Abstract
Chemotaxis toward organic acids has been associated with colonization fitness and virulence and the opportunistic pathogen Pseudomonas aeruginosa exhibits taxis toward several tricarboxylic acid intermediates. In this study, we used high-throughput ligand screening and isothermal titration calorimetry to demonstrate that the ligand binding domain (LBD) of the chemoreceptor PA2652 directly recognizes five C4-dicarboxylic acids with KD values ranging from 23 µM to 1.24 mM. In vivo experimentation showed that three of the identified ligands act as chemoattractants whereas two of them behave as antagonists by inhibiting the downstream chemotaxis signalling cascade. In vitro and in vivo competition assays showed that antagonists compete with chemoattractants for binding to PA2652-LBD, thereby decreasing the affinity for chemoattractants and the subsequent chemotactic response. Two chemosensory pathways encoded in the genome of P. aeruginosa, che and che2, have been associated to chemotaxis but we found that only the che pathway is involved in PA2652-mediated taxis. The receptor PA2652 is predicted to contain a sCACHE LBD and analytical ultracentrifugation analyses showed that PA2652-LBD is dimeric in the presence and the absence of ligands. Our results indicate the feasibility of using antagonists to interfere specifically with chemotaxis, which may be an alternative strategy to fight bacterial pathogens.
Introduction
A series of different signal transduction systems permit bacteria to sense changing environmental conditions and to generate adaptive responses. Next to one- and two-component systems, chemosensory pathways represent a major mechanism in bacterial signal transduction1–3. In these systems, the direct binding of chemoeffectors or chemoeffector-loaded periplasmic binding proteins to the ligand binding domain (LBD) of chemoreceptors4 generates a molecular stimulus that alters the autophosphorylation of the histidine kinase CheA and consequently the transphosphorylation of the CheY response regulator, which represents the pathway output2. Chemosensory pathways were shown to mediate chemotaxis and type IV pili-based motility, or are involved in regulating alternative cellular processes5–7.
The opportunistic pathogen Pseudomonas aeruginosa is an important model organism to investigate chemosensory pathways8. Its chemoreceptors feed into 4 different pathways. Two of these signalling cascades, che and che2, mediate flagellum-mediated taxis9,10. However, the wsp pathway controls c-di-GMP levels5 whereas the fourth pathway, chp, is responsible for type IV pili-mediated motility11,12 and the regulation of cAMP levels13. The function of most of the 26 P. aeruginosa chemoreceptors remains unknown but others have been characterized in depth, including the three paralogous receptors PctA, PctB and PctC for the chemotaxis to different amino acids14–17 and the CtpH and CtpL18,19 receptors that mediate chemoattraction to inorganic phosphate.
P. aeruginosa is a ubiquitous pathogen able to infect a broad range of different hosts such as human, animals, plants or fungi20. Part of our research interests consists in assessing how chemosensory signalling mechanisms compare in phylogenetically related species that have different lifestyles. To address this issue we study P. putida KT2440, a non-pathogenic and nutritionally versatile soil bacterium with saprophytic lifestyle21–23. The genome of P. putida KT2440 encodes 3 chemosensory pathways24 and 27 chemoreceptors, which is very similar to the number of chemoreceptors in P. aeruginosa PAO1. However, sequence analyses and functional data appear to indicate that these are not sets of homologous proteins with homologous function. Initial evidence suggests that chemoreceptors that mediate responses to different compound classes are rather different. One such example are chemoreceptors of KT2440 and PAO1 for tricarboxylic acid (TCA) cycle intermediates. In KT2440, three receptors, McpS, McpQ and McpR, have been shown to mediate responses to TCA cycle intermediates. McpS is a broad ligand range chemoreceptor that binds most of the TCA cycle intermediates25,26. Interestingly, McpS binds citrate, an abundant compound in plant tissues and root exudates, with only low affinity. However, McpS does not bind the metal ion complexed form of citrate27, which is the primary form of citrate in the environment. This may have been the reason for the evolution of McpQ, a chemoreceptor that binds specifically citrate in both its metal-free and metal-complexed forms28. In addition, McpR was found to mediate chemotaxis to malate and fumarate29. Remarkably, McpS and McpQ possess an helical bimodular (HBM) type sensor domain30 whereas McpR has a 4-helix bundle (4HB) domain31.
TCA cycle responsive chemoreceptors so far identified in P. aeruginosa are McpK, mediating specific responses to α-ketoglutarate32, as well as the malate specific receptor, PA265233. Whereas McpK has an HBM type LBD, PA2652 possess a sCACHE domain. CACHE domains are abundant sensor domains in chemoreceptors and sensor kinases34,35 and exist in two forms: (i) sCACHE (single CACHE), composed of a single structural module; and (ii) dCACHE (double CACHE), consisting of two CACHE modules in tandem. P. aeruginosa and P. putida contain a significant number of dCACHE containing chemoreceptors, namely 5 and 9, respectively. However, the genomes of both strains only encode a single sCACHE domain containing receptor. McpP, the sCACHE containing receptor of KT2440, was found to bind acetate, pyruvate, propionate and L-lactate36. McpP and PA2652 share 37% of sequence identity whereas the identity of their respective LBDs is only 23% (Supplementary Fig. S1), underling the important sequence divergence.
Here we report the characterization of the chemoreceptor PA2652 of P. aeruginosa. The study that reported its initial identification showed that a mutant in this gene did not respond to malate33, but it is unknown whether it binds malate directly or via periplasmic binding proteins. This study also demonstrated that PA2652 is involved in the response of P. aeruginosa to malate but not to other organic acids such as succinate, 2-oxoglutarate, citrate or acetate33. We used here high throughput approaches37,38 to define more precisely the chemoeffector range of PA2652.
Results
PA2652 binds several C2-substituted C4-dicarboxylic acids
To identify the LBD of PA2652, its sequence was analysed by the DAS transmembrane region prediction algorithm39. The receptor was found to possess two transmembrane regions (Supplementary Fig. S2) and the DNA fragment encoding the section in between both regions was cloned into an expression vector. The resulting protein, PA2652-LBD, was expressed in Escherichia coli and purified from the soluble fraction of the E. coli lysate by metal affinity chromatography.
To identify ligands that may bind to the LBD of PA2652, we conducted Differential Scanning Fluorimetry (DSF) based high throughput ligand screening assays as described previously37,38. DSF analyses permit the determination of melting temperature (Tm) values, which corresponds to the temperature at which half the protein is in its native conformation whereas the remaining half has undergone thermal unfolding. Since ligand binding typically enhances the thermal stability of proteins, increases in the Tm in the presence of ligands may be indicative of specific binding. We screened 450 different compounds available in five different ligand arrays from Biolog (Supplementary Fig. S3). The screened collection included different carbon and nitrogen sources, phosphorous and sulfurous compounds as well as different nutrient supplements. DSF assays evidenced a Tm of 45.5 °C for the ligand-free PA2652-LBD and Fig. 1 shows the Tm changes produced by the presence of each of the 95 compounds of the PM1 array of different carbon sources. Tm increases of more than 2 °C, an accepted threshold for significant hits, were observed for a mixture of L- and D-malic acid, D,L-bromosuccinic acid as well as L-malic acid, whereas D-malic acid did not cause significant increases. Screening of other arrays also resulted in several hits, including citraconic acid and racemic mixtures of citramalic acid (Supplementary Table S1).
Figure 1.
