Abstract
Objectives:
Inflammatory bowel diseases (IBD), comprised of Crohn’s disease (CD) and ulcerative colitis (UC), are characterized by a complex pathophysiology that is thought to result from an aberrant immune response to a dysbiotic luminal microbiota in genetically susceptible individuals. New technologies support the joint assessment of host-microbiome interaction.
Methods:
Using whole genome sequencing and shotgun metagenomics, we studied the clinical features, host genome, and stool microbial metagenome of 85 IBD patients, and compared the results to 146 control individuals. Genetic risk scores, computed on 159 single nucleotide variants, and human leukocyte antigen (HLA) types differentiated IBD patients from healthy controls.
Results:
Genetic risk was associated with the need for use of biologics in IBD and, modestly, with the composition of the gut microbiome. As compared with healthy controls, IBD patients had hallmarks of stool microbiome dysbiosis, with loss of a diversified core microbiome, enrichment and depletion of specific bacteria, and enrichment of bacterial virulence factors.
Conclusions:
We show that genetic risk may have a role in early risk stratification in the care of IBD patients and propose that expression of virulence factors in a dysbiotic microbiome may contribute to pathogenesis in IBD.
Introduction
The interaction between host genes and stool microbiome composition may contribute to the pathogenesis as well as clinical presentation of inflammatory bowel diseases (IBD); however, this relationship is incompletely understood. Genome-wide association studies (GWAS) have identified dozens of loci and genes associated with IBD.1, 2 Although some genetic loci are unique to Ulcerative Colitis (UC) or to Crohn’s Disease (CD), the majority are associated with both types of IBD.1 While the absolute effect of each genetic variant on the risk of IBD is quite small, GWAS inform the understanding of the pathogenesis of IBD and underscore the importance of the interaction between the host and the microbial community.1, 2, 3
Several studies have shown that there is a significant reduction in the diversity of the stool microbiome of individuals with IBD;4, 5, 6 furthermore, this reduction in diversity has been shown to occur early in the course of Crohn’s disease in a pediatric population, suggesting that the dysbiosis may not only be an effect of IBD but also contribute to ongoing pathogenesis.4, 5, 7, 8, 9 The microbiota of patients with IBD is characterized by depletions in bacteria with anti-inflammatory effects, including Bifidobacterium adolescentis, Faecalibacterium prausnitzii and other butyrate producing bacteria, and an expansion in pathogenic bacteria (pathobionts), including Proteobacteria such as adherent-invasive Escherichia coli.8, 10, 11 A recent study demonstrated in a large cohort of IBD patients that disease location was also a significant determinant of the microbiome.12
Technological advances in sequencing and data analysis have transformed the ability to sequence host genomes and microbiomes as well as metagenomes. Shotgun metagenomics enhances resolution of detecting and characterizing bacterial strains as compared to 16S ribosomal DNA sequencing13, 14, 15 and allows for the assessment of non-bacterial components of the microbiome, including fungi, viruses, and archaea.
We compared the host genome and microbiome in 83 well-characterized IBD patients to those from 146 representative population controls using whole genome sequencing of both the host and the stool microbiome. This study reveals novel associations between host genetic risk, stool microbiome, and clinical features of inflammatory bowel disease, demonstrating the power and value of technological advances in sequencing.
Methods
Study population
IBD cohort
Patients with a diagnosis of CD or UC who were seen at the University of California, San Diego at the Inflammatory Bowel Disease Center were recruited and consented into the IBD Biobank. Each patient’s clinical phenotype was assessed by an IBD specialist to define disease subtype (UC or CD), location, and phenotype (Table 1). Clinical data were collected prospectively, and phenotypes were confirmed by an IBD specialist physician. Missing values were imputed using missForest.16 The study participants provided written informed consent, and the study was approved by the Institutional Review Board of University of California, San Diego. All metadata are presented in Supplementary Table S1. We used principal component (PC) analysis to analyze the clinical metadata that include disease phenotype, current and prior treatments, disease activity and location, and complications as well as relevant covariates.
