Abstract
Accumulating evidences have illuminated that an amount of microRNAs are involved in human diseases including hepatocellular carcinoma (HCC). In this study, we found that the expression of miR-382 in HCC tissues was down-regulated compared with the non-cancerous tissues. Over-expression of miR-382 could significantly inhibit the migration and invasion of HCC cells in vitro and in vivo. Bioinformatic algorithms and luciferase reporter assays suggested that Golgi Membrane Protein 1 (GOLM1) was a direct target of miR-382. Interestingly, we found the down-regulation of GOLM1 in HCC cells could rescue these cells from miR-382-mediated suppression of migration and invasion. Our findings might demonstrate that miR-382 inhibited the metastasis of HCC by targeting GOLM1. Furthermore, cox proportional hazards analyses suggested that low expression of miR-382 was an independent prognostic factor for the HCC patients. In conclusion, our results highlighted that miR-382, a novel prognostic factor, target GOLM1 to inhibit metastasis of hepatocellular carcinoma.
Keywords: HCC, miR-382, migration, invasion, prognosis
Introduction
Hepatocellular carcinoma (HCC) has an extremely poor prognosis and it is the fifth most common cancer in men and the ninth in women worldwide [1,2]. It is also the second leading cause of cancer death in men and the sixth in women worldwide [3,4]. Although there has been a remarkable progress in the treatment of liver cancer, recurrence and metastasis still claim the lives of countless HCC patients annually [5,6].
MicroRNAs (miRNAs) are defined as small non-coding RNAs with approximately 19-25 nucleotides that participate in gene modulation via binding to the 3’-untranslated region (3’-UTR) of their target mRNAs, leading to mRNA transinhibition or mRNA degradation [7,8]. Through the attenuation of target mRNA, miRNA acts as a significant role at the posttranscriptional level and contributes to the regulation of diverse biological processes, including cellular differentiation, growth, proliferation, and apoptosis [8-10]. In particular, miRNAs are able to act either as tumor promoters or as tumor suppressors via targeting the given anti-oncogenes or oncogenes [11]. In the research field of HCC, previous studies have suggested the a great deal of miRNAs are involved in the pathogenesis of HCC including miR-331-3p, miR-26a-5p, miR-197, miR-484, and miR-1299 [12-16]. However, the exact role of miR-382 in HCC still remains elusive.
GOLM1, a novel defined oncogene, has been demonstrated as one of the leading genes correlated with the development of HCC [17,18]. Growing evidences indicate that GOLM1 is correlated with early recurrence, distant metastasis, and poor prognosis of HCC patients via selectively interacting with epidermal growth factor receptor (EGFR) and serveing as a specific cargo adaptor to assist EGFR/RTK anchoring on the trans-Golgi network (TGN). However, how the expression of GOLM1 is modulated in HCC cells still remains unknown.
In the current study, we found that miR-382 was significantly downregulated in HCC tissues and cells compared with the nontumorous tissues and cells. The expression levels of miR-382 were demonstrated to be related with the specific clinical characteristics including vascular invasion and Edmondson grade. Then, by performing a series of experiments in vitro and vivo, we found that miR-382 may act as a significant role in the development of HCC by inhibiting metastasis of HCC through targeting GOLM1. Furthermore, Kaplan-Meier analysis and Cox proportional regression analysis indicated that the low expression of miR-382 might be a potential biomarker for predicting the survival of HCC patients.
Materials and methods
Clinical sample
113 paired fresh HCC tissues, corresponding adjacent noncancerous tissues and related clinical information were obtained from the patients undergoing hepatectomy at Shandong provincial hospital affiliated to Shandong University. The study was approved by our Institutional Ethics Committee. All patients provided their written informed consent to participate in this study. The fresh tissue samples which were confirmed by the histopathological examination were collected in the operating room and processed immediately. Each sample was frozen and stored at liquid nitrogen.
Cell culture
The HepG2, SNU423, LM3, Hep3B, 97H, 97L, Huh7 human hepatoma cell lines and the human normal L02 cell line used in this study were obtained from KeyGen (Nanjing KeyGen Biotech Co. Ltd, China). All of the cells were cultured in DMEM medium (GIBCO, Carlsbad, USA) pre-treated with 10% fetal bovine serum, 80 U/ml of penicillin sodium at 37°C in humidified air containing 5% carbon dioxide.
Quantitative RT-PCR
Trizol reagent (Sigma) was used to isolate the total RNA from tissues and cells. Expression of miR-382 was determined using MicroRNA First-Strand Synthesis and miRNA Quantitation kits (Takara, Dalian, China) according to the manufacturer’s instructions. The Ct values of U6 and GAPDH were used as the internal control to normalize the relative expression of miR-382 and GOLM1, respectively.