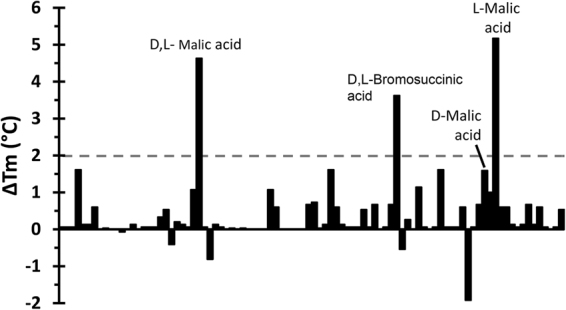
Differential Scanning Fluorimetry based high-throughput ligand screening of PA2652-LBD. Shown are Tm changes for each of the 95 compounds present in the Biolog PM1 compound array of carbon sources with respect to the Tm of the ligand-free protein of 45.5 °C. The dashed line indicates the threshold of 2 °C for significant hits.
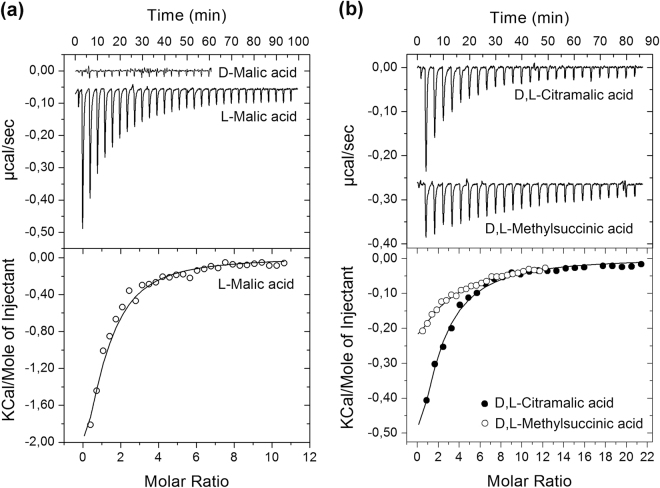
Thermal shift assays indicate but do not constitute proof of binding32,38. To unambiguously determine ligand binding, we conducted Isothermal Titration Calorimetry (ITC)40 binding studies with the purified protein. Figure 2a shows the microcalorimetric titration of PA2652-LBD with the L- and D-isomers of malic acid. L-malic acid bound with a KD value of 23 ± 1 µM, which is very similar to the affinities of ligands for McpP-LBD36. Binding was driven by both favourable enthalpy (ΔH = −4.2 ± 1.2 kcal) and entropy changes (TΔS = 2.1 ± 1 kcal/mol). In marked contrast, D-malic acid did not show binding (Fig. 2a) confirming the DSF data (Fig. 1).
Figure 2.
Isothermal titration calorimetry analysis of ligand binding to PA2652-LBD. (a) Titration with both malic acid isomers. (b) Titration with racemic mixtures of citramalic and methylsuccinic acids. The upper panels are the titration raw data for the injection of 9.6–14.4 μl aliquots of 1–2 mM ligand solution into 20–35 μM of protein. The lower panels are the integrated, dilution heat corrected and concentration normalized peak areas fitted with the “One binding site” model of ORIGIN.
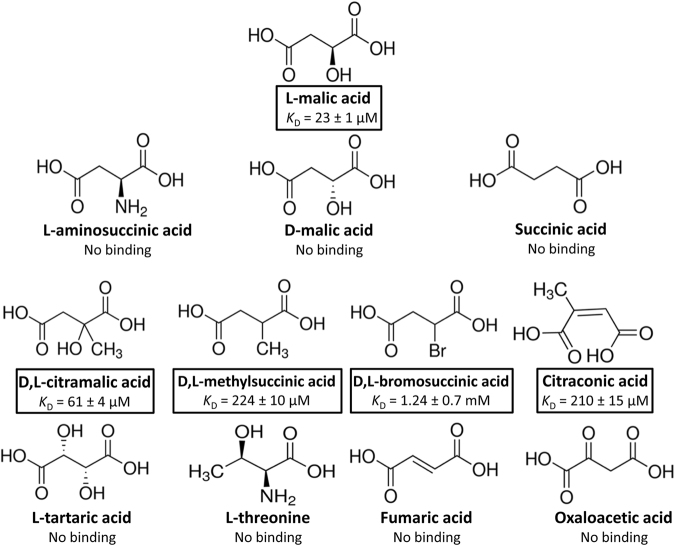
We subsequently verified by ITC the binding of other compounds that caused Tm increases of at least 2 °C and detected binding for citraconic acid and racemic mixtures of citramalic and bromosuccinic acids with KD values of 210 ± 15 µM, 61 ± 4 µM and 1.24 ± 0.7 mM, respectively (Figs 2b and 3). As a result, it became clear that receptor PA2652 binds different C4-dicarboxylic acids and, in order to complete the ligand profile of this receptor, we analysed the binding of additional structurally related compounds. These experiments resulted in the detection of binding for D,L-methylsuccinic acid (not present in the compound arrays; Fig. 2b) whereas assays using other C4-dicarboxylates (Supplementary Table S1) such as succinic, fumaric, oxalacetic, aminosuccinic or tartaric acids resulted in an absence of binding. We also investigate whether different commercially available D- stereoisomers of citramalic and methylsuccinic acids bind to PA2652-LBD. As observed for D-malic acid, no binding was observed (Supplementary Fig. S4), as an indication that the LBD of PA2652 specifically binds L-stereoisomers of organic acids. Importantly, the five ligands recognized by PA2652 are thus C2-substituted C4-dicarboxylic acids (Fig. 3).
Figure 3.
Summary of isothermal titration calorimetry studies. Shown are the structures of the five ligands that showed binding as well as of other compounds that were analysed but did not reveal binding. Dissociation constants are means and standard deviations from three experiments.
L-malic acid binding does not change the dimeric state of PA2652-LBD
Ligand-induced dimerization of the chemoreceptor ligand binding domain was proposed to be a necessary prerequisite for signalling41. Chemoreceptors employ different types of LBD4 and previous studies have assessed the effect of ligand binding to the oligomeric state of individual LBDs. These experiments have resulted in two different scenarios. On one hand, individual 4HB or HBM domains are primarily monomeric in its ligand-free state and ligand binding induces dimerization25,32,42,43. In contrast, dCACHE domains were found to be monomeric in the absence and presence of ligands15. However, no information is available on the effect of ligand binding on the oligomeric state of sCACHE domains.
To address this issue, PA2652-LBD was analysed by sedimentation velocity analytical ultracentrifugation (AUC). Initial experiments were conducted using concentrations of ligand-free protein ranging from 5 to 20 µM. These assays showed no significant differences in the sedimentation coefficients calculated for the species identified, which rules out hydrodynamic non-ideality behaviour. The analysis of single species in the sedimentation profile resulted in a standard sedimentation coefficient of sw,20 = 3.1 S and a frictional ratio of 1.4; the latter indicative of an elongated protein shape (Fig. 4). The molecular weight extracted from the sedimentation coefficient and the shape was of 41 kDa. Considering that the sequence-derived mass of the PA2652-LBD monomer is 21.5 kDa, the observed species is clearly a protein dimer. Since no shift to higher sedimentation coefficients was measured with increasing protein concentrations (data not shown), the dimer species can be considered stable over the protein concentration range analysed. The above experiments were also performed in the presence of saturating concentrations of L-malic acid. In these assays, the behaviour of PA2652-LBD was highly similar to that of the unliganded protein and data analysis resulted in the same sedimentation coefficient and frictional ratio as observed for the ligand-free protein (Fig. 4). It can therefore be concluded that PA2652-LBD forms stable dimers in the ligand-free state and that the binding of L-malic acid does not have any significant effects on the oligomerization state of the protein.