Table 1. Baseline IBD Patient Characteristics.
| Number of IBD patients (%) (Total N=86) | |
|---|---|
| Age in years | |
| Median (IQR) | 37 (26.8-52.5) |
| Sex | |
| Male | 41 (48%) |
| Female | 45 (52%) |
| Age at Diagnosis in years | |
| Median (IQR) | 25 (16.8-40.3) |
| Family history of IBD | 33 (38%) |
| Smoking | |
| Never | 60 (69.8%) |
| Prior smoker | 20 (23.2%) |
| Current smoker | 6 (7.0%) |
| Diagnosis | |
| Ulcerative colitis | 41 (48%) |
| Crohn’s disease | 45 (52%) |
| UC anatomic involvement at diagnosis (% UC) | |
| Proctitis | 11 (27%) |
| Left sided colitis | 12 (29%) |
| Extensive colitis | 18 (44%) |
| UC current anatomic involvement (% UC) | |
| Proctitis | 5 (12%) |
| Left sided colitis | 14 (34%) |
| Extensive colitis | 22 (54%) |
| CD location at diagnosis (% CD) | |
| Ileal | 10 (22%) |
| Colonic | 27 (60%) |
| Ileocolonic | 6 (13%) |
| Isolated upper gastrointestinal | 1 (2%) |
| CD location (% CD) | |
| Ileal | 14 (31%) |
| Colonic | 15 (33%) |
| Ileocolonic | 15 (33%) |
| Isolated upper gastrointestinal | 1 (2%) |
| CD Behavior (% CD) | |
| Inflammatory | 27 (60%) |
| Stricturing | 12 (27%) |
| Penetrating | 6 (13%) |
| Perianal Disease (% CD) | 10 (22%) |
| Prior Gastrointestinal Surgeries (% total) | |
| Colectomy | 11 (13%) |
| Ileocolonic resection | 7 (8%) |
| Small bowel resection | 5 (6%) |
| Partial colonic resection | 3 (3%) |
| Current medication use | |
| steroid | 21 (24%) |
| immune modulator | 26 (30%) |
| 5-aminosalicylate acid | 20 (23%) |
| Current biologic use | |
| Tumor necrosis factor antagonist | 38 (44%) |
| Integrin antagonist | 3 (3%) |
| p40 antagonist | 2 (2%) |
Controls
We enrolled active healthy adults >18 years old (without acute illness, activity-limiting unexplained illness or symptoms, or known active cancer) able to come to the Health Nucleus in La Jolla, CA, for on-site data collection including whole genome, microbiome, and other testing. The study participants provided written consent and the research protocol was approved by the Western Institutional Review Board.
Human genome and microbiome sequencing
Blood sample (approximately 10 mL) was collected from each subject for DNA extraction. The whole genome sequencing was carried out at Human Longevity, Inc., San Diego. Next Generation Sequencing library preparation was carried out using the TruSeq Nano DNA HT kit (Illumina Inc.) as described previously (Supplementary Materials) for sequencing on Illumina HiSeq X. More details can be found in Telenti et al.17
For microbiome analysis, participants collected stool samples at home, aliquoted, and frozen at −80C until DNA isolation. Nextera XT libraries were prepared manually and sequencing was performed on an Illumina HiSeq 2500 as described previously (Supplementary Materials). Microbiome sequence data were processed as previously described.18 Non-human reads were mapped to Human Longevity’s reference genome database, a collection of ~11 900 genomes of bacteria, archaea, viruses, and eukaryotes downloaded from NCBI GenBank including both complete and draft genomes. After read mapping, an in-house implementation of an expectation maximization algorithm19 was used to process the reads that were ambiguously mapped to multiple genomes to estimate the relative genome abundance (RGA). The genome coverage, which is the total length of mapped reads divided by the reference genome length, was calculated for each reference genome based on the expectation maximization’s assignment of reads to genomes. Open reading frames (ORFs) were predicted from genome scaffolds using MetaGene20 and were compared against the virulence factors database VFDB21 to identify virulence factor genes with over 90% sequence identity.