Cell transfection
Firstly, HCC cells (1×105) were implanted in 6-well plates. One day after the implantation, the cells were transfected with 25 nM of miR-382 mimic, anti-miR-382 inhibitor and the negative control (GenePharma, Shanghai, China), using Lipofectamine 2000 reagent (GIBCO, Carlsbad, USA) according to the manufacturer’s protocol. Meanwhile, all miR-382 and GOLM1 Ectopic expression and knockdown lentivirus as well as their negative control lentivirus were purchased from GeneChem (Shanghai, China). All lentiviral vectors expressed enhanced green fluorescent protein (GFP), the expression of miR-382 and GOLM1 in the treated cells was confirmed by realtime-PCR 48 h after the transfection.
Target prediction for microRNAs
Bioinformatics prediction of target genes and miRNA binding sites was performed using different databases: TargetScan (http://www.targetscan.org/), miRanda (http://www.microrna.org/). The overlapping target molecule for miRNA-382 from these databases was GOLM1 and was considered for further experimental analysis.
In vivo assay
BALB/C nude mice underwent anesthesia under ketamine (100 mg/kg, i.p.) and xylazine (20 mg/kg, i.p.). Pre-treated cells (Hep-G2-miR-382-ovexpression, Huh7-miR-382-downregulation and the control cells) were respectively suspended in 200 μl PBS and filtered through a sterile 70-μm nylon mesh filter (BD Falcon, NJ, USA) to form a single cell suspension. Then cells were injected into the tail vein of nude mice to establish peripheral intravascular implanted models (8 in each group). Mice were sacrificed after 6 weeks to check tumor metastasis in the pulmonary. Lung tissues were examined by H-E staining to identify the number of tumors.
Western blotting
With the use of RIPA buffer containing PMSF (Beyotime, Nantong, China), proteins were extracted from tissues or cells. The same amount of protein loading in each lane was determined by the GAPDH. The antibodies were used according to the manufacturer’s instructions, and were purchased from Santa Cruz Biotechnology (Santa Cruz, USA). After using the secondary antibodies (Santa Cruz, USA), the signals were detected by SuperSignal Chemiluminescent Substrate kit (Pierce, Rockford, USA) according to manufacturer’s instructions. The integrated density of the band was confirmed by the ImageJ software (NIH, Bethesda, MD, USA).
Migration and invasion assays
Cellular migration and invasion abilities were examined in 24-well plates using Millicell tissue culture plate well inserts (Millipore, Bedord, MA) for 12 hours and BD BioCoatMatrigel Invasion Chambers (Becton Dickson, Mountain View, CA) for 20 hours, respectively. 5×104 HCC cells were implanted in the serum-free medium inside the inserts and the 10% FBS medium was added in the lower chamber. Firstly, the HCC cells at the bottom of the membrane were fixation with methanol, then we stained the cells with 0.005% crystal violet in PBS for 1 hour, finally the number of migrated or invaded cells was confirmed by a microscope.
Proliferation assay
Cell proliferation ability was tested by using the Cell Counting Kit-8 (Nanjing KeyGen Biotech Co. Ltd, China). Firstly, the pre-transfected HCC cell lines were implanted into 96-well plates with the amount of 1×104 cells per well. 10 μl of the kit reagent was added to each well at 0, 24, 48, 72 h after implantation. The cell proliferation value of the plate was determined based on the resulting OD value.
Luciferase assay
DNA sequences containing the miR-382 binding site on the 3’-UTR of GOLM1 were cloned into the downstream of the firefly luciferase stop codon in a pmirGLO control vector (Promega, Milan, Italy). Huh7 cells were implanted in 24 well plates. The following day, the cells were respectively co-transfected with 25 nM of miR-382 mimic, miR-382 inhibitor or miR-NC (GenePharma, Shanghai, China) and 500 ng of 3’-UTR (WT or Mut) of GOLM1 pmirGLO recombinant vectors. Then the cells were harvested 24 h after transfection. Firefly luciferase activity was detected with Dual Luci-ferase Assay (Promega, Milan, Italy) in accordance with the manufacturer’s instructions.
Statistical analysis
All experimental assays were performed independently in triplicate. two-tailed Student t test was used to assess the statistical differences between groups. All statistical data were carried out using Statistical Program for Social Sciences 18.0 software (SPSS, USA) and presented with Graphpad prism 5.0 (GraphPad Software, CA). P value less than 0.05 was considered as significant.
