Abstract
We tested the hypothesis that, compared with subjects with no history of psychiatric illness (controls), changes in gene expression in the dorsolateral prefrontal cortex from two subgroups of subjects with schizophrenia, one with a marked deficit in muscarinic M1 receptors (muscarinic receptor-deficit schizophrenia (MRDS)), would identify different biochemical pathways that would be affected by their aetiologies. Hence, we measured levels of cortical (Brodmann area 9) mRNA in 15 MRDS subjects, 15 subjects with schizophrenia but without a deficit in muscarinic M1 receptors (non-MRDS) and 15 controls using Affymetrix Exon 1.0 ST arrays. Levels of mRNA for 65 genes were significantly different in the cortex of subjects with MRDS and predicted changes in pathways involved in cellular movement and cell-to-cell signalling. Levels of mRNA for 45 genes were significantly different in non-MRDS and predicted changes in pathways involved in cellular growth and proliferation as well as cellular function and maintenance. Changes in gene expression also predicted effects on pathways involved in amino acid metabolism, molecular transport and small-molecule biochemistry in both MRDS and non-MRDS. Overall, our data argue a prominent role for glial function in MRDS and neurodevelopment in non-MRDS. Finally, the interactions of gene with altered levels of mRNA in the cortex of subjects with MRDS suggest many of their affects will be upstream of the muscarinic M1 receptor. Our study gives new insight into the molecular pathways affected in the cortex of subjects with MRDS and supports the notion that studying subgroups within the syndrome of schizophrenia is worthwhile.
Introduction
The diagnosis of schizophrenia is made after observing a defined constellation of symptoms present over a specified period of time.1 The notion of the diagnosis of schizophrenia, from its inception, was that it would define a syndrome of disorders as indicated by its description: ‘schizophrenia does not appear to us as a disease in the narrower sense but as a disease group….One should therefore really speak of schizophrenias in the plural’.2 Although the diagnosis of schizophrenia has proven a long-lasting clinical tool, it is now recognised that continuing to study schizophrenia as a biological entity is hampering advances in understanding the aetiologies of the underlying disorders encapsulated within the syndrome.3
We have defined a subgroup within the syndrome of schizophrenia that can be separated into a distinct population because of a marked loss of cortical muscarinic M1 receptors (CHRM1) in Brodmann Area 9 (BA 9);4 we have termed the subgroup as being a muscarinic receptor deficit form of schizophrenia (MRDS). Our data suggest that ∼25% of subjects with schizophrenia will have a marked loss of cortical muscarinic M1 receptors, an estimate that is supported by a neuroimaging study on a small cohort of subjects with schizophrenia.5 Understanding the aetiology of MRDS has become of increased significance with the development of drugs specifically targeting the muscarinic M1 receptor as these drugs are proposed as novel new treatments for the disorder.6 The new muscarinic M1 receptor-specific drugs are novel in that they act at an allosteric site on the receptor to either directly influence receptor activity or modify the affinity of the orthosteric site, at which acetylcholine binds, to change the activity of the receptor.6 Independent of the mechanism of action of drugs targeting the muscarinic M1 receptor, a loss of 75% of those receptors in the cortex of subjects with MRDS could make these individuals nonresponsive to such therapies. Hence, continuing to increase understanding of the aetiology of MRDS is important as aid to hypothesis-driven drug design7 directed at developing drugs that would be effective in treating subjects with that disorder.
In practice, we have defined subjects with MRDS as having levels of [3H]pirenzepine binding in BA 9 of <90 fmol mg−1 estimated tissue equivalents, whereas non-MRDS have levels of >120 fmol mg−1 estimated tissue equivalents (Supplementary Figure 1). This capacity to subdivide subjects with schizophrenia opened the potential to identify the pathophysiologies of a more biologically homogenous subgroup within the syndrome. Studies on MRDS have shown they have widespread decreases in muscarinic receptors throughout the central nervous system (CNS),8, 9 similar to what has been reported in a neuroimaging study showing widespread decreases in muscarinic receptors in subjects with schizophrenia.5 We have also shown that subjects with MRDS have an altered CHRM1 promoter methylation pattern and high cortical levels of a microRNA that targets CHRM1 expression.10 Finally, using human cortex, we have shown that orthosteric agonist-induced CHRM1 G-protein recruitment is less effective,11 that allosteric agonist-induced CHRM1 G-protein recruitment is not affected11 and there is a decreased response to the allosteric modulator benzyl quinolone carboxylic acid12 in MRDS. Although these findings add validity to the notion that subjects with MRDS have a different aetiology and may respond differently to drugs that act at different sites on the muscarinic M1 receptor, they do not provide extensive information on the molecular basis of MRDS.
The ability to identify the potential molecular basis for disease aetiologies has been enhanced by the development of high-throughput technologies. Such technologies allow the probing of the human genome, transcriptome and proteome to look for the molecular footprint left by different diseases with the view of using this information to develop new therapeutics.13 In schizophrenia, this has resulted in efforts to define the molecular characteristics of the syndrome and, from such understanding, define the biochemical pathways affected by the aetiology of the disorder.
Schizophrenia has been recognised as occurring in subjects with a genetic predisposition who have encountered environmental factors that trigger the processes that lead to the onset of the symptoms of the disorder.14 It is now known that environmental factors act through epigenetic mechanisms to change gene expression,15 and it is expected that these changes in gene expression will bring about the changes in cellular and CNS function that result in the symptoms that define schizophrenia. Hence, changes in gene expression in the CNS from subjects with schizophrenia should be a valuable indicator as to the mechanism underpinning the aetiology of the disorder. Pursuing this hypothesis has led to a number of studies comparing levels of gene expression in the CNS from subjects with schizophrenia with that in subjects with no history of psychiatric disorders. One such study in BA 9 showed that the expression of genes involved in the regulation of presynaptic function was the most affected in tissue from subjects with schizophrenia.16 There have now been a significant number of studies that have examined levels of gene expression in the CNS from subjects with schizophrenia and they have implicated pathways involved in synaptic, mitochondrial, immune system, γ-aminobutyric acidergic and oligodendrocyte function in the aetiology of schizophrenia.17 This complexity of findings led to the suggestion that ‘our success in interpreting the molecular changes in schizophrenia will depend on our ability to understand the biology using innovative ideas’.17 Equally, such data could be more interpretable if it was not focussed on the syndrome of schizophrenia but on subgroups within the syndrome that could be identified using some biological measure. This notion was the basis for our study on gene expression in cortex from subjects with schizophrenia that included subjects with MRDS and subjects with forms of the disorder not associated with losses in cortical muscarinic receptors (non-MRDS) as well as age- and sex-matched control subjects.
Materials and methods
Post-mortem brains were collected with approval from the Ethics Committee of the Victorian Institute of Forensic Medicine and all tissue was collected by this Institute after gaining written consent from the nearest next of kin. This study was approved by the Human Ethics Committee of Melbourne Health. All tissue was obtained from the Victorian Brain Bank at the Florey Institute for Neuroscience and Mental Health.
Case history review
Case history reviews were conducted using the DIBS (Diagnostic Instrument for Brain Studies),18, 19 a post-mortem clinical assessment tool that enables a diagnostic consensus to be reached using DSM-IV (Fourth Edition of the Diagnostic and Statistical Manual of Mental Disorders) criteria.20 Using data gathered during the case history reviews, duration of illness was calculated as the time from first hospital admission to death, the final recorded dose of antipsychotic drugs was converted to a standardised drug dose (chlorpromazine equivalents per day).21 Whether a case had been treated with anticholinergics and/or benzodiazepines was also recorded. In addition, toxicology reports were reviewed to determine whether alcohol or nonprescribed drugs that are not readily available from pharmacists or supermarkets were detected in blood or urine.