Genetic risk of IBD
The IBD genetic risk was estimated from the whole-genome sequence data of the IBD patients and Human Longevity population based on the single nucleotide polymorphisms (SNPs) reported to be significantly associated with IBD.2 SNPs that had p-value>5e-8 or were inconsistent between human reference genome versions hg19 and hg38 were excluded from calculation of the genetic risk. A total of 159 SNPs were incorporated in the estimation of the IBD risk (Supplementary Table S2). HLA class I and II four-digit typing was done from short read whole genome sequencing using the xHLA algorithm.22
Association between host genetics, clinical parameters and microbiome
Associations between the clinical metadata and the IBD genetic risk were tested using regression analysis under a generalized linear model. Smoking, gender, and age were included as covariates. Associations between the stool microbiome taxonomic abundance (represented by the principal components) and the IBD genetic risk types were tested using logistic regression under a generalized linear model with binomial distribution, including the sex, age, ancestry, and diagnosis (UC or CD) as covariates. Sensitivity analysis was performed that excluded patients with early IBD onset (age of diagnosis <18), use of antibiotics, and J pouch.
Additional statistical analyses
For dimensionality reduction of the clinical and microbiome profiles, Principal Component Analysis (PCA) was performed using the R function `prcomp`. For differential taxonomic abundance of the microbiome, pairwise non-parametric Mann–Whitney U test was performed using the R function `wilcox.test` then the resulted P-value was corrected for multiple testing using the R function `p.adjust`.
Results
Clinical characteristics of IBD patients
The study included 85 patients with a diagnosis of UC (n=45) or CD (n=40) (Table 1). The cohort included 44 females and 41 males with age range between 18 and 83 and a median age of 37 years (interquartile range [IQR] 26.8-52.5). There was a median disease duration of 8 years (IQR 3-14). The age at diagnosis ranged from 7 to 77 with a median of 25 years. Of the patients with ulcerative colitis, 12% have proctitis, 34% have left sided ulcerative colitis, and 54% have extensive colitis. Of the patients with Crohn’s disease, 31% have ileal disease, 33% have colonic disease, and 33% have ileocolonic disease, and 2% have isolated upper gastrointestinal disease. 76 of the 85 patients were of European (EUR) ancestry. Additionally, there were smaller groups of other ancestries, specifically: 3 Admixed American (AMR), 2 Central South Asian (CSA), 2 East Asian (EAS), 1 African (AFR), and 1 MDE (Middle Eastern).
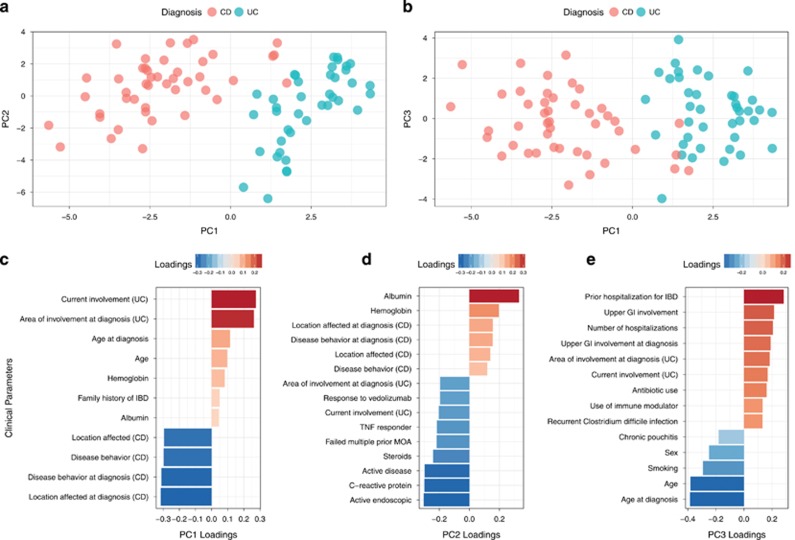