Results
MiR-382 was aberrantly down-regulated in HCC tissues and cell lines
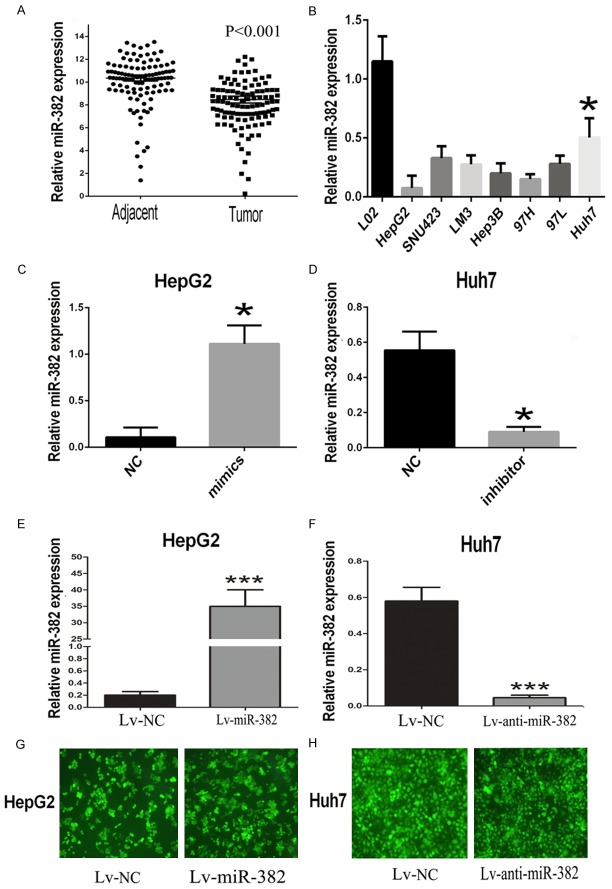
By utilizing realtime-PCR, the expression of miR-382 in 113 pairs of human HCC tissues was found to be obviously restrained in comparison with the corresponding adjacent tissues (Figure 1A). Similarly, we checked the expression of miR-382 in human L02 normal liver cells and seven human HCC cell lines: HepG2, SNU423, LM3, Hep3B, MHCC97H, MHCC97L, Huh7. The results showed that miR-382 was down-regulated in the HCC cells compared with the human normal liver cells (Figure 1B). Furthermore, we divided 113 tissue samples of HCC patients into two groups: miR-382 low-expression group (n=56) and high-expression group (n=57) based on the expression levels of miR-382. Then the Chi-square analysis was performed to explore the potential correlations between miR-382 and the clinical characteristics, the findings suggested that miR-382 expression was significantly related with the vascular invasion and Edmondson grade, which might imply the abnormal expression of miR-382 contributed to the development of HCC (Table 1).
Figure 1.
The aberrant expression of miR-382 in HCC tissues and cells. A. The expression of miR-382 in 113 pairs of human HCC tissues were demonstrated to be significantly down-regulated compared with the the corresponding adjacent tissues. B. We further confirmed the miR-382 expression was down-regulated in the HCC cell lines in comparison with the human normal L02 cells. C, D. By using the transfection with miR-382 mimic and inhibitor, we respectively up-regulated the expression of miR-382 in HepG2 and down-regulated the miR-382 in Huh7. E. We transfected HepG2 with the ectopic expressions lentivirus of miR-382 to perform further in vivo assay. F. We also transfected Huh7 with the knockdown lentivirus of miR-382 to perform further in vivo assay. G, H. All lentiviral vectors expressed enhanced green fluorescent protein, we used the fluorescence microscope to test the transfection efficiency of the lentivirus in HepG2 and Huh7. Data are presented as means ± SEM. (*P<0.05, ***P<0.0001)
Table 1.
Correlation between miR-382 expression and clinicopathological characteristics of HCC patients (n=113)
| Characteristics | All Patients | miR-382 low expression (< Mediana) | miR-382 high expression (≥ Mediana) | p Chi-squared test p-value |
|---|---|---|---|---|
| No. | 113 | 56 | 57 | |
| Age (years) | ||||
| < 60 | 87 | 41 | 46 | 0.344 |
| ≥ 60 | 26 | 15 | 11 | |
| Gender | ||||
| Male | 89 | 46 | 43 | 0.384 |
| Female | 24 | 10 | 14 | |
| HbeAg | ||||
| Negative | 34 | 16 | 18 | 0.727 |
| Positive | 79 | 40 | 39 | |
| Cirrhosis | ||||
| Absent | 36 | 16 | 20 | 0.457 |
| Present | 77 | 40 | 37 | |
| ALT (U/L) | ||||
| ≤ 45 | 39 | 18 | 21 | 0.599 |
| > 45 | 74 | 38 | 36 | |
| AFP (ng/ml) | ||||
| ≤ 13.6 | 33 | 15 | 18 | 0.575 |
| > 13.6 | 80 | 41 | 39 | |
| Tumor size (cm) | ||||
| ≤ 5 | 38 | 22 | 16 | 0.207 |
| > 5 | 75 | 34 | 41 | |
| Vascular invasion | ||||
| Absent | 87 | 38 | 49 | 0.022* |
| Present | 26 | 18 | 8 | |
| Edmondson grade | ||||
| I + II | 71 | 22 | 49 | 0.000* |
| III + IV | 42 | 34 | 8 |
The median expression level of miR-382 was used as the cutoff.
Indicates p value <0.05.