From the data collected using the DIBS, post-mortem interval (PMI) was calculated as the time from death to autopsy. Where death was not witnessed, tissue was only collected from subjects who had been seen alive up to 5 h before being found dead; in these instances, the PMI was taken as the midpoint between the person being found and being last seen alive. In all cases, cadavers were refrigerated within 5 h of being found and, after removal at autopsy, brain tissue was rapidly processed and frozen to −70 °C using a standardised procedure.22 Following these two processes acts to minimise autolytic effects.23 The pH of the brain tissue was measured as described previously24 as this provides a good measure of overall tissue preservation.25 Finally, levels of cotinine in the cortex of cases to be used for the microarray array study were measured using NicAlert (Nymox, Hasbrouck Heights, NJ, USA) strips, the results from which have been validated by measuring cotinine using gas chromatography–nitrogen phosphorus detection.26 NicAlert scores of >3 are indicative of nicotine intake within 48 h of specimen collection.
As part of this study design, the cases used in the microarray experiment were matched as closely as possible for age, sex, PMI and brain pH across all diagnostic cohorts. Rates of suicide, prescription of anticholinergic and benzodiazepines, presence of nonprescription drugs in blood and urine, frequency of NicAlert scores of >3 as well as the types of antipsychotic received were also matched as closely as possible between MRDS and non-MRDS (Table 1 and Supplementary Table 1).
Table 1. A summary of demographic, CNS collection and treatment data (mean±s.e.m.) for control subjects, subjects with Sz as a whole as well as subjects with MRDS and non-MRDS used in this study.
| n | Age (year) | Sex (M/F) | PMI (h) | pH | RIN | DoI (year) | FRADD | Sui (Y/N) | AChol (Y/N) | Benz (Y/N) | THC (Y/N) | Alc (Y/N) | Other (Y/N) | NicAlert >3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (A) qPCR | |||||||||||||||
| Controls | 30 | 49±3.0 | 24/6 | 43.6±2.9 | 6.36±0.03 | 8.92±0.14 | |||||||||
| Sz | 30 | 48±3.0 | 24/6 | 40.5±2.4 | 6.30±0.04 | 8.82±0.10 | 22±2.8 | 561±96 | 8/22 | 13/17 | 12/18 | 9/21 | 13/17 | 2/30 | |
| P | 0.91 | 1.00 | 0.41 | 0.14 | 0.56 | ||||||||||
| (B) Microarray | |||||||||||||||
| Controls | 15 | 49±4.4 | 12/3 | 46.1±3.9 | 6.40±0.04 | 8.53±0.15 | 8/15 | ||||||||
| MRDS | 15 | 48±4.1 | 13/2 | 39.4±2.7 | 6.29±0.04 | 8.82±0.10 | 22±4.1 | 642±154 | 4/11 | 7/8 | 7/8 | 4/11 | 6/9 | 0/15 | 8/15 |
| Non-MRDS | 15 | 48±4.1 | 11/4 | 41.5±4.0 | 6.29±0.06 | 8.85±0.10 | 22±4.0 | 480±118 | 4/11 | 6/9 | 5/10 | 5/10 | 7/8 | 2/13 | 8/15 |
| F | 0.009 | 0.69 | 0.9 | 0.59 | 1.34 | ||||||||||
| d.f. | 2,42 | 2,42 | 2,42 | 2,42 | |||||||||||
| P | 0.99 | 0.41 | 0.56 | 0.27 | 0.95 | 0.41 | 1.00 | 0.71 | 0.46 | 0.69 | 0.71 | 0.14 | 1.00 | ||
Abbreviations: AChol, prescribed anticholinergic drugs; Alc, alcohol; Benz, prescribed benzodiazepines; CNS, central nervous system; DoI, duration of illness; F, female; FRADD, final recorded antipsychotic drug dose given as chlorpromazine equivalents/day; M, male; MRDS, muscarinic receptor-deficit schizophrenia; N, no; non-MRDS, subjects with schizophrenia who do not have a deficit in muscarinic receptors; PMI, post-mortem interval; qPCR, quantitative PCR; RIN, RNA Integrity Number; Sui, suicide; Sz, schizophrenia; THC, cannabinoids; Y, yes.
Results from toxicology screens of urine or blood after death identified the presence of cannabinoids, alcohol and other nonprescribed medications that are not freely available from pharmacies or supermarkets.
CNS processing
BA 9 was excised from the lateral surface of the frontal lobe from an area that includes the middle frontal gyrus superior to the inferior frontal sulcus of the left hemisphere from 30 subjects who were listed as having schizophrenia in reports of death. Using data from a previous study in BA 9,4 we included 15 subjects with MRDS, 15 subjects with non-MRDS (as defined in the Introduction) and 30 subjects with no history of psychiatric illness (controls), 15 of the controls being used in the microarray study (Table 1 and Supplementary Table 1).
Total RNA was isolated from ∼100 mg of frozen grey matter using 1.0 ml TRIzol reagent (Life Technologies, Scoresby, VIC, Australia) and, following homogenisation and phase separation as per the manufacturer’s instructions, the aqueous phase was added to an equal volume of 70% ethanol. RNA was then isolated using RNeasy mini kits (Qiagen, cat. no. 74104, Chadstone Centre, VIC, Australia) with all samples being treated with DNase using on column digestion; the absence of DNA contamination was confirmed by PCR using primers specific for genomic DNA. RNA quantity and quality were analysed by spectrophotometry (NanoDrop; Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) and by obtaining RNA integrity numbers (RINs) using an Agilent 2100 bioanalyser (Agilent Technologies, Santa Clara, CA, USA). All samples used for the microarray study had RINs of ⩾7.00 as this is a good predictor of a sample suitable for microarray hybridisation (Supplementary Table 1).27
The Affymetrix Human Exon 1.0 ST Array (Affymetrix, Santa Clara, CA, USA) was used to measure levels of RNA in BA 9 in 45 cortical samples (Table 1 and Supplementary Table 1). The arrays were processed by the Australian Genome Research Facility (AGRF; Melbourne, Australia) where ribosomal RNA was eliminated and random priming used to generate cRNA that was end labelled with biotin using the Affymetrix synthesis and labelling kit. Samples that passed the quality checkpoints were prepared for hybridisation using a standard probe cocktail. Each sample was loaded into an Affymetrix Human Exon 1.0 ST Array and hybridised overnight. Following post-hybridisation washes, the chips were scanned and the fluorescent signals converted into a DAT file. After visual confirmation of the scans and quality control analysis, these files were used to generate subsequent cell intensity (CEL) and chip (CHP) files for analysis. Data files are currently held by the CRC for Mental Health and will be made available to bona fide researchers upon request.