Figure 4.

Sedimentation velocity analytical ultracentrifugation analysis of PA2652-LBD. The sedimentation coefficient profile is shown for the protein at 20 µM in the absence and in presence of 1 mM L-malic acid. Values shown are expressed at the conditions of the experiment, namely at a temperature of 7 °C and PIPES buffer.
PA2652 ligands act as attractants and antagonists
To evaluate the physiological relevance of the ligands identified, we first addressed the question of whether they could support bacterial growth as sole carbon source. To this end, we conducted growth experiments of the wild type (wt) strain and a mutant defective in PA2652 in minimal medium supplemented with the different ligands as sole carbon sources. As shown in Supplementary Fig. S5, L-malic acid and racemic mixtures of citramalic and methylsuccinic acids were able to efficiently sustain growth of both PAO1 strains. Alternatively, D,L-bromosuccinic acid poorly supported bacterial growth, whereas citraconic acid could not be used as sole carbon source (Supplementary Fig. S5).
Secondly, we assessed the capacity of the ligands identified to induce chemotaxis. To this end, we carried out quantitative capillary chemotaxis assays using the wt strain and PA2652 ligands at concentrations ranging from 10 µM to 100 mM. Our results showed that L-malic acid induces strong chemotaxis responses over the concentration range of 100 µM to 10 mM (Fig. 5). Significant responses were also observed for racemic mixtures of bromosuccinic and citramalic acids, with a maximum chemotactic response at a concentration of 10 mM. Minor but statistically significant responses were also observed for D,L-methylsuccinic acid, whereas citraconic acid, the only ligand that did not support growth (Supplementary Fig. S5), did not cause any chemotactic response (Fig. 5).
Figure 5.
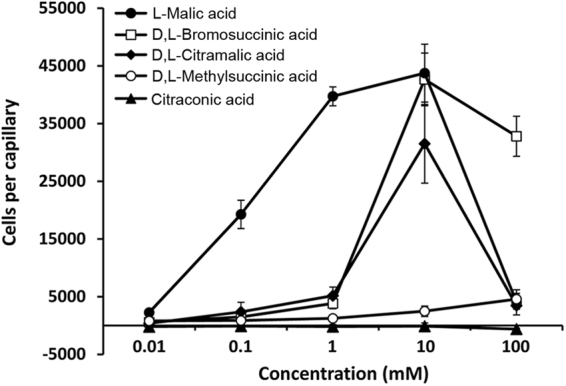
Quantitative capillary chemotaxis assays of Pseudomonas aeruginosa PAO1 toward different organic acids. Data are means and standard deviations from three biological replicates conducted in triplicate. Data were corrected with the number of cells that swam into buffer containing capillaries (1713 ± 231).
To determine the role of the PA2652 receptor in the observed tactic responses, we characterized phenotypically a mutant defective in the corresponding gene. Initial control experiments involved the measurement of chemotaxis of the wt and a PA2652 mutant strain toward casamino acids, which is mediated by the three paralogous receptors PctA, PctB and PctC14,15. Both the wt and the mutant strain showed similar responses to casamino acids (Supplementary Fig. S6) indicating that the mutation of PA2652 did not result in any undesired secondary effects. Quantitative chemotaxis assays of the mutant strain toward 10 mM ligand solutions showed a dramatic reduction in chemotaxis for all ligands (Fig. 6), indicating that PA2652 is the primary receptor for these C4-dicarboxylic acids. The minor responses in the mutant strain may be potentially due to a secondary receptor. Importantly, in trans expression of PA2652 in a mutant defective in PA2652 resulted in the complementation of the chemotaxis defect toward C4-dicarboxylic acids (Supplementary Fig. S7).
Figure 6.

Quantitative capillary chemotaxis assays of Pseudomonas aeruginosa PAO1 and its mutant defective in PA2652 to different PA2652 chemoeffectors. In all cases, chemoeffectors were used at a final concentration of 10 mM. Data were corrected with the number of cells that swam into buffer containing capillaries (1565 ± 327). Data are means and standard deviations from three biological replicates conducted in triplicate.
Attractants and antagonists compete for binding at PA2652-LBD in vitro and in vivo
The above results suggest that chemoattractants (L-malic, D,L-bromosuccinic and L-citramalic acids) and antagonists (L-methylsuccinic and citraconic acids) may compete for binding to PA2652-LBD. In order to understand the mechanistic role of these antagonistics in the chemotaxis behaviour of P. aeruginosa, we first carried out ITC binding studies. In these assays, we measured the affinity of L-malic acid for PA2652-LBD in the presence of different concentrations of citraconic and D,L-methylsuccinic acids. Our data showed that heat released from the binding of L-malic acid was reduced as the antagonist concentration increased. The apparent affinity of L-malic acid decreased approximately by a factor of two in the presence of 2 mM antagonists and increasing the concentration of antagonists to 20 mM led to a further reduction in the apparent affinity for L-malic acid (Fig. 7 and Supplementary Table S2). Control assays titrating buffer or buffer/antagonist solutions with L-malic acid or L-malic acid/antagonist solutions resulted in small and uniform peaks indicative of dilution heats (Supplementary Fig. S8). Considering the close structural similarity of antagonists and L-malic acid (Fig. 3) it is likely that these compounds compete for binding at the same site at PA2652.
Figure 7.
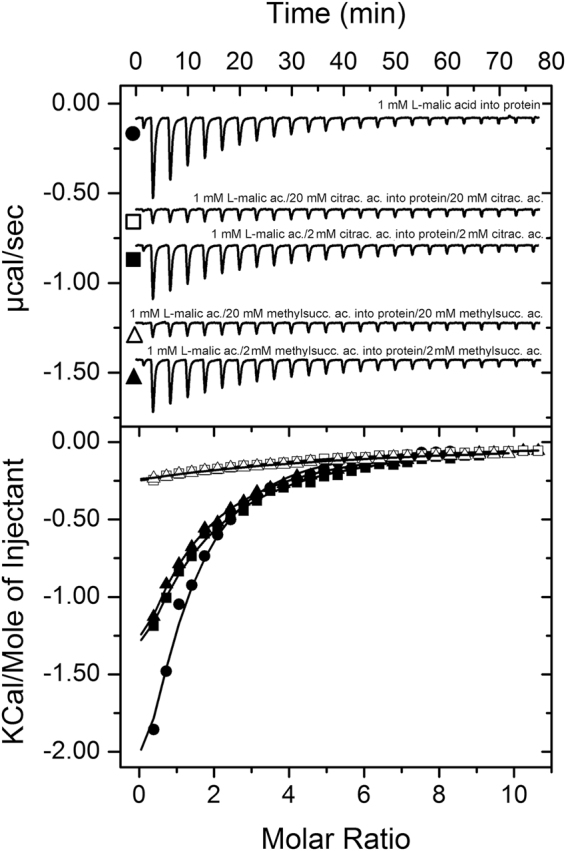
Attractants and antagonists compete for binding at PA2652-LBD in vitro. Isothermal titration calorimetry analysis of the binding of L-malic acid to PA2652-LBD in the absence and presence of 2 or 20 mM of the antagonists, citraconic and D,L-methylsuccinic acids. Upper panel: Titration raw data for the injection of 9.6 μl aliquots of 1 mM of L-malic acid into 20 μM of protein in the absence and presence of antagonists (present both in the injector syringe and sample cell). Lower panel: Integrated, dilution heat corrected and concentration normalized peak areas fitted with the “One binding site” model of ORIGIN. The apparent binding constants are listed in Supplementary Table S2.