We used principal component (PC) analysis to analyze the clinical parameters that include disease phenotype, current and prior treatments, disease activity, and complications (Table 1 and Supplementary Table S1). PC1 describes the separation between CD and UC (Figure 1). The major clinical parameters contributing to PC1 were UC- and CD-specific characteristics. The UC-related characteristics included anatomic degree of colonic involvement at the time of diagnosis and current involvement based on the Montreal classification.23 The CD-associated features included anatomic location of CD based on Montreal classification system and disease behavior defined as inflammatory, stricturing, and penetrating at the time of diagnosis as well as current location and behavior. The main factors contributing to PC2 included factors indicating disease activity, including presence of active endoscopic disease, clinical symptoms of IBD, C-reactive protein, albumin, and steroid use. PC3 was influenced by certain clinical features that can be associated with severity of IBD, including prior IBD-related hospitalization, number of hospitalizations, age at diagnosis, upper gastrointestinal involvement, area of involvement on diagnosis in UC, recurrent Clostridium difficile infection, and chronic pouchitis. Antibiotic use, previously shown to alter the microbiome in patients with IBD,8 contributed to PC3. Sensitivity analysis excluding early onset IBD, use of antibiotics, and J pouch maintained clean separation of UC and CD (Supplementary Figures). The PCA served to inspect the effect of the microbiome and host genetic contributions on disease phenotype and clinical features.
Figure 1.
Analysis of clinical metadata of IBD patients. (a,b). Principal component analysis. PC1 explains 16% of the variance. PC2 explains 10% of the variance. PC3 explains 6% of the variance. The patients are color-coded according to clinical diagnosis: Crohn’s Disease (CD) and Ulcerative Colitis (UC). (c-e). Loadings of the clinical metadata on the principal components. Failed multiple MOA=failed multiple prior drugs with different mechanisms of action.
IBD Genetic Risk
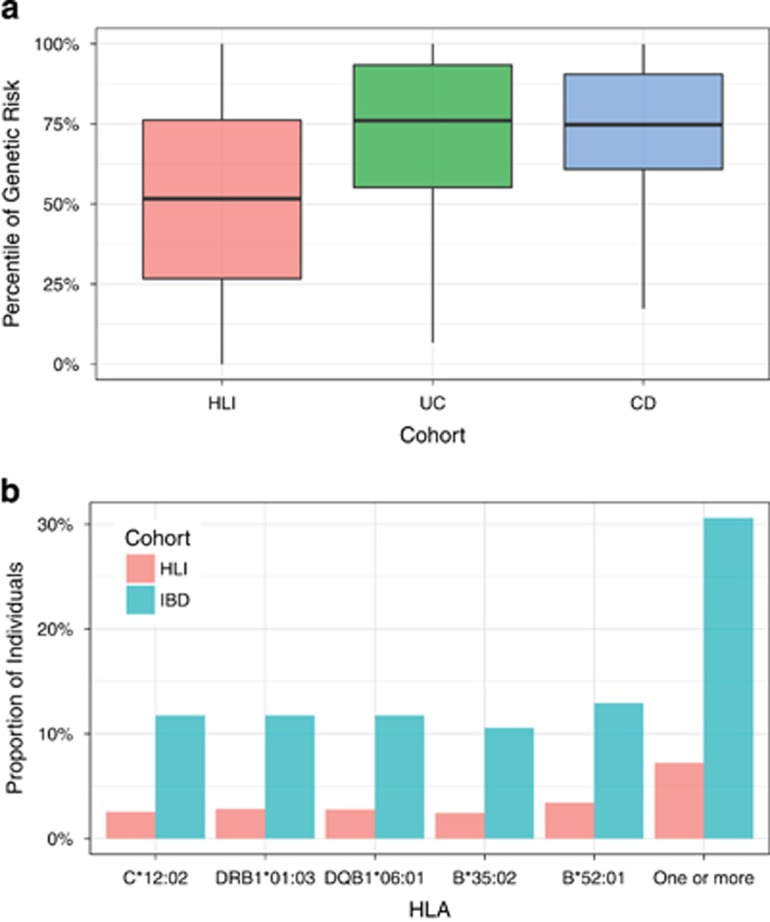
Analysis of host genetic risk was performed by computing on 159 SNPs known to be significantly associated with IBD genetic risk in a recent large GWAS analysis.2 The genetic risk was found significantly elevated in the IBD participants compared to that of a large population (n=10,545) of individuals without a diagnosis of IBD,17 p-value=5.6e-10 (Figure 2a). The various SNPs used in the calculation of the genetic risk score did not discriminate individuals with UC from those with CD (Supplementary Figure S1).