Microarray data analyses
All CEL files were imported into JMP Genomics 5.1 (SAS, Cary, NC, USA) at the gene level, collapsing the exon-level data onto known transcripts. To control for array-to-array variation, the data were normalised using the Robust Multichip Average (RMA) algorithm. The data were then log2 transformed and, because they were to be analysed across multiple groups, normalised for between-group comparisons using the ‘Least Square means’ method (see: http://support.sas.com/onlinedoc/913/getDoc/en/statug.hlp/glm_sect34.htm) and then imported using metaprobe set and probe set list files. Analyses were first performed at the syndrome level using schizophrenia as the primary comparator to controls. Planned secondary analysis comparing data from subjects with MRDS, those with non-MRDS and the control group were then completed. Following standard practice,28 we used a nonstringent group-wise P-value cutoff of 0.01 with a fold change of 20% as we had shown this fold change can be validated using real-time quantitative PCR (qPCR) which is an indication of a viable change in gene expression.29 Data were filtered using these criteria to contain only the expression profiles that belonged to transcripts for which there was ‘core’ level confidence (see http://media.affymetrix.com/support/technical/whitepapers/exon_probeset_trans_clust_whitepaper.pdf).
Gene expression pathway analyses
The genes with altered levels of expression in BA 9 from subjects with schizophrenia, MRDS and non-MRDS were analysed using Ingenuity Pathway Analysis build version 366632M (IPA: http://www.ingenuity.com/products/ipa) filtered only to include relationships that have been experimentally observed. The IPA core analysis identifies molecular and cellular functions and canonical pathways that are significantly affected based on Fisher’s right-tailed exact test and calculates the –log(P-value). The number of genes with different expression profiles within that classification is reported as a ratio of the total number of genes in the classification. The core analysis also provides a score for identified biological networks in the format of –log(Fisher’s exact test result) of the hypergeometric distribution of the data that is used to rank networks according to their relevance to the data set.
Gene expression: replication with qPCR
Levels of mRNA for the seven genes with the largest differences in levels of cortical mRNA at the level of syndrome diagnosis (Schizophrenias vs control), measured using expression microarrays, were selected for analysis using qPCR. For the qPCR, 15 additional control subjects were included to balance sample sizes and add power to the analyses, with the RNA extracted using the same protocol used for the microarray study (Supplementary Table 1).
For qPCR, first-strand complementary DNA was synthesised from 2 μg RNA using 100 units M-MLV-RT (Applied Biosystems, Scoresby, VIC, Australia)30 and used as a template for qPCR. Potential primer sequences were elucidated using Beacon Designer software (PREMIER Biosoft, Palo Alto, CA, USA); primers (Supplementary Table 2) were taken as acceptable if they gave reaction efficiencies, calculated from standard curves run on each plate constructed from a 10-fold dilution series of the human CNS complementary DNA, of between 90 and 110% and that the amplicon generated by these primers gave the expected gene sequence.31 Where it was possible, primers were designed to span intronic regions within a gene.
Six genes were identified using expression array data that appeared to have characteristics required for reference genes and their levels measured in the post-mortem BA 9 samples. Therefore, levels of mRNA for each of these genes and Carrier Family 39 (Zinc Transporter), Member 12 (SLC39A12), Solute Carrier Organic Anion Transporter Family, Member 1C1 (SLCO1C1), ATPase 13A4 (ATP13A4), Fatty Acid Elongase 2 (ELOVL2), Methyltransferase Like 7A (METTL7A), Histone Cluster 1, H4a (HIST1H4A) and Rap Guanine Nucleotide Exchange Factor 1 (RAPGEF1) were measured in BA 9 using SYBR green detection in a Bio-Rad iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the data being acquired using IQ5 optical system 2.0 software (Bio-Rad). Relative expression was determined using the Pfaffl method32 when reaction efficiencies, calculated from standard curves run on each plate, fell between 95 and 105%. Analyses of the data from the potential reference genes using geNorm (https://genorm.cmgg.be/) suggested glyceraldehyde-3-phosphate dehydrogenase (GAPDH: M=1.398), peptidylprolyl isomerase A (PPIA: M =1.386) and α-synuclein (SNCA: M =1.407) as the best reference genes. Hence, levels of mRNA for genes of interest were expressed as the geometric mean of the three reference genes to provide more stable data for analyses.31
Statistical analyses: qPCR
Student’s t-test or one-way analysis of variance (followed by Holm–Sidak multiple comparisons test to identify the source of variance) were used to compare demographic and pharmacological data between groups. Student’s t-test was also used to compare qPCR data across diagnoses but results were expressed as the ratio of mRNA in schizophrenia compared with controls to be in a comparable format to our microarray data. Correlations between levels of mRNA measured using qPCR and potential confounding factors were determined by Pearson’s correlation, with an r-value of >0.5 denoting a moderate relationship and >0.7 a strong relationship.33, 34, 35 These analyses were completed using GraphPad Prism 6.02 (GraphPad Software, La Jolla, CA, USA) or Minitab Statistical Software release 16 (Minitab, Sydney, NSW, Australia).
Results
Demographics and sample collection data
At the level of the syndrome of schizophrenia, there were no significant differences in age, gender ratio, PMI, CNS pH or RINs between the diagnostic groups (Table 1a and Supplementary Table 1). In addition, there were no significant variations in these variables between MRDS, non-MRDS or controls (Table 1b and Supplementary Table 1). In addition, duration of illness, final recorded dose of antipsychotic drugs, rates of suicide completion, frequency of prescription of anticholinergic drugs or benzodiazepine, frequency of detection of nonprescription drugs or frequency of NicAlert scores of >3 did not differ between subjects with MRDS and non-MRDS.
Gene expression arrays
Levels of mRNA for 233 genes were altered at P<0.01 in BA 9 from the schizophrenias with mRNA for 187 genes being higher in the cortex of subjects with the disorder; using the ±Δ0.2-fold criteria, levels of mRNA for 56 genes were taken as varying between the schizophrenias and controls (Supplementary Table 3). For MRDS, levels of mRNA for 330 genes varied from controls at a significance of P<0.01, 277 being higher in the cortex of subjects with MRDS. Applying the ±Δ0.2-fold criteria meant that levels of mRNA for 65 genes were taken as varying in the cortex of subjects with MRDS compared with controls (Supplementary Table 4). For non-MRDS, levels of mRNA for 157 genes were changed in BA 9 when compared with controls at the level of P<0.01; levels of mRNA for 117 genes were higher in non-MRDS. The ±Δ0.2-fold criteria meant that levels of mRNA for 45 genes were taken as varying between non-MRDS and controls (Supplementary Table 5).