MiR-382 inhibited metastasis of HCC cells both in vitro and vivo
Based on the above results, we furthered our research by up-regulating the expression of miR-382 in HepG2. The expression of miR-382 was checked by using realtime-PCR 48 hours after transfection. The results showed miR-382 was obviously overexpressed in HepG2 after the transfection with miR-382 mimic compared with the miR-NC transfected HepG2 (Figure 1C). Then a series of functional assays were performed to explore the role of miR-382 in HCC. Results of transwell invasion assays showed that miR-382 overexpression could prevent the invasive ability of HepG2 cells compared with the miR-NC groups (Figure 2A). In order to further verify the effects of miR-382 in HCC, Huh7, which has a relative high miR-382 expression, was transfected with miR-382 inhibitor to prevent the miR-382 expression (Figure 1D). Interestingly, transwell invasion assays showed that down-regulation of miR-382 could promote the invasion of Huh7 cells compared with the control (Figure 2A). Interestingly, the transwell migration assays also suggested that, compared with the miR-382 low-expression groups, less number of HCC cells in miR-382 high-expression groups moved through the high-magnification migration chamber (Figure 2B). Taken together, our results from the transwell assays confirmed miR-382 could restrain the migratory and invasive ability of human HCC cells. To further confirm the inhibiting effect of metastasis induced by miR-382, the tail vein xenograft models were built. By transfecting with the ectopic expression and knockdown lentivirus, we respectively overexpressed the miR-382 expression in HepG2 and suppressed the expression of miR-382 in Huh7 (Figure 1E-H). Then the treated cells were injected into the nude mice from the tail vein. 42 days after the injection, we found more lung metastases were formed in the mice injected with the miR-382 low-expression cells compared with the mice groups with miR-382 high-expression (Figure 2C and 2D), and the results were confirmed by the histological examination (Figure 2E and 2F).
Figure 2.
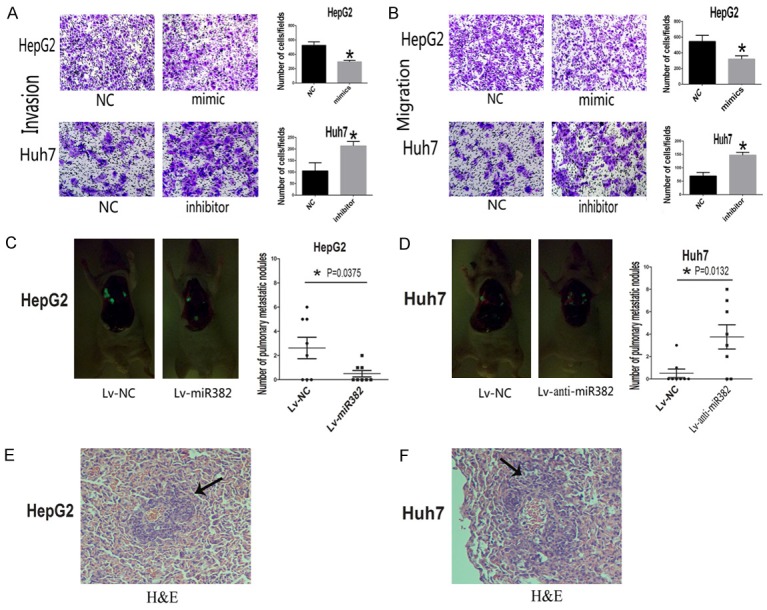
MiR-382 prevented metastasis both in vitro and vivo. A. The results of transwell invasion assay showed that overexpression of miR-382 could prevent the invasive ability of HCC cells compared with the miR-382 low expression groups. B. Up-regulation of miR-382 could also inhibit the migration in HCC cells. C, D. The results of tail vein xenograft models further confirmed the inhibitory effects of miR-382 on the metastasis of HCC cells, more lung metastases were formed in the mice injected with the miR-382 low-expression cells compared with the mice of miR-382 high-expression groups. E, F. Histological examination showed the pathological features of the lung metastases. Data are presented as means ± SEM. (*P<0.05).
MiR-382 had no effects on proliferation, apoptosis and cell cycle
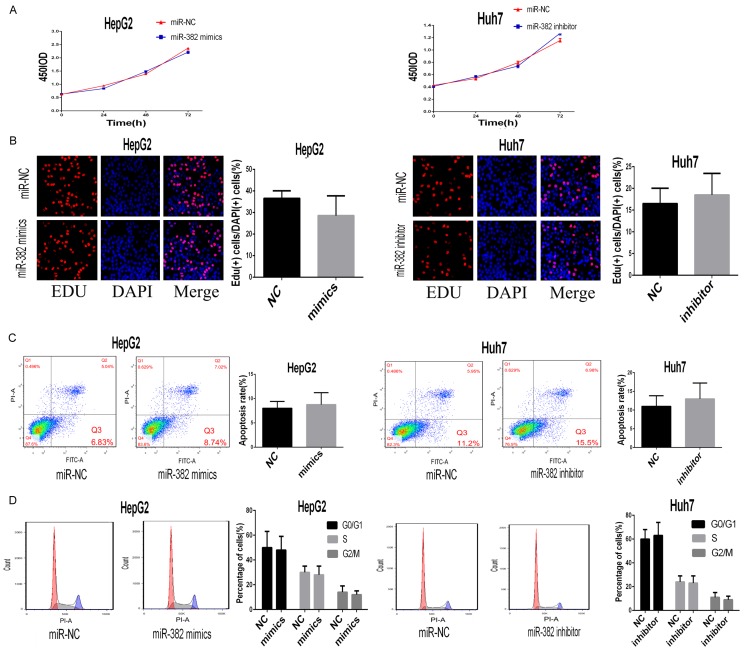
To further explore the effects of miR-382 on HCC, we also performed EDU assays, apoptosis assays and cell cycle assays by using the pre-treated HCC cells. However, we found miR-382 had no significant effects on proliferation, apoptosis and cell cycle of HCC cells (Figure 3A-D).