Following the demonstration of competition in vitro we have assessed the influence of antagonists on the chemotaxis toward L-malic acid. To this end we conducted chemotaxis assays toward L-malic acid in the absence or presence of 10, 20 and 40 mM of citraconic and D,L-methylsuccinic acids (that were added to both, the bacterial suspension and the chemoattractant solution). Our results showed that antagonists reduced the chemotaxis toward L-malic acid, with higher decreases in the chemotactic response as the concentration of the antagonist was increased, therefore indicating that citraconic and methylsuccinic acids are inhibiting the activation of the chemotaxis signalling cascade triggered by L-malic acid (Fig. 8). To verify whether the presence of antagonists may have a global inhibitory effect on the chemotactic properties of P. aeruginosa we performed assays toward L-alanine, a chemoattractant recognized by the receptors PctA and PctB15, in the presence antagonists. As shown in Supplementary Fig. S9, the presence of 20 or 40 mM citraconic and D,L-methylsuccinic acids did not significantly alter the chemotaxis toward 1 mM L-alanine.
Figure 8.
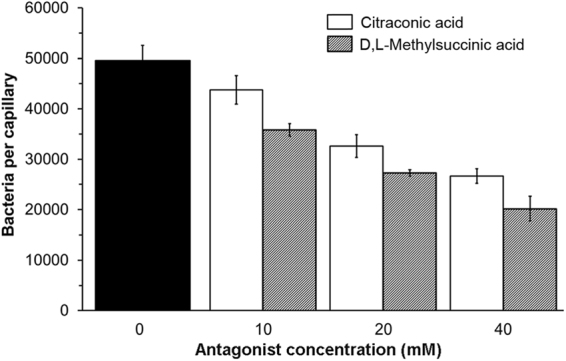
Antagonists reduce the magnitude of chemotaxis toward L-malic acid. Quantitative capillary chemotaxis assays of P. aeruginosa PAO1 toward 1 mM L-malic acid in the absence (black bars) and presence of different antagonist concentrations. Data are means and standard deviations from three biological replicates conducted in triplicate. Data were corrected with the number of cells that swam into buffer containing capillaries (4266 ± 1133).
The PA2652 chemoreceptor signals through the che chemosensory pathway
As mentioned in the introduction, P. aeruginosa has two chemosensory pathways that were found to be involved in chemotaxis, namely the che and che2 pathways9,10. A recent bioinformatic study has predicted that PA2652 signals through the che pathway44. To verify this prediction we conducted chemotaxis assays to L-malic acid using the wt strain as well as mutants in cheA1 and cheA2, encoding respectively the histidine kinases of the che and che2 pathways. As shown in Supplementary Fig. S10, mutation of cheA1 abolished chemotaxis to L-malic acid, whereas the response of the cheA2 mutant was almost identical to that of wt strain, confirming bioinformatic predictions44.
The effect of PA2652 in plant root colonization
P. aeruginosa is an ubiquitous pathogen and also able to colonize and infect different plants20,45,46. Previous studies using a number of different bacterial species have shown that chemotaxis to root exudates is an important prerequisite for efficient plant root colonization47. Taken together data from different plant species, malate and citrate are considered the most abundant organic acids in plant tissues and root exudates, and they can represent up to 25% of total photosynthate exuded by the plant48,49. Therefore, considering the important concentrations of malate in root exudates and using maize as model plants, we carried out competitive root colonization assays of the kanamycin resistant P. aeruginosa strain PAO1-Km and a mutant defective in PA2652. The assays showed that the fitness of the mutant in PA2652 was comparable to that of the wild type strain (Supplementary Fig. S11).
Discussion
Sensing of environmental signals by one- and two-component systems as well as chemotaxis signalling pathways occurs through sensor domains. Remarkably, the development of next generation sequencing technologies has allowed to determine that these three transduction mechanisms share a significant number of sensor domain types4,50. Additionally, the combination of computational and experimental approaches enabled the identification of inhibiting compounds, known as antagonists, that are sensed by one- and two-component systems. These molecules compete for binding to the sensor domains and block the agonist-induced response51–54. Importantly, there is increasing evidence indicating that signal antagonists can also act on chemoreceptors. Thus, it was shown that the binding of several compounds to the 4HB type LBDs of the Tar and MCP2201 chemoreceptors, from E. coli and Comamonas testosteroni respectively, did not cause any chemotactic responses55–57.
In this study we identified four new ligands for PA2652, which all occur naturally58–61. However, significant chemotaxis was only observed for two of them, bromosuccinic and citramalic acids - to our knowledge the first report of bacterial chemotaxis to both of these compounds. In contrast, no or very minor responses were observed for citraconic and methylsuccinic acids, respectively (Fig. 5). Additional in vitro experimentation showed that citraconic and methylsuccinic acids bind to the sCACHE domain of PA2652 and act as antagonists by competing for binding with chemoattractants (Fig. 7). Consequently, taxis of P. aeruginosa toward chemoattractants was reduced in the presence of these two antagonists (Fig. 8), as observed previously for other chemoreceptor antagonists55,56. Interestingly, malate was shown to be an antagonist of MCP2201 and its binding to the LBD of this chemoreceptor reduced the chemotactic behavior of C. testosteroni toward diverse aromatic compounds56. Taken together, our data illustrate that the action of antagonists is not only restricted to 4HB LBDs, but also occurs at chemoreceptors with a different LBD type. Could these findings be of applied interest? The increasing emergence of multidrug-resistant bacteria is challenging human health and new targets for the development of antibiotics are urgently needed62. Chemotaxis has been shown to be required for the full virulence of multiple human pathogens63 and drugs targeting chemosensory signalling pathways constitute promising approaches for the discovery of new antibiotics64. Since many drugs are based on antagonists65,66, the identification of compounds interfering with chemotactic transduction pathways may potentially enable the rational design of drugs inhibiting chemotaxis-associated processes.
CACHE domains are the most abundant sensor domains in chemoreceptors and sensor kinases34,35. Whereas P. aeruginosa PAO1 and P. putida KT2440 have an elevated number of dCACHE containing chemoreceptors, both strains have a single sCACHE domain containing chemoreceptor. Although both receptors bind organic acids, their ligand profiles are different. Whereas McpP binds several C2- and C3-carboxylic acids36, we show here that PA2652 binds several C2-substituted C4-dicarboxylic acids. Interestingly, the homologous chemoreceptor in the plant pathogen P. syringae pv. actinidiae was found to have a ligand profile that is very similar to that of P. putida KT244067.