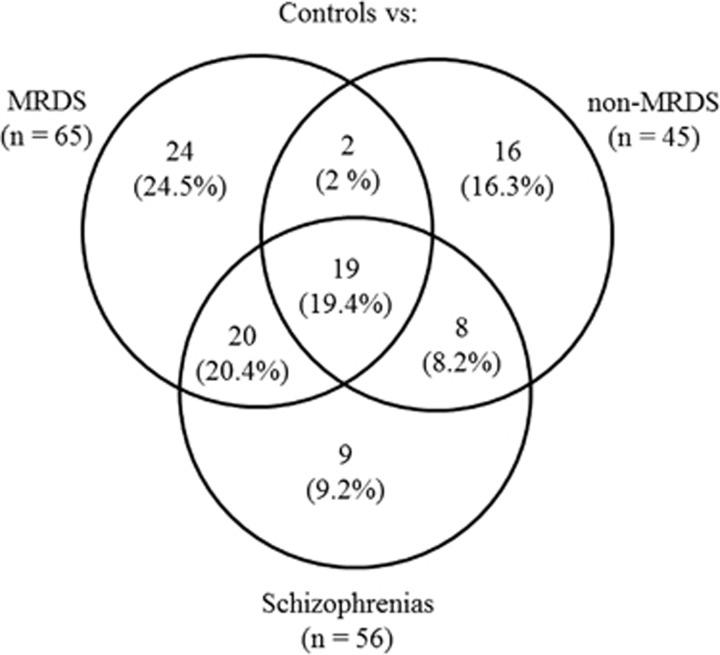
A novel aspect of this study is that we could compare gene expression in BA 9 from subjects with schizophrenia at the level of the syndrome and in two subgroups within the syndrome divided according to whether or not they had a marked loss of muscarinic receptors in the cortical region. Using this approach our data showed that, compared with controls, levels of mRNA for 19 genes varied in the schizophrenias before and after it was dissected in MRDS and non-MRDS (Table 2 and Figure 1). In addition, levels of mRNA for 20 genes varied in the schizophrenias and in subjects with MRDS, levels of mRNA for 8 genes varied in the schizophrenias and non-MRDS, whereas levels of mRNA for 2 genes varied in MRDS and non-MRDS compared with controls. Finally, compared with controls, levels of mRNA for 9 genes varied only in cortex from subjects with the schizoprhenias, 24 only varied in MRDS and 16 varied only in non-MRDS.
Table 2. Genes for which levels of mRNA in BA 9, when compared with controls, differed at the level of P<0.01 and a ±Δ0.2-fold in the syndrome of Sz as a whole as well as in MRDS and non-MRDS (All), in schizophrenia and MRDS (Sz+MRDS), schizophrenia and non-MRDS (Sz+nMRDS), MRDS and non-MRDS (MRDS+nMRDS), schizophrenia only (Sz), MRDS only and non-MRDS only.
| All | Sz+MRDS | Sz+nMRDS | MRDS+nMRDS | Sz | MRDS | nMRDS |
|---|---|---|---|---|---|---|
| ATP13A4 | AGT | GRAMD1C | HEPH | EDNRB | APOE | BBOX1 |
| BMPR1B | AGXT2L1 | KCNJ10 | TST | FGF2 | ATP1A2 | GEMC1 |
| CLDN10 | AQP4 | MT1X | GABRG1 | AXL | GPR125 | |
| CYBRD1 | AZGP1P1 | PCBP2 | GLIS3 | CRB1 | HSPD1 | |
| GJA1 | EFEMP1 | PON2 | GPAM | DDIT4 | LBR | |
| HGF | ELOVL2 | PRAMEF7 | HIST1H4F | DDIT4L | LGR4 | |
| HIST1H4A | FAM74A3 | RAPGEF1 | HSP90AB3P | HEPACAM | LIFR | |
| HSD17B6 | GJB6 | RFTN2 | MT4 | KRTAP8-1 | MDM4 | |
| IL17RB | GPR37L1 | RANBP3L | MEGF10 | MT1M | ||
| METTL7A | HIST1H4C | MLC1 | MT4 | |||
| OR52N5 | HIST1H4D | NAB1 | PXMP4 | |||
| SLC15A2 | NOTCH2 | NHP2 | RALGAPA2 | |||
| SLC1A3 | PRSS35 | OR4D5 | RHOQ | |||
| SLC39A12 | SELENBP1 | POLR3G | RPL14 | |||
| SLC4A4 | SLC16A9 | PRELP | SMAD4 | |||
| SLC7A2 | SLC7A11 | RPL21 | VDAC3 | |||
| SLCO1C1 | SSPN | SDC4 | ||||
| STK17B | TGFB2 | SLC44A2 | ||||
| TLR4 | TP53BP2 | SYT15 | ||||
| TPP1 | TAF1A | |||||
| TECRL | ||||||
| TLCD1 | ||||||
| TMEM47 | ||||||
| VTI1B |
Abbreviations: BA 9, Brodmann Area 9; MRDS, muscarinic receptor-deficit schizophrenia; Sz, schizophrenia.
Figure 1.
A Venn diagram showing the division of the 65 genes differentially expressed in the cortex of muscarinic receptor-deficit schizophrenia (MRDS), the 45 genes differentially expressed in subjects with schizophrenia but without a deficit in cortical muscarinic receptors (non-MRDS) and 56 genes differentially expressed at the level of schizophrenia as a syndrome into (1) the number of genes that were changed in only one of the three groups, (2) the number of genes where changed gene expression overlapped in the different subgroups and schizophrenia or (3) the number of genes that were changed in all three groups (n=19).
Pathway analysis
All of the transcripts that differed at the level of the schizophrenias (n=56) and non-MRDS (n =45) mapped to pathways in IPA compared with 47 of 65 transcripts for MRDS. Hence, mapped genes were used to identify the canonical pathway most likely affected by the changes in gene expression. Thus, in the schizophrenias and MRDS the most likely canonical pathways to be affected were involved in hepatic fibrosis/hepatic stellate cell activation (Table 3), whereas in non-MRDS the most likely affected pathways were involved in cardiomyocyte differentiation via bone morphogenetic protein receptors. Encouragingly, the changes in gene expression in the schizophrenias, MRDS and non-MRDS were most consistent with those that would occur in neurological disorders.
Table 3. Ingenuity pathway analyses of the impact of genes that are differentially expressed in Brodmann Area 9 from subjects with schizophrenia at the level of the syndrome (Schizophrenias) and those within the MRDS.