Figure 3.
MiR-382 had no effects on proliferation, apoptosis and cell cycle. A, B. According to results of CCK8 and EDU assay, we found miR-382 had no effects on proliferation in vitro. C. By using apoptosis assay, we miR-382 could not affect the apoptosis rate of the treated cells compared with the control. D. The results from the cell cycle assay indicated that miR-382 had no effects on the cell cycle of HCC cells. Data are presented as means ± SEM.
GOLM1 was a direct downstream target of miR-382
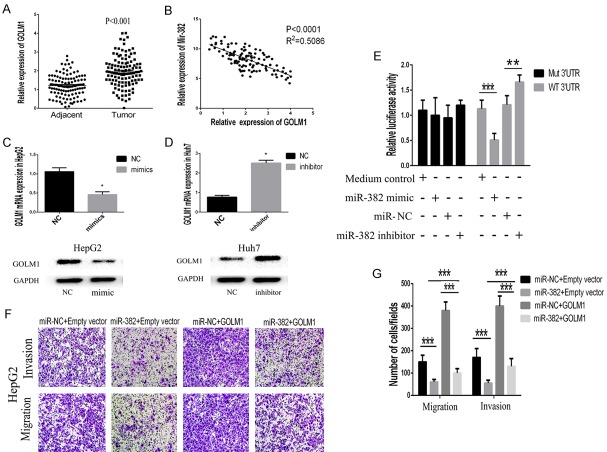
To identify which target gene was responsible for the effects of miR-382 on HCC metastasis, the target genes of miR-382 were predicted by using two different Bioinformatics databases: Target Scan (http://www.targetscan.org) and miRanda (http://www.microrna.org/). The prediction results suggested the overlapping target molecule for miRNA-382 was GOLM1 whose 3’-UTR contained a conserved putative target site for miR-382. Meanwhile, GOLM1 was found to be obviously increased in HCC tissues compared with the control (Figure 4A). With the use of Pearson correlation analysis, we found the negatively correlations between the expression of miR-382 and GOLM1 (Figure 4B). Therefore, we constructed luciferase vectors carrying the 3’-UTR of GOLM1 behind the firefly luciferase gene coding region. As shown in Figure 4E, miR-382 mimic significantly suppressed luciferase activity of GOLM1 containing a wild-type 3’-UTR, but showed no effect on activity of GOLM1 with a mutant 3’-UTR, meanwhile miR-382 inhibitor could promote the luciferase activity of GOLM1. Additionally, real-time PCR and western blot also confirmed the miR-382 could suppress GOML1 expression in HCC cells (Figure 4C and 4D).
Figure 4.
MiR-382 inhibited cell invasion and migration via targeting GOLM1. A. We demonstrated that GOLM1 was significantly increased in HCC tissues compared with the control. B. By utilizing the Pearson correlation analysis, we found miR-382 expression was negatively correlated with the expression of GOLM1 in HCC tissues. C, D. The findings from the realtime-PCR and western blot suggested that overexpression of miR-382 could suppress the expression of GOLM1 in HCC cells. E. MiR-382 mimic significantly suppressed luciferase activity of GOLM1 containing a wild-type 3’-UTR, but showed no effect on activity of GOLM1 with a mutant 3’-UTR, meanwhile treatment with miR-382 inhibitor increased luciferase activity of GOLM1. Collectively, the results above indicated GOLM1 was a direct target gene of miR-382. F, G. By performing transwell invasion and migration assay, the results showed that up-regulation of GOLM1 could partially neutralize the inhibiting effects of miR-382 on migration and invasion in HepG2. Data are presented as means ± SEM. (*P<0.05, **P<0.01, ***P<0.0001).
MiR-382 inhibited cell invasion and migration via targeting GOLM1
To clarify the role of GOLM1 underlying the miR-382-mediated effects in HCC cells, we respectively transfected the cells of miR-382 overexpression groups and control groups with GOLM1-expressing lentivirus. By performing transwell invasion and migration assays, we found GOLM1 overexpression could partially neutralize the inhibiting effects of miR-382 on migration and invasion in HepG2 (Figure 4F and 4G). Our findings demonstrated that the inhibitory effects of miR-382 on migration and invasion of human HCC cells could be partially counteracted by the up-regulation of GOLM1. Our findings might illuminate miR-382 could be a significant upstream molecule which modulated the expression of GOLM1.