Several previous studies have shown that the affinity of ligands for the LBD correlate with the magnitude of the chemosensory output16,43. However, this correlation was not observed for the PA2652 ligands (Figs 3 and 5). In the initial study of PA2652, chemotaxis assays were performed using malate samples containing both isomers33. Here we show that PA2652 binds exclusively the L- but not the D-isomer of malic, citramalic and methylsuccinic acids (Fig. 2a and Supplementary Fig. S4). In this respect, clear parallels exist to McpP that binds only L-lactate but not D-lactate36. Many bacteria are able to synthesize D-malate68,69 and Pseudomonas species were found to metabolize both isomers70. However, L-malate is a common carboxylic acid whereas its D-isomer is less frequent71. In the context of sensory mechanisms for the regulation of the metabolism of organic acids, it has been proposed that common carboxylic acids are sensed in the periplasm whereas uncommon acids are sensed in the cytosol69,71. For example the DcuS sensor kinase, comprising a periplasmic LBD, senses L-malate72 whereas the cytosolic transcriptional regulator DmlR senses D-malate73. This differentiation appears to also apply to L-malate chemoreceptors. Chemoreceptors can sense their ligands either in the cytosol or the extracytoplasmic space74. A significant number of malate responsive chemoreceptors have been identified in a variety of different species and the corresponding information is summarized in Table 1. Whereas in some studies mixtures of D- and L-malate were used, other reports study the individual malate isomers. Interestingly, next to PA2652, the McpS receptor of P. putida KT244026, McpM of Ralstonia pseudosolanacearum75 as well as the Pfl01_0728 and Pfl01_3768 of P. fluorescens76 were found to mediate specifically L-malate chemotaxis. All these receptors are predicted to possess a LBD in the periplasmic space confirming the hypothesis of extracytoplasmic sensing of common organic acids. The response of R. pseudosolanacearum to D-malate has also been investigated and was attributed to an fortuitous response of L-malate sensing receptors77. In contrast to other malate specific receptors, the LBD of both malate chemoreceptors of R. pseudosolanacearum have a 4HB domain (Table 1).
Table 1.
Summary of information available on malate responsive chemoreceptors.
| Name | Species | Ligands | Binding mode | LBD typea | Predicted LBD locationb | Reference |
|---|---|---|---|---|---|---|
| PA2652 | P. aeruginosa PAO1 | L-malic, citramalic, citraconic, bromosuccinic and methylsuccinic acids | direct | sCACHE | periplasm | This work |
| McpS (PP4658) | P. putida KT2440 | L-malic, oxalacetic, citric, isocitric, succinic, fumaric and butyric acids | direct | HBM | periplasm | 25,26 |
| McpM (GenBank accession no. LC005239) | Ralstonia pseudosolanacearum Ps29 | L-malic, D-malic, D-tartaric, succinic, and fumaric acids | unknown | 4HB | periplasm | 75,77 |
| McpT (GenBank accession no. LC005228) | Ralstonia pseudosolanacearum Ps29 | D-malic acid, D-tartaric and L-tartaric acids | unknown | 4HB | periplasm | 77 |
| McfS (Pput_4520) | P. putida F1 | succinic, malic, citric and fumaric acids | unknown | HBM | periplasm | 29 |
| McfR (Pput_0339) | P. putida F1 | succinic, malic and fumaric acids | unknown | 4HBB | periplasm | 29 |
| McpS (Pfl01_0728) | P. fluorescens Pf0-1 | L-malic and succinic acids | unknown | HBM | periplasm | 76 |
| McpT (Pfl01_3768) | P. fluorescens Pf0-1 | L-malic and succinic acids | unknown | sCACHE | periplasm | 76 |
| CcmL (Tlp3) | Campylobacter jejuni 11168-O | chemoattractants: malic and fumaric acids, Ile, purine; chemorepellents: Lys, Arg, glucosamine, succinic acid, thiamine | direct | dCACHE | periplasm | 85 |
| MCP2201 (CtCNB1_2201) | Comamonas testosteroni CNB-1 | malic acid (inhibitor of taxis to other organic acids), oxaloacetic, citric, isocitric, α-ketoglutaric, succinic and fumaric acids | direct | 4HB | periplasm | 56 |
The inspection of information available on the different malate responsive chemoreceptors (Table 1) also shows that these receptors differ in their LBD type. In general, chemoreceptor LBDs can be classified according to their size into clusters I and II78. Interestingly, malate responsive receptors include cluster I (sCACHE, 4HB) as well as cluster II (dCACHE, HBM) LBDs, and direct malate binding has been observed for all 4 LBD types (Table 1). This diversity in the molecular architecture of malate responsive receptors underlines the important physiological relevance of this ligand. Thus, as an example, the importance of malate chemotaxis has been reflected in R. pseudosolanacearum since a mutant defective in mcpM exhibits reduced virulence as compared to the wt strain in tomato plants75. Additionally, taxis to organic acids has been shown to be important for the colonization of the gastrointestinal tract of chicken by Campylobacter jejuni79.
Based on the hypothesis that ligand induced chemoreceptor dimerization is a prerequisite for signalling41, the effect of ligands on the oligomeric state of different LBD types has been investigated in the past15,19,25,28,42,43. The individual 4HB domains of receptors Tar, CtpH and PcaY_PP19,42,43 and HBM LBDs (receptors McpS and McpQ)25,28 were found to be largely monomeric in their ligand free state whereas in all cases the binding of the ligand induced complete LBD dimerization. This is due to the fact that ligands bind at the dimer interface and that amino acids from both monomers of the dimer establish contacts with the bound ligand26,80. In marked contrast, dCACHE LBDs of the PctA and PctB chemoreceptors were entirely monomeric in the absence and presence of ligands. Importantly, no information was available on the oligomeric state of sCACHE domains and we show here that yet another scenario applies to PA2652-LBD. Thus, in the absence of ligand, PA2652-LBD was entirely dimeric over the concentration range tested and L-malic acid did not have any effect on its oligomeric state (Fig. 4).
P. aeruginosa has two chemosensory pathways involved in chemotaxis. Chemotaxis to L-malic acid in the cheA2 mutant strain was indistinguishable from that of the wt, whereas no response was observed in a mutant defective in cheA1. These results confirm the bioinformatic predictions44 but also demonstrate the exclusivity of the che pathway in mediating L-malic acid responses.
Methods
Bacterial Strains, Culture Media, and Growth Conditions
Bacterial strains used in this study are listed in Supplementary Table S3. Pseudomonas aeruginosa PAO1 and its derivative strains were routinely grown at 37 °C in Luria Broth (LB; 5 g yeast extract l−1, 10 g Bacto tryptone l−1 and 5 g NaCl l−1) or M9 medium supplemented with 1 mM MgSO4, 6 mg l−1 Fe-citrate, trace elements81 and 15 mM glucose as carbon source. For growth experiments to assess the capacity of PA2652-LBD ligands to support growth a sole C-source, PAO1 cells were pre-cultured overnight in M9 medium supplemented with 10 mM glucose and washed twice with M9 medium salts, prior to the inoculation of M9 medium containing10 mM of the different carbon sources. When necessary, the pH of the medium was adjusted to 7.0 prior to inoculation. Bacterial growth over the time was monitored using Bioscreen Microbiological Growth Analyser (Oy Growth Curves Ab Ltd, Helsinki, Finland). Escherichia coli strains were grown at 37 °C in LB. Escherichia coli DH5α was used as a host for gene cloning. When appropriate, antibiotics were used at the following final concentrations (in µg ml−1): kanamycin, 25 (E. coli strains) and 100 (Pseudomonas strains); tetracycline, 40.
Construction of expression plasmid for PA2652-LBD
The DNA fragment encoding the LBD of PA2652 (Lys34–Thr205) was amplified from genomic DNA of P. aeruginosa PAO1 using primers 5′-TAATCATATGAAAAAACAGGCTGATGCCGA-3′ and 5′-TAATGTCGACTCAGGTGCCGATACGCTCGTC-3′ containing restriction sites for NdeI and SalI, respectively (underlined). The resulting PCR product was digested with NdeI and SalI and cloned into pET28b(+) using the same enzymes. The resulting plasmid, termed pET28b-PA2652LBD, was verified by DNA sequencing of the insert and flanking regions.