| Top canonical pathways | P |
Top disease and biofunction |
|||||
|---|---|---|---|---|---|---|---|
| Diseases and disorders | P | Molecular and cellular functions | P | Physiological system development and function | P | ||
| Schizophrenias | |||||||
| Hepatic fibrosis/hepatic stellate cell activation | 1.32E−05 | Neurological disease | 2.99E−03 to 8.85E−09 | Amino acid metabolism | 2.99E−03 to 1.06E−09 | Tissue development | 2.99E−03 to 7.55E−07 |
| Triacylglycerol biosynthesis | 4.11E−03 | Organismal injury and abnormalities | 2.99E−03 to 2.51E−07 | Molecular transport | 2.99E−03 to 1.06E−09 | Organismal development | 2.99E−03 to 1.29E−06 |
| Hepatic cholestasis | 1.15E−02 | Skeletal and muscular disorders | 2.99E−03 to 6.30E−06 | Small-molecule biochemistry | 2.99E−03 to 1.06E−09 | Organ development | 2.99E−03 to 1.42E−06 |
| Regulation of the epithelial–mesenchymal transition pathway | 1.57E−02 | Inflammatory disease | 2.63E−03 to 8.77E−06 | Cellular development | 2.99E−03 to 7.55E−07 | Reproductive system development and function | 2.99E−03 to 1.42E−06 |
| FGF signalling | 2.62E−02 | Inflammatory response | 2.99E−03 to 8.77E−06 | Cell morphology | 2.99E−03 to 1.01E−06 | Nervous system development and function | 2.99E−03 to 1.84E−06 |
| MRDS | |||||||
| Hepatic fibrosis/hepatic stellate cell activation | 2.58E−03 | Neurological disease | 6.54E−03 to 1.29E−08 | Cellular movement | 5.07E−03 to 1.05E−06 | Tissue morphology | 6.54E−03 to 3.32E−07 |
| LXR/RXR activation | 6.83E−03 | Organismal injury and abnormalities | 6.54E−03 to 3.32E−07 | Molecular transport | 6.54E−03 to 2.00E−06 | Cardiovascular system development and function | 6.54E−03 to 1.06E−05 |
| IL-12 signalling and production in macrophages | 8.17E−03 | Cardiovascular disease | 6.54E−03 to 1.06E−05 | Amino acid metabolism | 5.86E−03 to 2.61E−06 | Organismal development | 4.91E−03 to 1.06E−05 |
| Thiosulfate disproportionation III (rhodanese) | 9.80E−03 | Inflammatory disease | 6.54E−03 to 1.06E−05 | Small-molecule biochemistry | 6.54E−03 to 2.61E−06 | Tissue development | 6.54E−03 to 1.76E−05 |
| Hepatic cholestasis | 1.48E−02 | Inflammatory response | 6.54E−03 to 1.06E−05 | Cell-to-cell signalling and interaction | 6.54E−03 to 1.06E−05 | Organ development | 6.53E−03 to 5.02E−05 |
| Non-MRDS | |||||||
| Cardiomyocyte differentiation via BMP receptors | 7.77E−04 | Neurological disease | 8.71E−03 to 8.01E−07 | Molecular transport | 6.54E−03 to 4.46E−06 | Organ development | 8.71E−03 to 6.56E−06 |
| Role of NANOG in mammalian embryonic stem cell pluripotency | 1.55E−03 | Cancer | 8.71E−03 to 1.41E−05 | Cellular growth and proliferation | 8.71E−03 to 1.19E−05 | Reproductive system development and function | 8.71E−03 to 6.56E−06 |
| Hepatic fibrosis/hepatic stellate cell activation | 6.52E−03 | Gastrointestinal disease | 8.71E−03 to 1.41E−05 | Amino acid metabolism | 1.46E−03 to 1.35E−05 | Nervous system development and function | 8.71E−03 to 3.75E−05 |
| L-carnitine biosynthesis | 6.54E−03 | Organismal injury and abnormalities | 8.71E−03 to 1.41E−05 | Small-molecule biochemistry | 6.54E−03 to 1.35E−05 | Tissue development | 8.71E−03 to 2.08E−04 |
| Thiosulfate disproportionation III (rhodanese) | 6.54E−03 | Renal and urological disease | 8.71E−03 to 5.53E−05 | Cellular function and maintenance | 8.71E−03 to 1.39E−05 | Skeletal and muscular system development and function | 8.71E−03 to 2.82E−04 |
Abbreviations: BMP, bone morphogenetic protein; FGF, fibroblast growth factor; IL-12, interleukin 12; LXR/RXR, liver X receptor/ retinoid X receptor; MRDS, muscarinic receptor-deficit schizophrenia.
Analysis at the level of molecular and cellular functions suggested that changes in levels of gene expression in the schizophrenias, MRDS and non-MRDS would most affect pathways involved in amino acid metabolism, molecular transport and small-molecule biochemistry. In contrast, in the schizophrenias, changes in gene expression would most affect pathways involved in cellular development and cell morphology. Exploring the changes in gene expression and their impact on molecular and cellular functions further showed that pathways involved in cellular movement and cell-to-cell signalling appeared likely to be uniquely affected in MRDS, whereas pathways involved in cellular growth and proliferation as well as cellular function and maintenance would be uniquely affected in non-MRDS.
At the level of physiological system development and functions, changes in gene expression in the schizophrenias suggested that pathways involved in tissue development would most likely be affected; this compared with pathways involved in tissue morphology being most affected in MRDS, whereas pathways being involved in organ development being most affected in non-MRDS (Table 2). At the level of physiological system and development and function, pathways involved in tissue morphology as well as cardiovascular system development and function appeared to be most likely affected in MRDS, whereas pathways involved in skeletal and muscular system development and function were most likely affected in non-MRDS.
qPCR
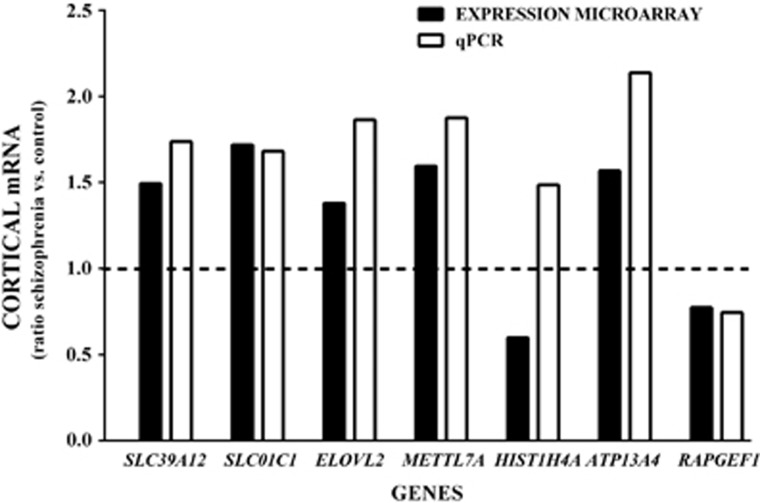
We have measured levels of mRNA for seven genes in BA 9 from subjects with schizophrenia and controls. Of the seven genes, microarray data suggest that levels of five of the genes (SLC39A12, SLCO1C1, ATP13A4, ELOVL2, METTL7A and ATP13A4) should be higher in the schizophrenias with levels of HIST1H4A and RAPGEF1 being decreased (Figure 2). Using qPCR we confirmed that levels of SCL39A12 (P<0.001), SLCO1C1 (P<0.01), ELOVL2 (P<0.001), METTL7A (P<0.001) and ATP13A4 I (P<0001) were higher in the schizophrenias. We also confirmed that levels of mRNA for RAPGEF1 (P<0.05) was lower in those subjects. In contrast, qPCR data suggested that levels of HIST1H4A (P<0.01) mRNA was higher in the schizophrenias compared with controls, and this was not consistent with the microarray data.
Figure 2.
Levels of mRNA in the cortex of subjects with schizophrenia, expressed as a ratio of controls, measured in Brodmann Area 9 using the Affymetrix Human Exon 1.0 ST Array (expression microarray) and quantitative PCR (qPCR). The degree of significance for data from microarray and qPCR are SLC39A12 P<0.0001 and P=0.007; SLC01C1 P=0.0002 and P=0092; ELOVL2 P=0.0018 and P<0.0006; METTL7A P=0.0010 and P<0.0001; HIST1H4A P=0.0004 and P=0.0067; ATP13A4 P=0.0037 and P<0.0001; and RAPGEF1 P=0.0043 and P=0.04, respectively.
Levels of mRNA measured using qPCR did not show strong correlations with age, PMI, pH or RIN (2.73 × 105< r2>0.19; Supplementary Table 6). There were no correlations between duration of illness or final recorded dose of antipsychotic drugs and levels of mRNA for the genes in subjects with schizophrenia (0.0006< r2>0.11; see Supplementary Table 6).
Discussion
In this study we have measured levels of mRNA in the dorsolateral prefrontal cortex from a cohort of subjects with schizophrenia containing two subsets of subjects we defined because they have (MRDS) or do not have (non-MRDS) a marked loss of cortical muscarinic receptors.4 This approach has allowed us to identify changes in levels of mRNA for genes that are present only in subjects with MRDS or only in non-MRDS that were not detectable within the syndrome of schizophrenia as a whole (Figure 1 and Table 2). It would seem reasonable to conclude that the genes encoding the mRNA that are only changed in MRDS or non-MRDS are making a unique contribution to the pathophysiologies of these subgroups within the syndrome of schizophrenia. In contrast, changes in gene expression at the level of the syndrome that can be detected in both subgroups may be associated with the pathways in the final stages of the genesis of symptoms that will be common to all subgroups within the syndrome or be an effect of the antipsychotic drug treatment given to both subgroups. Notably, despite differences in gene expression between subjects with MRDS and non-MRDS, all changes in gene expression appear to be in pathways that are associated with neurological disease.