Low expression of miR-382 was significantly associated with prognosis of HCC
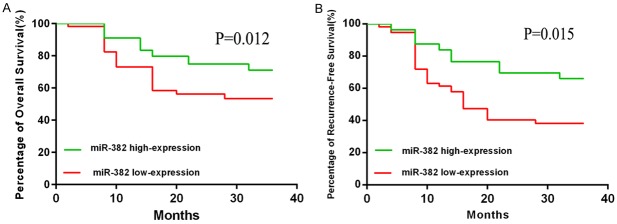
We performed Kaplan-Meier analysis to examine whether miR-382 was related with overall survival (OS) and relapse-free survival (RFS) of the HCC patients (Figure 5A and 5B). 113 HCC samples were divided into miR-382 high expression group and low expression group, median was used as cut off. The findings of Kaplan-Meier analyses showed HCC patients with low-expression of miR-382 had a better RFS and OS than the high-expression group. The difference was statistically significant. In addition, we performed the Cox proportional regression analyses and the results showed that miR-382 was an independent prognostic factor for the HCC patients (adjusted hazard ratio (AHR): 0.216 [95% Confidence Interval (CI): 0.121-0.401]; P¼=0.027) (Table 2).
Figure 5.
miR-382 was significantly associated with prognosis of HCC. The findings of Kaplan-Meier analysis indicated that patients with low expression of miR-382 had a better OS and RFS than the miR-382 high expression group. The difference was statistically significant. Data are presented as means ± SEM.
Table 2.
Cox proportional hazards analyses
| Clinical factor | P value | Univariable analysis | |
|
| |||
| HR | 95% CI | ||
|
| |||
| MiR-382 | 0.027* | 0.216 | 0.121-0.401 |
| Age | 0.429 | 1.529 | 0.746-3.069 |
| Gender | 0.248 | 0.541 | 0.301-1.423 |
| TNM | 0.001 | 13.064 | 3.133-32.466 |
| AFP | 0.237 | 1.517 | 0.576-2.615 |
| HBV/HCV infection | 0.467 | 1.319 | 0.598-2.231 |
| Vascular invasion | 0.001 | 4.115 | 1.819-13.394 |
|
| |||
| Clinical factor | P value | Multivariable analysis | |
|
| |||
| AHR | 95% CI | ||
|
| |||
| MiR-382 | 0.002* | 0.316 | 0.293-0.776 |
| Age | 0.399 | 1.416 | 0.655-2.615 |
| Gender | 0.219 | 1.377 | 0.549-2.717 |
| TNM | 0.000 | 17.361 | 4.358-21.387 |
| AFP | 0.154 | 0.316 | 0.216-1.231 |
| HBV/HCV infection | 0.775 | 0.698 | 0.357-1.229 |
| Vascular invasion | 0.000* | 10.322 | 8.265-12.651 |
Indicates P<0.05.
HR: Hazard ratio. AHR: Adjusted hazard ratio. CI: Confidence interval.
Discussion
Hepatocellular carcinoma, the fifth most common cancer worldwide, is confirmed to be a severe threat to public health [5]. Despite there has been a huge progress in the development of modern medical treatments for HCC, metastasis and recurrence still result in poor prognosis of countless HCC patients [19,20]. Hence, identifying the sensitive biomarkers for early detection and exploring their potential crucial role in the development of HCC are desirable and urgently needed. MicroRNAs are defined as a kind of small conserved RNA molecules, approximately 19-25 nucleotides in length [21]. On account of their crucial roles in physiology and pathology of human, more and more attentions have been paid on miRNAs in the past decade. Increasing evidences have showed that miR-382 acts as a tumor suppressor and is involved in the pathogenesis of various malignancies including osteosarcoma, ovarian cancer and colorectal cancer [22-24]. For instance, Xu et al have reported that miR-382 is able to function as the tumor suppressor in osteosarcoma. And the up-regulation of miR-382 is a potential effective treatment to suppress tumor metastasis and inhibit CSC-induced relapse in osteosarcoma [23]. Moreover, Tan et al in their study find miR-382 prevents invision and migration via binding the 3’-UTR of ROR1 through modulating the EMT pathway in ovarian cancer [22]. However, the specific role of miR-382 in the pathogenesis of HCC and the underlying mechanisms still remain elusive.