Overexpression and purification of PA2652-LBD
Escherichia coli BL21 (DE3) containing pET28b-PA2652LBD was grown in 2 L Erlenmeyer flasks containing 400 ml LB medium supplemented with 50 μg ml−1 kanamycin at 30 °C. Once the culture reached an OD600 of 0.6, protein overexpression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a concentration of 0.1 mM. Growth was continued at 16 °C overnight prior to cell harvest by centrifugation at 10 000 × g for 30 min at 4 °C. Cell pellets were resuspended in buffer A (20 mM Tris/HCl, 0.1 mM EDTA, 300 mM NaCl, 10 mM imidazole, 5% (v/v) glycerol, pH 8.0) and broken by French press treatment at 1000 psi. After centrifugation at 20 000 × g for 1 h, the supernatant was loaded onto a 5 ml HisTrap column (Amersham Bioscience), previously equilibrated with five column volumes of buffer A, washed with buffer A containing 35 mM of imidazole and eluted with a 35–300 mM imidazole gradient in buffer A. Protein-containing fractions were pooled and dialyzed for immediate analysis.
Thermal Shift Assay based high-throughput ligand screening
Thermal shift assays were performed on a MyIQ2 Real-Time PCR instrument (BioRad). Ligands from the different compound arrays (Biolog, Hayward, CA, USA; see Supplementary Fig. S3) were dissolved in 50 μl of MilliQ water, which, according to the manufacturer, corresponds to a concentration of 10–20 mM. Screening was performed using 96 wells plates. Each well contained 20 μM of protein dialyzed into TNG buffer (20 mM Tris/HCl, 150 mM NaCl, 10% (v/v) glycerol, pH 6.7), 2.5 μl of the resuspended compounds and SYPRO Orange (Life Technologies) at 5× concentration. In a single well (ligand free protein) the compound was substituted by water. Samples were heated from 23 °C to 85 °C at a scan rate of 1 °C/min. The protein unfolding curves were obtained by following the changes in SYPRO Orange fluorescence. Melting temperatures were determined using the first derivative values from the raw fluorescence data.
Isothermal titration calorimetry binding studies
Experiments were conducted on a VP-microcalorimeter (Microcal, Amherst, MA, USA) at 20 °C. PA2652-LBD was dialyzed overnight against TNG buffer, adjusted to a concentration of 20–35 μM and placed into the sample cell of the instrument. The protein was titrated by the injection of 9.6–14.4 μl aliquots of 1–20 mM ligand solutions that were prepared in TNG buffer (20 mM Tris/HCl, 150 mM NaCl, 10% (v/v) glycerol, pH 6.8) immediately before use. The mean enthalpies measured from the injection of ligands into buffer were subtracted from raw titration data prior to data analysis with the MicroCal version of ORIGIN. Data were fitted with the “One binding site model”.
Analytical ultracentrifugation studies
Experiments were performed on a Beckman Coulter Optima XL-A analytical ultracentrifuge (Beckman-Coulter, Palo Alto, CA, USA) equipped with UV-visible absorbance detection system, using an An50Ti 8-hole rotor and 12 mm path-length charcoal-filled epon double-sector centrepieces. The experiments were carried out at a rotor speed of 48 000 rpm and 7 °C using 400 µL samples of proteins dialyzed into in PIPES buffer (20 mM PIPES, pH 7.0). Protein was at 5–20 μM and L-malic acid (stock solution made up in dialysis buffer) was added at a final concentration of 1 mM. Dialysis buffer with and without ligand were used as reference. Light at a wavelength of 234 nm was recorded in the absorbance optics mode. A least squares boundary modelling of the data was used to calculate sedimentation coefficient distributions with the size-distribution c(s) method implemented in the SEDFIT v11.71 software82. The Svedberg equation allowed us to estimate the experimental molecular weight from the sedimentation and diffusion coefficients obtained. Buffer density (ρ = 1.0015 g/mL) and viscosity (η = 0.01449 Poise) at 7 °C were calculated from the buffer composition using SEDNTERP software83. This software was also used to calculate the partial specific volume (0.721 ml/g) and the molecular weight (21.5 kDa) of PA2652-LBD from its sequence.
Plasmid construction for genetic complementation assays
For the construction of the complementing plasmid, a full copy of the PA2652 gene was amplified by PCR using the primers PA2652-NdeI-F (5′-TAATCATATGATGCGTCTGACCCTGAAATCC-3′) and PA2652-BamHI-R (5′- TAATGGATCCGACAGGAAGGCTCTGTGGCG-3′). Restriction sites for NdeI and BamHI are underlined. The resulting fragment was digested with NdeI and BamHI and cloned into the same sites in pBBR1MCS2_START to generate pBBR2652f4. The insert was confirmed by PCR and sequencing, and pBBR2652f4 was used to transform the PA2652 defective mutant by electroporation.
Quantitative Capillary Chemotaxis Assays
Overnight cultures of P. aeruginosa strains were diluted to an OD660 of 0.05 in MS medium (30 mM Na2HPO4, 20 mM KH2PO4, 25 mM NH4NO3, 1 mM MgSO4) supplemented with 6 mg l−1 Fe-citrate, trace elements81 and 15 mM glucose as carbon source, and grown at 37 °C with orbital shaking (200 rpm). At an OD660 of 0.4 (early stationary phase of growth) cultures were centrifuged at 1,700 × g for 5 min and the resulting pellet was washed twice with chemotaxis buffer (50 mM potassium phosphate, 20 mM EDTA, 0.05% (v/v) glycerol, pH 7.0). Subsequently, the cells were resuspended in the same buffer, adjusted to an OD660 of 0.1 and 230 µl aliquots of the bacterial cultures were placed into 96-well plates. For the quantitative assays, one-microliter capillary tubes (Microcaps, Drummond Scientific, Ref. P1424) were heat-sealed at one end and filled with either the chemotaxis buffer (negative control) or chemotaxis buffer containing the chemoeffectors to test. The capillaries were immersed into the bacterial suspensions at its open end. After 30 min at room temperature, capillaries were removed from the bacterial suspensions, rinsed with sterile water and the content expelled into 1 ml of M9 medium salts. Serial dilutions were plated onto M9 minimal medium (containing the appropriate antibiotics) supplemented with 15 mM glucose as carbon source. The number of colony forming units was determined after overnight incubation. In all cases, data were corrected with the number of cells that swam into buffer containing capillaries.
To determine the effect of antagonists in the chemotactic properties of PAO1, the assay was performed as previously described with two minor modifications: (i) Capillary tubes were filled with chemotaxis buffer containing 1 mM of the chemoattractants (L-malic acid or L-alanine) and 10–40 mM of the antagonists (methylsuccinic or citraconic acids); (ii) Bacterial cultures were washed with chemotaxis buffer and cells were finally resuspended in the same buffer but containing equimolar concentrations of methylsuccinic/citraconic acids to those present in the capillary tubes.
Competitive Root Colonization Assays
Sterilization and germination of maize seeds was carried out as described previously84. Subsequently, 10 mL of M9 salts containing a 106 CFU/ml 1:1 mixture of PAO1-Km (wild type) and PAO-PA2652 (mutant) were added to 50 ml Sterilin tubes containing 40 g of sterile washed silica sand. Thereafter, one sterile seed was planted per Sterilin tube containing the inoculated silica sand. Plants were maintained at 24 °C with a daily light period of 16 h. After 6 days, bacterial cells were recovered from the rhizosphere and serial dilutions were plated on LB-agar medium supplemented with kanamycin or tetracycline to select PAO1-Km or the PA2652 mutant strain, respectively.