At the level of schizophrenia as a syndrome, our study supports findings from previous studies for a role for inflammatory-related pathways,29, 36, 37, 38, 39, 40 metabolic pathways,16, 41, 42 molecular transport43, 44 and small-molecule biochemistry29 in the aetiology of schizophrenia. In addition, our connectome data implicating TP53 (see below) in the pathways affected in schizophrenia could implicate abnormalities in chromosome maintenance.44, 45 This overall concordance in data on changes in the CNS transcriptome reflects consistency in pathways that are predicted to be affected within the aetiology of the syndrome of schizophrenia.17
Following established practice, we used qPCR to show there are higher levels of SLC39A12, SLC01C1, ELOVL2, METTL7A, HIST1H4A and ATP13A4, as well as lower levels of RAPGEF1 mRNA in BA 9 from the schizophrenias. Except for HIST1H4A, the data from qPCR agree with data on the same mRNAs from the expression microarray; we can provide no explanation as to why data on HIST1H4A differed between qPCR and microarray. Adding to these data, we have published data to show there are higher levels of mRNA from SLC39A12 (ref. 46) and SELENBP1,47 another gene with higher levels of mRNA in BA 9 according to microarray data, in multiple cortical regions from the schizophrenias that are not present in the cortex of subjects with mood disorders or in the cortex of rats after treatment with antipsychotic drugs. Thus, our data from qPCR would suggest that at least 88% of the changes in gene expression in the cortex of subjects with MRDS, non-MRDS and at the level of the schizophrenias should be replicable using other approaches to measuring levels of mRNA. In addition, changes in levels of mRNA identified using microarrays should have some selectivity for schizophrenia and/or subgroups within the syndrome.
As in most studies that do not use measurements from treatment-naive subjects, there are a number of potential confounds relating to our study. Hence, at the level of the schizophrenias, we could not control for the impact of antipsychotic drug treatments when comparing results with controls. However, we have shown that levels of mRNA for SLC39A2 (ref. 46) and SELENBP1,47 which are increased in the schizophrenias, are not altered in the cortex of subjects with bipolar disorder, some of whom were treated with antipsychotic drugs, or the cortex of rats after treatment with antipsychotic drugs. In addition, expression array data from the cortex of rats treated with antipsychotic drugs do not show that such treatments target the pathways that our data suggest are altered in the cortex from subjects with schizophrenia.48 All these data argue that the changes in gene expression we report in the cortex of the schizophrenias is not likely to be wholly due to a simple drug effect. At the level of MRDS versus non-MRDS, we closely matched the two groups at the level of types and doses of prescribed antipsychotic drugs, the frequency of prescription of benzodiazepines and anticholinergic drugs, the frequency of intake of nicotine within 48 h of death as well as the frequency of alcohol and nonprescription drugs not readily available from pharmacies of supermarkets in urine at autopsy. Thus, our carefully considered study design should minimise the likelihood that findings on changed cortical gene expression unique to one of our subgroups is simply due to the effect of the potential confounds to which both subgroups are likely exposed. However, despite careful matching, the mean final recorded antipsychotic drug dose is 34% higher in the subjects with MRDS and therefore it is not possible to totally exclude the effects of antipsychotic drugs as a confounding factor when comparing cortical gene expression in subjects with MRDS and non-MRDS.
This study is unique in that we have measured gene expression in the cortex from subjects within syndrome of schizophrenia who have been divided into two subgroups, the MRDS and non-MRDS sub-groups, based on a loss of cortical muscarinic receptors.4 Significantly, compared to controls, levels of mRNA for more genes are altered in MRDS than are altered in non-MRDS. We postulate this is because we have increased the signal-to-noise ratio for gene expression by studying MRDS, a more biologically homogenous group. In examining changes in gene expression in dorsolateral prefrontal cortex from these subgroups we found that levels of mRNA from 24 genes were only altered in MRDS. Taking into account all genes with changed levels of mRNA in the cortex of subjects with MRDS, our analyses suggest pathways involved in cellular movement and cell-to-cell signalling should be significantly affected in this subgroup. Significantly, for both cellular movement and cell-to-cell signalling, the functional processes most affected are those involved in the formation of glial scars. Glial scars are formed after CNS injury and are thought to inhibit CNS repair.49 There appears to be little evidence to support the notion that glial scars significantly contribute to the aetiology of schizophrenia, but the involvement in glia is suggested.50, 51 Thus, our data may be indicative that glia have a more significant role in the aetiology of MRDS than they do in non-MRDS.
In non-MRDS, the changes in the levels of mRNA were for genes involved in pathways affecting cellular growth and proliferation as well as cellular function and maintenance. Notably, an early study of mRNA levels in the cortex suggested that cellular growth and proliferation would be affected in schizophrenia and bipolar disorder.52 Although we are not aware of cellular function and maintenance being highlighted as being affected in human CNS studies, one study using blood cells has reported an overrepresentation of genes in a number of functional categories that included cellular function and maintenance.53 Thus, in the non-MRDS group the impact of changes in gene focus on pathways such as cellular growth and proliferation and cellular function and maintenance might suggest this group has a stronger neurodevelopmental component to the aetiology of their disorder. There are significant amounts of data involving changes in neurodevelopment in the aetiology of the syndrome of schizophrenia.54
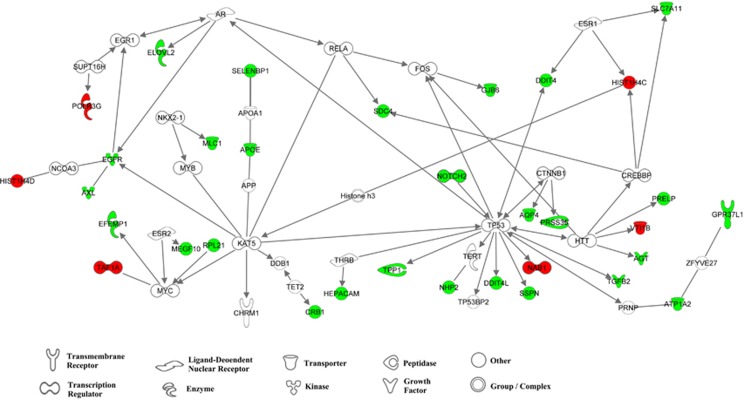
We have identified genes that are only altered in either MRDS or non-MRDS that has given us some insight into their aetiologies. A significant component of this study was to further understand the aetiology of a subgroup within the syndrome of schizophrenia that had MRDS. Hence, we have examined the interactome between genes with altered levels of mRNA in the cortex of subjects with MRDS and how they might interact with CHRM1 expression. This showed that many of the genes with altered levels of mRNA in the cortex of subjects with MRDS interacted upstream of CHRM1 (Figure 3) with the interactive link being K(Lysine) acetyltransferase 5 (KAT5). KAT5 belongs to the MYST family of histone acetyl transferases55 and has a role in DNA repair and apoptosis and is thought to play an important role in signal transduction. Although levels of KAT5 mRNA levels were not different in subjects with MRDS, lower levels of KAT5 mRNA have been reported in another study using BA 8 and BA 9 from subjects with schizophrenia.43 The interactome between genes with altered levels of expression in MRDS reveals an upstream link from CHRM1 via KAT 5 to SELENBP1 through APOE. This is of interest because we have reported levels of SELENBP1 mRNA47 and APOE protein56, 57 to be higher in BA 9 from subjects with the syndrome of schizophrenia. Another significant node in the interactome appears to be tumour protein p53, a tumour suppressor protein that contains transcriptional activation, DNA binding and oligomerisation domains.58 Significantly, this gene has been suggested as a gene that is involved in the aetiology of schizophrenia through modulation of apoptosis.59
Figure 3.