In this study, we foucused on the effects of miR-382 on the malignant phenotype of HCC cells including proliferation, invasion, migration, apoptosis and cell cycle. The results suggested the expression of miR-382 was significantly down-regulated in tumor tissues compared with the corresponding adjacent nontumorous tissues. Furthermore, we confirmed the expression of miR-382 was inhibited in HCC cells. These findings indicated the abnormal expression of miR-382 has the potential for the prediction of HCC risk and early detection of HCC. Then the Chi-square analysis was performed to explore the correlations between the aberrant expression of miR-382 and the clinical characteristics of HCC patients. The results suggested that the expression level of miR-382 significantly associated with the tumor metastasis and the Edmondson grade. The above results implied the potential effects of miR-382 on the development of HCC, so we transfected the HepG2 cells which has a relative low expression of miR-382 with miR-382 mimic to over-express miR-382 and the Huh7 which has a relative high expression with miR-382 inhibitor to downregulate the expression of miR-382, both the two cells were respectively transfected with miR-NC. Based on the well prepared cells above, a series of functional assays including invasion, migration, proliferation, apoptosis and cell cycle were performed in the two human HCC cell lines. The results from the functional assays showed that miR-382 inhibited invasion and migration of HCC cells but had no effect on the proliferation, apoptosis and cell cycle. We also performed the tail vein xenograft model and the results confirmed the inhibitory effects of miR-382 on metastasis. Collectively, the findings we attained demonstrated miR-382 was a potential tumor suppressor of HCC.
The results above have preliminarily demonstrated miR-382 could act as a crucial role in the development of HCC, however, the precise mechanisms underlying its effects were still unknown. By utilizing a bioinformatics prediction programme, we found that the 3’-UTR of GOLM1 contained a conserved putative target site for miR-382. Interestingly, GOLM1 was reported to be a leading gene relating to HCC metastasis. Specifically, GOLM1 was demonstrated to be correlated with early recurrence, metastasis, and poor survival of HCC patients. Both gain- and loss-of-function studies evaluated that GOLM1 could act as a key oncogene by promoting HCC growth and metastasis. Coincidentally, the influences of GOLM1 on HCC seemed to associate with the effects of miR-382, so we tried to explore the potential correlations between miR-382 and GOLM1. With the use of Pearson correlation analysis, we found the miR-382 expression in HCC tissues was negatively corrrelated with the expression of GOLM1, which implied miR-382 was a key upstream regulatory molecule of GOLM1. Hence, we checked the expression of GOLM1 in the cells transfected with miR-382 inhibitor, mimic, miR-NC and found GOLM1 expression could be negatively regulated by the miR-382. Above all, the results of luciferase reporter assay confirmed GOLM1 was a direct target gene of miR-382. To further explore whether the effects of miR-382 on HCC cells were entirely or partially through modulating the GOLM1, we upregulated the GOLM1 expression in HepG2 with overexpression of miR-382 and performed the invasion and migration assay. The data we got indicated that overexpression of GOLM1 could rescue the invasion and migration inhibitory effects of miR-382. Taken together, we demonstrated that GOLM1 was an oncogene in HCC as a target of miR-382. Furthermore, Cox proportional regression analysis indicated that the miR-382 was an independent prognostic factor for the HCC patients.
As with previous reports, we confirm that miR-382 act as a tumor suppressor miRNA in HCC tumorigenesis and progression. MiR-382 prevents metastasis in HCC cells partially through targeting GOLM1. Moreover, Cox proportional regression analysis suggest that miR-382 is an independent prognostic factor for the HCC patients. Taken together, all findings in this study are in favor of understanding the mechanisms of miRNA in HCC carcinogenesis. The better understanding of the carcinogenic mechanisms of miRNA can enable us to explore novel prognosis markers and potential targets for cancer therapy in the near future.
Acknowledgements
This work was supported by a grant from the Natural Science Foundation of Shangdong (No. 2014GSF118007, 2017WSA10029, ZR2013HM061).
Disclosure of conflict of interest
None.