Electronic supplementary material
Acknowledgements
We acknowledge financial support from FEDER funds and Fondo Social Europeo through grants from the Junta de Andalucía (grant CVI-7335) and the Spanish Ministry for Economy and Competitiveness (grants BIO2013-42297 and BIO2016-76779-P). M.A.M. was supported by the Spanish Ministry of Economy and Competitiveness Postdoctoral Research Program, Juan de la Cierva (JCI-2012-11815). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author Contributions
D.M.-M., A.O., F.J.P.-M. and M.A.M. conducted experiments and analysed data. T.K. and and M.A.M. designed experiments and wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20283-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 4.Matilla MA, Krell T. Chemoreceptor-based signal sensing. Curr. Opin. Biotechnol. 2017;45:8–14. doi: 10.1016/j.copbio.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 5.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 7.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato J, Kim HE, Takiguchi N, Kuroda A, Ohtake H. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J. Biosci. Bioeng. 2008;106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 9.Guvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol. Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 10.Ferrandez A, Hawkins AC, Summerfield DT, Harwood CS. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J. Bacteriol. 2002;184:4374–4383. doi: 10.1128/JB.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol. Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 12.Whitchurch CB, et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 2004;52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 13.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol. Microbiol. 2010;76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology. 1997;143:3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 15.Rico-Jimenez M, et al. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA) Mol. Microbiol. 2013;88:1230–1243. doi: 10.1111/mmi.12255. [DOI] [PubMed] [Google Scholar]
- 16.Reyes-Darias JA, Yang Y, Sourjik V, Krell T. Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa. Mol. Microbiol. 2015;96:513–525. doi: 10.1111/mmi.12953. [DOI] [PubMed] [Google Scholar]
- 17.Reyes-Darias JA, et al. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol. Microbiol. 2015;97:488–501. doi: 10.1111/mmi.13045. [DOI] [PubMed] [Google Scholar]
- 18.Wu H, et al. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 2000;182:3400–3404. doi: 10.1128/JB.182.12.3400-3404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rico-Jimenez M, et al. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci. Rep. 2016;6:28967. doi: 10.1038/srep28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahme LG, et al. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA. 2000;97:8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espinosa-Urgel M, Kolter R, Ramos JL. Root colonization by Pseudomonas putida: love at first sight. Microbiology. 2002;148:341–343. doi: 10.1099/00221287-148-2-341. [DOI] [PubMed] [Google Scholar]
- 22.Regenhardt D, et al. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 2002;4:912–915. doi: 10.1046/j.1462-2920.2002.00368.x. [DOI] [PubMed] [Google Scholar]
- 23.Bagdasarian M, et al. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Fontana C, et al. High specificity in CheR methyltransferase function: CheR2 of Pseudomonas putida is essential for chemotaxis, whereas CheR1 is involved in biofilm formation. J. Biol. Chem. 2013;288:18987–18999. doi: 10.1074/jbc.M113.472605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacal J, et al. Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J. Biol. Chem. 2010;285:23126–23136. doi: 10.1074/jbc.M110.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pineda-Molina E, et al. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc. Natl. Acad. Sci. USA. 2012;109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacal J, Garcia-Fontana C, Callejo-Garcia C, Ramos JL, Krell T. Physiologically relevant divalent cations modulate citrate recognition by the McpS chemoreceptor. J. Mol. Recognit. 2011;24:378–385. doi: 10.1002/jmr.1101. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Mora D, et al. McpQ is a specific citrate chemoreceptor that responds preferentially to citrate/metal ion complexes. Environ. Microbiol. 2016;18:3284–3295. doi: 10.1111/1462-2920.13030. [DOI] [PubMed] [Google Scholar]
- 29.Parales RE, et al. Pseudomonas putida F1 has multiple chemoreceptors with overlapping specificity for organic acids. Microbiology. 2013;159:1086–1096. doi: 10.1099/mic.0.065698-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega A, Krell T. The HBM domain: introducing bimodularity to bacterial sensing. Protein Sci. 2014;23:332–336. doi: 10.1002/pro.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulrich LE, Zhulin IB. Four-helix bundle: a ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics. 2005;21:iii45–48. doi: 10.1093/bioinformatics/bti1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Mora D, et al. Identification of a Chemoreceptor in Pseudomonas aeruginosa That Specifically Mediates Chemotaxis Toward alpha-Ketoglutarate. Front. Microbiol. 2016;7:1937. doi: 10.3389/fmicb.2016.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarez-Ortega C, Harwood CS. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl. Environ. Microbiol. 2007;73:7793–7795. doi: 10.1128/AEM.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. Cache Domains That are Homologous to, but Different from PAS Domains Comprise the Largest Superfamily of Extracellular Sensors in Prokaryotes. PLoS Comput. Biol. 2016;12:e1004862. doi: 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J. Mol. Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia V, et al. Identification of a Chemoreceptor for C2 and C3 Carboxylic Acids. Appl. Environ. Microbiol. 2015;81:5449–5457. doi: 10.1128/AEM.01529-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKellar JL, Minnell JJ, Gerth ML. A high-throughput screen for ligand binding reveals the specificities of three amino acid chemoreceptors from Pseudomonas syringae pv. actinidiae. Mol. Microbiol. 2015;96:694–707. doi: 10.1111/mmi.12964. [DOI] [PubMed] [Google Scholar]
- 38.Krell T. Tackling the bottleneck in bacterial signal transduction research: high-throughput identification of signal molecules. Mol. Microbiol. 2015;96:685–688. doi: 10.1111/mmi.12975. [DOI] [PubMed] [Google Scholar]
- 39.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 40.Krell T. Microcalorimetry: a response to challenges in modern biotechnology. Microb. Biotechnol. 2008;1:126–136. doi: 10.1111/j.1751-7915.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stock J. Receptor signaling: dimerization and beyond. Curr. Biol. 1996;6:825–827. doi: 10.1016/S0960-9822(02)00605-X. [DOI] [PubMed] [Google Scholar]
- 42.Milligan DL, Koshland DE., Jr. Purification and characterization of the periplasmic domain of the aspartate chemoreceptor. J. Biol. Chem. 1993;268:19991–19997. [PubMed] [Google Scholar]
- 43.Fernandez M, Matilla MA, Ortega A, Krell T. Metabolic Value Chemoattractants Are Preferentially Recognized at Broad Ligand Range Chemoreceptor of Pseudomonas putida KT2440. Front. Microbiol. 2017;8:990. doi: 10.3389/fmicb.2017.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ortega DR, et al. Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2017;114:12809–12814. doi: 10.1073/pnas.1708842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bais HP, Walker TS, Schweizer HP, Vivanco JM. Root-specific elicitation and antimicrobial activity of rosmarinic acid in hairy root cultures of Ocimum basilicum. Plant. Physiol. Biochem. 2002;40:983–995. doi: 10.1016/S0981-9428(02)01460-2. [DOI] [Google Scholar]