The interactome for genes with changed levels of mRNA in the cortex of subjects with muscarinic receptor-deficit schizophrenia (MRDS) generated using Ingenuity Pathway Analyses using only direct interactions and all data sources. Green symbols are genes with increased levels of mRNA and red symbols show genes with decreased levels of mRNA in the cortex of subjects with MRDS; the symbols define the current functional classification of the gene. Confidence levels were set on experimentally observed or high (predicted) interactions; species was set at mammalian with stringent filtration. All cell lines were included with stringent filtration. Mutations were included and all types of relationships were allowed.
In conclusion, our study has provided new insights into the potential mechanisms leading to the decrease in cortical muscarinic M1 receptors in subjects with MRDS. In particular, our data suggest that changes in glial function may be particularly involved in the aetiology of MRDS, whereas neurodevelopment may be more strongly implicated in the aetiology of MRDS. Being able to draw such conclusions means that our data support the notion that being able to study the aetiologies of subgroups of subjects within the syndrome of schizophrenia should allow their pathophysiologies to be more rapidly elucidated.3 Moreover, a growing insight into the aetiology of MRDS should prove to be valuable in influencing hypothesis-driven drug design7 aimed at developing new treatments to MRDS. This could become critical if it is shown that subjects with MRDS prove to be resistant to treatment with the new drugs that target muscarinic M1 receptors that are proposed to be useful in treating schizophrenia.6 Given this concern, it would seem timely to extend existing neuroimaging data showing lower levels of muscarinic receptors in the CNS from subjects with schizophrenia5 to determine whether this approach can identify living subjects with MRDS.
Acknowledgments
We gratefully acknowledge Geoff Pavey for his technical assistance and curation of the human brain tissue. This project was supported by the National Health and Medical Research Council (Australia; Project Grant 566967, Fellowship (BD) APP1002240), the Australian Research Council (Fellowship (ES) FT100100689) and the Victorian Government’s Operational Infrastructure Support Programme. Tissue was sourced from the Victorian Brain Bank, supported by the Mental Health Research Institute, The Alfred, Victorian Forensic Institute of Medicine, The University of Melbourne and funded by Australia’s National Health & Medical Research Council, Helen Macpherson Smith Trust, Parkinson’s Victoria and Perpetual Philanthropic Services.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
The authors declare no conflict of interest.
Supplementary Material
References
- American Psychiatric AssociationDiagnostic and Statistical Manual of Mood Disorders Fifth Edition vol. 5. American Psychiatric Association: Arlington, 2013. [Google Scholar]
- Bleuler E. Demetia Praecox or the Groups of Schizophrenias. International Press: New York, 1950. [Google Scholar]
- Tamminga CA. Accelerating new knowledge in schizophrenia. Am J Psychiatry 2008; 165: 949–951. [DOI] [PubMed] [Google Scholar]
- Scarr E, Cowie TF, Kanellakis S, Sundram S, Pantelis C, Dean B. Decreased cortical muscarinic receptors define a subgroup of subjects with schizophrenia. Mol Psychiatry 2009; 14: 1017–1023. [DOI] [PubMed] [Google Scholar]
- Raedler TJ, Knable MB, Jones DW, Urbina RA, Gorey JG, Lee KS et al. In vivo determination of muscarinic acetylcholine receptor availability in schizophrenia. Am J Psychiatry 2003; 160: 118–127. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Jones CK, Lindsley CW. Subtype-selective allosteric modulators of muscarinic receptors for the treatment of CNS disorders. Trends Pharmacol Sci 2009; 30: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Roth BL. The pipeline and future of drug development in schizophrenia. Mol Psychiatry 2007; 12: 904–922. [DOI] [PubMed] [Google Scholar]
- Gibbons AS, Scarr E, Boer S, Money T, Jeon WJ, Felder C et al. Widespread decreases in cortical muscarinic receptors in a subset of people with schizophrenia. Int J Neuropsychopharmacol 2013; 16: 37–46. [DOI] [PubMed] [Google Scholar]
- Dean B, Thomas N, Lai CY, Chen WJ, Scarr E. Changes in cholinergic and glutamatergic markers in the striatum from a sub-set of subjects with schizophrenia. Schizophr Res 2015; 169: 83–88. [DOI] [PubMed] [Google Scholar]
- Scarr E, Craig JM, Cairns MJ, Seo MS, Galati JC, Beveridge NJ et al. Decreased cortical muscarinic M1 receptors in schizophrenia are associated with changes in gene promoter methylation, mRNA and gene targeting microRNA. Transl Psychiatry 2013; 3: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah-Uddin H, Scarr E, Pavey G, Harris K, Hagan JJ, Dean B et al. Altered M(1) muscarinic acetylcholine receptor (CHRM1)-Galpha(q/11) coupling in a schizophrenia endophenotype. Neuropsychopharmacology 2009; 34: 2156–2166. [DOI] [PubMed] [Google Scholar]
- Dean B, Hopper S, Conn PJ, Scarr E. Changes in BQCA allosteric modulation of [3H]NMS binding to human cortex within schizophrenia and by divalent cations. Neuropsychopharmacology 2016; 41: 1620–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Yamamoto M, Izuhara Y. From molecular footprints of disease to new therapeutic interventions in diabetic nephropathy. Ann NY Acad Sci 2005; 1043: 740–749. [DOI] [PubMed] [Google Scholar]
- Tsuang M. Schizophrenia: genes and environment. Biol Psychiatry 2000; 47: 210–220. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci 2007; 8: 355–367. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron 2000; 28: 53–67. [DOI] [PubMed] [Google Scholar]
- Horvath S, Mirnics K. Schizophrenia as a disorder of molecular pathways. Biol Psychiatry 2015; 77: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Keks N, Roberts S, Opeskin K, Dean B, Mackinnon A et al. Problem of diagnosis in postmortem brain studies of schizophrenia. Am J Psychiatry 1996; 153: 533–537. [DOI] [PubMed] [Google Scholar]
- Roberts SB, Hill CA, Dean B, Keks NA, Opeskin K, Copolov DL. Confirmation of the diagnosis of schizophrenia after death using DSM-IV: a Victorian experience. Aust NZ J Psychiatry 1998; 32: 73–76. [DOI] [PubMed] [Google Scholar]
- American Psychiatric AssociationDiagnostic and Statistical Manual of Mental Disorders Fourth Edition vol. 4. American Psychiatric Association: Washington, DC, 1994. [Google Scholar]
- Remington GJ. Antipsychotics (Neuroleptics). In: B-Bjai JJ (ed). Clincal Handbook of Psychotropic Drugs (15th Update). Hogrefe & Huber: Seattle, Toronto, Gottingen, Bern, 1999. pp 55–84. [Google Scholar]
- Dean B, Pavey G, Chai SY, Mendelsohn FAO The localisation and quantification of molecular changes in the human brain using in situ radioligand binding and autoradiography. In: Dean B, Kleinman JE, Hyde TM (eds). Using CNS Tissue in Psychiatric Research: A Practical Guide. Harwood Academic Press: Amsterdam, 1999. pp 67–83. [Google Scholar]