References
- 1.Ma C, Kesarwala AH, Eggert T, Medina-Echeverz J, Kleiner DE, Jin P, Stroncek DF, Terabe M, Kapoor V, ElGindi M, Han M, Thornton AM, Zhang H, Egger M, Luo J, Felsher DW, McVicar DW, Weber A, Heikenwalder M, Greten TF. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–257. doi: 10.1038/nature16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, Sinha S, Iannelli F, Radaelli E, Dos Santos A, Rapoud D, Guettier C, Samuel D, Natoli G, Carninci P, Ciccarelli FD, Garcia-Perez JL, Faivre J, Faulkner GJ. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong JZ, Wang LP, Zhang SN, Zou ZL, Lu MQ. Erlotinib might be a double-edged sword in HCC. Hepatology. 2015;61:729. doi: 10.1002/hep.27205. [DOI] [PubMed] [Google Scholar]
- 4.Mazzaferro V. Squaring the circle of selection and allocation in liver transplantation for HCC: an adaptive approach. Hepatology. 2016;63:1707–1717. doi: 10.1002/hep.28420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francini E, Bianco V. Tolerability of single-agent sorafenib in the treatment of elderly patients with hepatocellular carcinoma (HCC) Hepatology. 2014;60:764–765. doi: 10.1002/hep.26921. [DOI] [PubMed] [Google Scholar]
- 6.Ma W, Wong CC, Tung EK, Wong CM, Ng IO. RhoE is frequently down-regulated in hepatocellular carcinoma (HCC) and suppresses HCC invasion through antagonizing the Rho/Rhokinase myosin phosphatase target pathway. Hepatology. 2013;57:152–161. doi: 10.1002/hep.25987. [DOI] [PubMed] [Google Scholar]
- 7.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, Iliopoulos D. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moy RH, Cole BS, Yasunaga A, Gold B, Shankarling G, Varble A, Molleston JM, tenOever BR, Lynch KW, Cherry S. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell. 2014;158:764–777. doi: 10.1016/j.cell.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of HCC by modulating m6 A-dependent primary miRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 12.Cao Y, Zhang J, Xiong D, Wang D, Wu T, Huang A, Tang H. Hsa-miR-331-3p inhibits VHL expression by directly targeting its mRNA 3’-UTR in HCC cell lines. Acta Biochim Pol. 2015;62:77–82. doi: 10.18388/abp.2014_779. [DOI] [PubMed] [Google Scholar]
- 13.Chang L, Li K, Guo T. miR-26a-5p suppresses tumor metastasis by regulating EMT and is associated with prognosis in HCC. Clin Transl Oncol. 2017;19:695–703. doi: 10.1007/s12094-016-1582-1. [DOI] [PubMed] [Google Scholar]
- 14.Dai W, Wang C, Wang F, Wang Y, Shen M, Chen K, Cheng P, Zhang Y, Yang J, Zhu R, Zhang H, Li J, Zheng Y, Lu J, Zhou Y, Xu L, Guo C. AntimiR-197 inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem Biophys Res Commun. 2014;446:541–548. doi: 10.1016/j.bbrc.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Lechel A, Gougelet A. Early HCC treatment: a future strategy against interferon/miR-484 axis to revert precancerous lesions? Gut. 2016;65:1073–1074. doi: 10.1136/gutjnl-2016-311446. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Wang G, Zhou X, Song X, Gao H, Ma C, Chang H, Li H, Liu FF, Lu J, Ma J. MiR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed Pharmacother. 2016;83:792–797. doi: 10.1016/j.biopha.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 17.Ye QH, Zhu WW, Zhang JB, Qin Y, Lu M, Lin GL, Guo L, Zhang B, Lin ZH, Roessler S, Forgues M, Jia HL, Lu L, Zhang XF, Lian BF, Xie L, Dong QZ, Tang ZY, Wang XW, Qin LX. GOLM1 Modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;30:444–458. doi: 10.1016/j.ccell.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu W, Qin L. GOLM1-regulated EGFR/RTK recycling is a novel target for combating HCC metastasis. Sci China Life Sci. 2017;60:98–101. doi: 10.1007/s11427-016-0311-x. [DOI] [PubMed] [Google Scholar]
- 19.Egawa M, Yoshida Y, Ogura S, Kurahashi T, Kizu T, Furuta K, Kamada Y, Chatani N, Hamano M, Kiso S, Hikita H, Tatsumi T, Eguchi H, Nagano H, Doki Y, Mori M, Takehara T. Increased FoxM1 expression is associated with clinicopathological features and confers a poor prognosis in human hepatocellular carcinoma. Hepatol Res. 2017;47:1196–1205. doi: 10.1111/hepr.12854. [DOI] [PubMed] [Google Scholar]
- 20.Peng L, Yang G, Wu C, Wang W, Wu J, Guo Z. Mutations in hepatitis B virus small S genes predict postoperative survival in hepatocellular carcinoma. Onco Targets Ther. 2016;9:7367–7372. doi: 10.2147/OTT.S121785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogel GB, Kai ZS, Zargar S, Hinton A, Jones GA, Wong AS, Ficici SG, Lopez AD, King CC. MicroRNA dynamics during human embryonic stem cell differentiation to pancreatic endoderm. Gene. 2015;574:359–370. doi: 10.1016/j.gene.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 22.Tan H, He Q, Gong G, Wang Y, Li J, Wang J, Zhu D, Wu X. miR-382 inhibits migration and invasion by targeting ROR1 through regulating EMT in ovarian cancer. Int J Oncol. 2016;48:181–190. doi: 10.3892/ijo.2015.3241. [DOI] [PubMed] [Google Scholar]
- 23.Xu M, Jin H, Xu CX, Sun B, Song ZG, Bi WZ, Wang Y. miR-382 inhibits osteosarcoma metastasis and relapse by targeting Y box-binding protein 1. Mol Ther. 2015;23:89–98. doi: 10.1038/mt.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou B, Song J, Han T, Huang M, Jiang H, Qiao H, Shi J, Wang Y. MiR-382 inhibits cell growth and invasion by targeting NR2F2 in colorectal cancer. Mol Carcinog. 2016;55:2260–2267. doi: 10.1002/mc.22466. [DOI] [PubMed] [Google Scholar]