- 46.Attila C, et al. Pseudomonas aeruginosa PAO1 virulence factors and poplar tree response in the rhizosphere. Microb. Biotechnol. 2008;1:17–29. doi: 10.1111/j.1751-7915.2007.00002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bardy, S. L., Briegel, A., Rainville, S. & Krell, T. Recent advances and future prospects in bacterial and archaeal locomotion and signal transduction. J. Bacteriol., 10.1128/JB.00203-17 (2017). [DOI] [PMC free article] [PubMed]
- 48.Jones DL. Orgaic acids in the rhizosphere - a critical review. Plant Soil. 1998;205:25–44. doi: 10.1023/A:1004356007312. [DOI] [Google Scholar]
- 49.Johnson JF, Allan DL, Vance CP, Weiblen G. Root Carbon Dioxide Fixation by Phosphorus-Deficient Lupinus albus (Contribution to Organic Acid Exudation by Proteoid Roots) Plant Physiol. 1996;112:19–30. doi: 10.1104/pp.112.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38:D401–407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Busch A, Lacal J, Martos A, Ramos JL, Krell T. Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proc. Natl. Acad. Sci. USA. 2007;104:13774–13779. doi: 10.1073/pnas.0701547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klein T, et al. Identification of small-molecule antagonists of the Pseudomonas aeruginosa transcriptional regulator PqsR: biophysically guided hit discovery and optimization. ACS Chem. Biol. 2012;7:1496–1501. doi: 10.1021/cb300208g. [DOI] [PubMed] [Google Scholar]
- 53.Silva-Jimenez H, et al. Study of the TmoS/TmoT two-component system: towards the functional characterization of the family of TodS/TodT like systems. Microb. Biotechnol. 2012;5:489–500. doi: 10.1111/j.1751-7915.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swem LR, et al. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell. 2009;35:143–153. doi: 10.1016/j.molcel.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bi S, et al. Discovery of novel chemoeffectors and rational design of Escherichia coli chemoreceptor specificity. Proc. Natl. Acad. Sci. USA. 2013;110:16814–16819. doi: 10.1073/pnas.1306811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ni B, Huang Z, Fan Z, Jiang CY, Liu SJ. Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds. Mol. Microbiol. 2013;90:813–823. doi: 10.1111/mmi.12400. [DOI] [PubMed] [Google Scholar]
- 57.Yu D, Ma X, Tu Y, Lai L. Both piston-like and rotational motions are present in bacterial chemoreceptor signaling. Sci. Rep. 2015;5:8640. doi: 10.1038/srep08640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuchs G, Berg IA. Unfamiliar metabolic links in the central carbon metabolism. J. Biotechnol. 2014;192:314–322. doi: 10.1016/j.jbiotec.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 59.Khorassani R, et al. Citramalic acid and salicylic acid in sugar beet root exudates solubilize soil phosphorus. BMC Plant Biol. 2011;11:121. doi: 10.1186/1471-2229-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, Yang D, Li M, Shi L. Metabolic Profiles Reveal Changes in Wild and Cultivated Soybean Seedling Leaves under Salt Stress. PLoS One. 2016;11:e0159622. doi: 10.1371/journal.pone.0159622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arun AB, et al. Pseudoxanthobacter soli gen. nov., sp. nov., a nitrogen-fixing alphaproteobacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2008;58:1571–1575. doi: 10.1099/ijs.0.65206-0. [DOI] [PubMed] [Google Scholar]
- 62.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 63.Matilla, M. A. & Krell, T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev. 42, 10.1093/femsre/fux052 (2018). [DOI] [PubMed]
- 64.Erhardt M. Strategies to Block Bacterial Pathogenesis by Interference with Motility and Chemotaxis. Curr. Top Microbiol. Immunol. 2016;398:185–205. doi: 10.1007/82_2016_493. [DOI] [PubMed] [Google Scholar]
- 65.Kenakin T. New concepts in drug discovery: collateral efficacy and permissive antagonism. Nat. Rev. Drug Discov. 2005;4:919–927. doi: 10.1038/nrd1875. [DOI] [PubMed] [Google Scholar]
- 66.Kenakin T, Williams M. Defining and characterizing drug/compound function. Biochem. Pharmacol. 2014;87:40–63. doi: 10.1016/j.bcp.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 67.Brewster JL, et al. Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae. Sci. Rep. 2016;6:35198. doi: 10.1038/srep35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Werf MJ, van den Tweel WJ, Hartmans S. Screening for microorganisms producing D-malate from maleate. Appl. Environ. Microbiol. 1992;58:2854–2860. doi: 10.1128/aem.58.9.2854-2860.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Unden, G., Strecker, A., Kleefeld, A. & Kim, O. B. C4-Dicarboxylate Utilization in Aerobic and AnaerobicGrowth. EcoSal Plus7, 10.1128/ecosalplus.ESP-0021-2015 (2016). [DOI] [PMC free article] [PubMed]
- 70.Hopper DJ, Chapman PJ, Dagley S. Metabolism of l-Malate and d-Malate by a Species of Pseudomonas. J. Bacteriol. 1970;104:1197–1202. doi: 10.1128/jb.104.3.1197-1202.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scheu PD, Kim OB, Griesinger C, Unden G. Sensing by the membrane-bound sensor kinase DcuS: exogenous versus endogenous sensing of C(4)-dicarboxylates in bacteria. Future Microbiol. 2010;5:1383–1402. doi: 10.2217/fmb.10.103. [DOI] [PubMed] [Google Scholar]
- 72.Cheung J, Hendrickson WA. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 2008;283:30256–30265. doi: 10.1074/jbc.M805253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lukas H, Reimann J, Kim OB, Grimpo J, Unden G. Regulation of aerobic and anaerobic D-malate metabolism of Escherichia coli by the LysR-type regulator DmlR (YeaT) J. Bacteriol. 2010;192:2503–2511. doi: 10.1128/JB.01665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salah Ud-Din AI, Roujeinikova A. Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell Mol. Life Sci. 2017;74:3293–3303. doi: 10.1007/s00018-017-2514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hida A, et al. Identification of the mcpA and mcpM genes, encoding methyl-accepting proteins involved in amino acid and l-malate chemotaxis, and involvement of McpM-mediated chemotaxis in plant infection by Ralstonia pseudosolanacearum (formerly Ralstonia solanacearum phylotypes I and III) Appl. Environ. Microbiol. 2015;81:7420–7430. doi: 10.1128/AEM.01870-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oku S, Komatsu A, Nakashimada Y, Tajima T, Kato J. Identification of Pseudomonas fluorescens Chemotaxis Sensory Proteins for Malate, Succinate, and Fumarate, and Their Involvement in Root Colonization. Microbes Environ. 2014;29:413–419. doi: 10.1264/jsme2.ME14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tunchai M, et al. Identification and characterization of chemosensors for d-malate, unnatural enantiomer of malate, in Ralstonia pseudosolanacearum. Microbiology. 2017;163:233–242. doi: 10.1099/mic.0.000408. [DOI] [PubMed] [Google Scholar]
- 78.Lacal J, Garcia-Fontana C, Munoz-Martinez F, Ramos JL, Krell T. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010;12:2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 79.Chandrashekhar K, et al. Transducer like proteins of Campylobacter jejuni 81-176: role in chemotaxis and colonization of the chicken gastrointestinal tract. Front. Cell Infect. Microbiol. 2015;5:46. doi: 10.3389/fcimb.2015.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Milburn MV, et al. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 81.Abril MA, Michan C, Timmis KN, Ramos JL. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laue, T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds Harding, S. E., Rowe, A. J., & Horton, J. C) 90–125 (Royal Society of Chemistry, 1992).
- 84.Matilla MA, Espinosa-Urgel M, Rodriguez-Herva JJ, Ramos JL, Ramos-Gonzalez MI. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 2007;8:R179. doi: 10.1186/gb-2007-8-9-r179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rahman H, et al. Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog. 2014;10:e1003822. doi: 10.1371/journal.ppat.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Finn RD, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.