Figure 2.
Genetic predisposition to IBD in patients compared to the general population. (a). Genetic risk score. The y-axis is the percentile of the IBD genetic risk. The general reference population has 10,545 individuals, composed of 8,253 EUR, 1,394 AFR, 315 MDE, 238 EAS, 212 AMR, and 132 CSA. (b). The frequency of HLA alleles significantly associated with IBD. The top panel shows the frequencies (on the y-axis) of the IBD-associated HLA alleles (on the x-axis)
Human Leukocyte Antigen (HLA) types have been previously associated with genetic risk in IBD based on large genome-wide studies.1, 24, 25, 26 We therefore investigated the association between HLA class I and II and IBD in the study. Patients with IBD carried different 165 HLA class I and II alleles. We observed an enrichment of five HLA alleles compared to the reference population:17 HLA-C*12:02 (p-value=2.5e-4, FDR=0.02), DRB1*01:03 (p-value=3.4e-4, FDR=0.02), DQB1*06:01 (5.2e-4, FDR=0.02), B*35:02 (p-value=6.4e-4, FDR=0.02), and B*52:01 (p-value=7.2e-4, FDR=0.02). Overall, 26 out of 85 individuals (~31%) carried one or more of the risk HLA alleles in the IBD cohort as compared to 756 out of 10545 individuals (~7%) in the reference population (Figure 2b). Sensitivity analysis excluding early onset IBD, use of antibiotics, and J pouch confirmed the effect of genetic risk (Supplementary Figures). The HLAs we identified as being associated with IBD have been previously reported.24, 25, 26
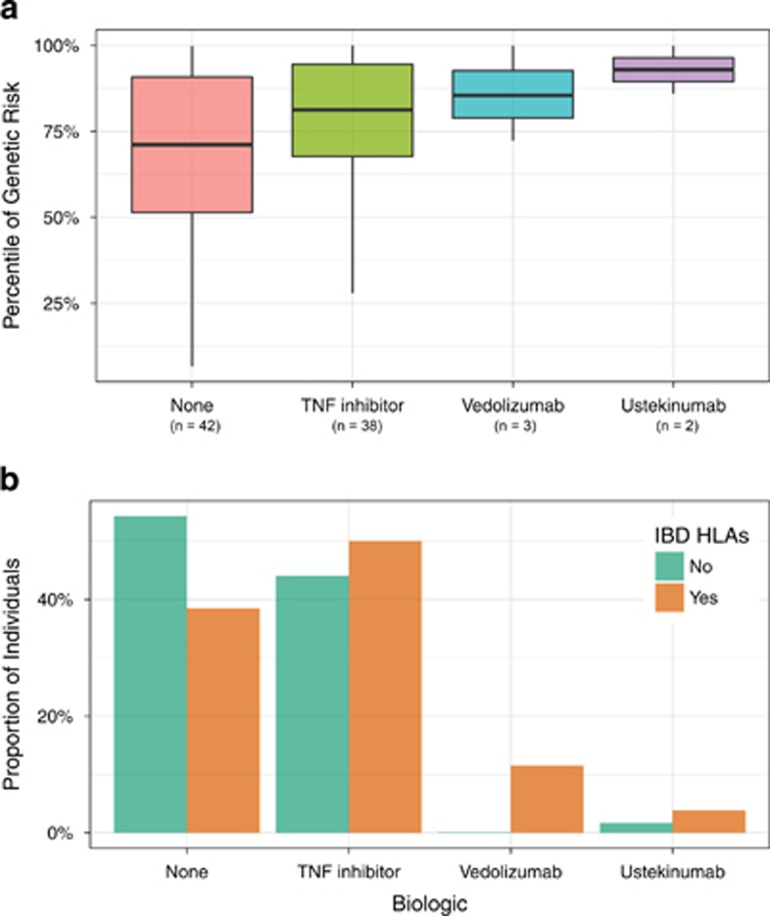
Genetic risk correlated with the use of biologics in IBD. Specifically, patients with IBD who were biologic-naïve had the lowest genetic risk scores. Patients treated with tumor necrosis factor (TNF) antagonists, considered the first-line biologic for moderate to severely active IBD, had higher genetic risk scores as compared to biologic-naïve patients. Individuals treated with other more recently approved biologics for IBD, including vedolizumab, an alpha-4 beta-7 integrin antagonist, and ustekinumab, an anti-IL-12/23 p40 antagonist, had the highest genetic risk scores. Though the number of patients was small (n=5), all of the patients clinically responded to these 2nd line agents supporting their clinical use (Figure 3a). Similarly, patients treated with the newest biologics carried a