- Ferrer I, Santpere G, Arzberger T, Bell J, Blanco R, Boluda S et al. Brain protein preservation largely depends on the postmortem storage temperature: implications for study of proteins in human neurologic diseases and management of brain banks: a BrainNet Europe Study. J Neuropathol Exp Neurol 2007; 66: 35–46. [DOI] [PubMed] [Google Scholar]
- Kingsbury AE, Foster OJ, Nisbet AP, Cairns N, Bray L, Eve DJ et al. Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Brain Res Mol Brain Res 1995; 28: 311–318. [DOI] [PubMed] [Google Scholar]
- Stan AD, Ghose S, Gao XM, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ et al. Human postmortem tissue: what quality markers matter? Brain Res 2006; 1123: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke F, Bullen C, Whittaker R, McRobbie H, Chen MH, Walker N. Diagnostic accuracy of NicAlert cotinine test strips in saliva for verifying smoking status. Nicotine Tob Res 2008; 10: 607–612. [DOI] [PubMed] [Google Scholar]
- Kiewe P, Gueller S, Komor M, Stroux A, Thiel E, Hofmann WK. Prediction of qualitative outcome of oligonucleotide microarray hybridization by measurement of RNA integrity using the 2100 Bioanalyzer capillary electrophoresis system. Ann Hematol 2009; 88: 1177–1183. [DOI] [PubMed] [Google Scholar]
- MAQC Consortium Shi L Reid LH Jones WD Shippy R Warrington JA et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 2006; 24: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan S, Tang B, Head SR, Gilmartin TJ, Sutcliffe JG, Dean B et al. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res 2008; 1239: 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udawela M, Scarr E, Hannan AJ, Thomas EA, Dean B. Phospholipase C beta 1 expression in the dorsolateral prefrontal cortex from patients with schizophrenia at different stages of illness. Aust NZ J Psychiatry 2011; 45: 140–147. [DOI] [PubMed] [Google Scholar]
- Dean B, Udawela M, Scarr E. Validating reference genes using minimally transformed qpcr data: findings in human cortex and outcomes in schizophrenia. BMC Psychiatry 2016; 16: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi V, Ramani R, Andes D, Diekema DJ, Pfaller MA, Ghannoum MA et al. Multilaboratory testing of two-drug combinations of antifungals against Candida albicans, Candida glabrata, and Candida parapsilosis. Antimicrob Agents Chemother 2011; 55: 1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Hu M, Wang C, Wang C, Zhang F, Han Q et al. Coordinated gene expression of Th17- and Treg-associated molecules correlated with resolution of the monophasic experimental autoimmune uveitis. Mol Vis 2011; 17: 1493–1507. [PMC free article] [PubMed] [Google Scholar]
- Udovičić M, Baždarić K, Bilić-Zulle L, Petrovečki M. What we need to know when calculating the coefficient of correlation? Biochem Med 2007; 17: 10–15. [Google Scholar]
- Rao JS, Kim HW, Harry GJ, Rapoport SI, Reese EA. Increased neuroinflammatory and arachidonic acid cascade markers, and reduced synaptic proteins, in the postmortem frontal cortex from schizophrenia patients. Schizophr Res 2013; 147: 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetre P, Emilsson L, Axelsson E, Kreuger J, Lindholm E, Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry 2007; 7: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Leonardi-Essmann F, Durrenberger PF, Parlapani E, Schneider-Axmann T, Spanagel R et al. Regulation of immune-modulatory genes in left superior temporal cortex of schizophrenia patients: a genome-wide microarray study. World J Biol Psychiatry 2011; 12: 201–215. [DOI] [PubMed] [Google Scholar]
- Tang B, Capitao C, Dean B, Thomas EA. Differential age- and disease-related effects on the expression of genes related to the arachidonic acid signaling pathway in schizophrenia. Psychiatry Res 2012; 196: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry 2007; 62: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci 2002; 22: 2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry 2004; 9: 684–697. [DOI] [PubMed] [Google Scholar]
- Maycox PR, Kelly F, Taylor A, Bates S, Reid J, Logendra R et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry 2009; 14: 1083–1094. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Dean B, Schork NJ, Thomas EA. Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Res 2010; 20: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Hwang H, Lee HW, Suk K. Microglia signaling as a target of donepezil. Neuropharmacology 2010; 58: 1122–1129. [DOI] [PubMed] [Google Scholar]
- Scarr E, Udawela M, Greenough MA, Neo J, Seo MS, Money TT et al. Altered expression of the zinc transporter SLC39A12 suggests a breakdown in zinc cortical homeostasis as part of the pathophysiology of schizophrenia. NPJ Schizophr 2016; 2: Article number: 16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udawela M, Money TT, Neo J, Seo MS, Scarr E, Dean B et al. SELENBP1 expression in the prefrontal cortex of subjects with schizophrenia. Transl Psychiatry 2015; 5: e615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ML, Eaton ME, Dudman JT, Konradi C. Antipsychotic drugs elevate mRNA levels of presynaptic proteins in the frontal cortex of the rat. Biol Psychiatry 2005; 57: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch MT, Silver J. CNS injury, glial scars, and inflammation: Inhibitory extracellular matrices and regeneration failure. Exp Neurol 2008; 209: 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowska G, Halaris A, Selemon LD. Reductions in neuronal and glial density characterize the dorsolateral prefrontal cortex in bipolar disorder. Biol Psychiatry 2001; 49: 741–752. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: a postmortem morphometric study of schizophrenia and Huntington disease. Arch Gen Psychiatry 1998; 55: 215–224. [DOI] [PubMed] [Google Scholar]
- Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry 2008; 64: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Martin MV, Rollins B, Moon EA, Bunney WE, Macciardi F et al. Mitochondrial mutations and polymorphisms in psychiatric disorders. Front Genet 2012; 3: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan MJ, Andrieux A, Bartzokis G, Cadenhead K, Dazzan P, Fusar-Poli P et al. Altering the course of schizophrenia: progress and perspectives. Nat Rev Drug Discov 2016; 15: 485–515. [DOI] [PubMed] [Google Scholar]
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 2007; 26: 5395–5407. [DOI] [PubMed] [Google Scholar]
- Dean B, Laws SM, Hone E, Taddei K, Scarr E, Thomas EA et al. Increased levels of apolipoprotein E in the frontal cortex of subjects with schizophrenia. Biol Psychiatry 2003; 54: 616–622. [DOI] [PubMed] [Google Scholar]
- Digney A, Keriakous D, Scarr E, Thomas E, Dean B. Differential changes in apolipoprotein E in schizophrenia and bipolar I disorder. Biol Psychiatry 2005; 57: 711–715. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Achatz MIW, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 2007; 26: 2157–2165. [DOI] [PubMed] [Google Scholar]
- Catts VS, Catts SV. Apoptosis and schizophrenia: is the tumour suppressor gene, p53, a candidate susceptibility gene? Schizophr Res 2000; 41: 405–415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.