughput sequencing. Front Microbiol. 2016;7:1479. doi: 10.3389/fmicb.2016.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheperjans F, Aho V, Pereira PA, et al. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov Disord. 2015;30(3):350–358. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 28.Unger MM, Spiegel J, Dillmann KU, et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson's disease and age-matched controls. Parkinsonism Relat Disord. 2016;32:66–72. doi: 10.1016/j.parkreldis.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Sampson TR, Debelius JW, Thron T, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. 2016;167(6):1469–1480. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl. 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangalam A, Shahi SK, Luckey D, et al. Human gut-derived commensal bacteria suppress CNS inflammatory and demyelinating disease. Cell Rep. 2017;20(6):1269–1277. doi: 10.1016/j.celrep.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salehipour Z, Haghmorad D, Sankian M, et al. Bifidobacterium animalis in combination with human origin of Lactobacillus plantarum ameliorate neuroinflammation in experimental model of multiple sclerosis by altering CD4+ T cell subset balance. Biomed Pharmacother. 2017;95:1535–1548. doi: 10.1016/j.biopha.2017.08.117. [DOI] [PubMed] [Google Scholar]
- 33.Kwon HK, Kim GC, Kim Y, et al. Amelioration of experimental autoimmune encephalomyelitis by probiotic mixture is mediated by a shift in T helper cell immune response. Clin Immunol. 2013;146(3):217–227. doi: 10.1016/j.clim.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Jaarsma D, Haasdijk ED, Grashorn JA, et al. Human Cu/Zn superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiol Dis. 2000;7(6 Pt B):623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- 35.Gurney ME, Pu H, Chiu AY, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264(5166):1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 36.Ryu H, Smith K, Camelo SI, et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. 2005;93(5):1087–1098. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- 37.Stilling RM, van de Wouw M, Clarke G, Stanton C, Dinan TG, Cryan JF. The neuropharmacology of butyrate: the bread and butter of the microbiota–gut–brain axis? Neurochem Int. 2016;99:110–132. doi: 10.1016/j.neuint.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y, Fan C, Li P, Lu Y, Chang X, Qi K. Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci Rep. 2016;6:37589. doi: 10.1038/srep37589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huuskonen J, Suuronen T, Nuutinen T, Kyrylenko S, Salminen A. Regulation of microglial inflammatory response by sodium butyrate and short-chain fatty acids. Br J Pharmacol. 2004;141(5):874–880. doi: 10.1038/sj.bjp.0705682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thangaraju M, Cresci GA, Liu K, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69(7):2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braniste V, Al-Asmakh M, Kowal C, et al. The gut microbiota influences blood–brain barrier permeability in mice. Sci Transl Med. 2014;6(263):263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devos D, Lebouvier T, Lardeux B, et al. Colonic inflammation in Parkinson's disease. Neurobiol Dis. 2013;50:42–48. doi: 10.1016/j.nbd.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Wu W, Sun M, Chen F, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017;10(4):946–956. doi: 10.1038/mi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crabbe PA, Bazin H, Eyssen H, Heremans JF. The normal microbial flora as a major stimulus for proliferation of plasma cells synthesizing IgA in the gut. The germ-free intestinal tract. Int Arch Allergy Appl Immunol. 1968;34(4):362–375. doi: 10.1159/000230130. [DOI] [PubMed] [Google Scholar]
- 45.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 46.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 47.Hilton D, Stephens M, Kirk L, et al. Accumulation of alpha-synuclein in the bowel of patients in the pre-clinical phase of Parkinson's disease. Acta Neuropathol. 2014;127(2):235–241. doi: 10.1007/s00401-013-1214-6. [DOI] [PubMed] [Google Scholar]
- 48.Mertsalmi TH, Aho VTE, Pereira PAB, et al. More than constipation—bowel symptoms in Parkinson's disease and their connection to gut microbiota. Eur J Neurol. 2017;24(11):1375–1383. doi: 10.1111/ene.13398. [DOI] [PubMed] [Google Scholar]
- 49.Cao H, Liu X, An Y, et al. Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci Rep. 2017;7(1):10322. doi: 10.1038/s41598-017-10835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shukla R, Ghoshal U, Dhole TN, Ghoshal UC. Fecal microbiota in patients with irritable bowel syndrome compared with healthy controls using real-time polymerase chain reaction: an evidence of dysbiosis. Dig Dis Sci. 2015;60(10):2953–2962. doi: 10.1007/s10620-015-3607-y. [DOI] [PubMed] [Google Scholar]
- 51.Dalmasso M, Strain R, Neve H, et al. Three new Escherichia coli phages from the human gut show promising potential for phage therapy. PLOS ONE. 2016;11(6):e0156773. doi: 10.1371/journal.pone.0156773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Wright K, Davis JM, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8(1):43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marietta EV, Murray JA, Luckey DH, et al. Suppression of Inflammatory Arthritis by Human Gut-Derived Prevotella histicola in humanized mice. Arthritis Rheumatol. 2016;68(12):2878–2888. doi: 10.1002/art.39785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 498 kb)