greater proportion of HLA alleles associated with IBD. We built a logistic model to assess the independent contributions of genetic risk computed from 159 variants and the IBD-associated HLA alleles (Table 2). The model retained HLA-DRB1*01:03, CD diagnosis and genetic risk as statistically significant. HLA-DRB1*01:03 also associated with some manifestations of disease activity (perianal disease, P-value=0.007; antibiotic use, P-value=0.003; and trend association for hospitalization, P-value=0.08) using the same regression models. In summary, genetic risk and HLA alleles may have a role in risk stratification and may also predict the need for second-line therapies in IBD.
Figure 3.
Correlation between IBD genetic risk and biologic use. (a). The x-axis represents the genetic risk of IBD (expressed as population percentile) and the y-axis represents the biologic used to treat IBD. (b). Frequency of HLA alleles significantly associated with IBD in adjusted univariate analyses. The top panel shows the frequencies (on the y-axis) of the IBD-associated HLA alleles (on the x-axis).
Table 2. Association between use of biologics and genetic risk and HLA types.
| Parameter | Estimate | Std. Error | z value | p-value |
|---|---|---|---|---|
| HLA DRB1*01:03 | 2.56 | 0.81 | 3.16 | 0.002 |
| Diagnosis | −1.32 | 0.49 | −2.70 | 0.007 |
| Genetic risk score | 0.02 | 0.01 | 2.51 | 0.012 |
| HLA DQB1*06:01 | −2.48 | 2.22 | −1.12 | 0.264 |
| Smoking | 0.43 | 0.40 | 1.08 | 0.281 |
| HLA C*12:02 | 2.42 | 2.61 | 0.93 | 0.353 |
| Age | −0.01 | 0.02 | −0.46 | 0.649 |
| Ancestry | −0.14 | 0.30 | −0.45 | 0.650 |
| Sex | 0.17 | 0.46 | 0.37 | 0.711 |
| HLA B*52:01 | 0.35 | 1.41 | 0.25 | 0.805 |
| HLA B*35:02 | −0.15 | 0.72 | −0.21 | 0.832 |
The logistic regression model includes clinical diagnosis (UC or CD) and demographic covariates in addition to the genetic risk score and top HLA alleles. The parameters are sorted by the statistical significance.
Microbiome
Microbiome dysbiosis in IBD
83 microbiome samples (out of 85) successfully passed quality control assessments. The total number of reads per sample ranged from 10 to 68 million reads with a median of 25 million reads for the IBD patients (83 samples), compared to a range of 10 to 60 million reads and median of 18 million reads for the healthy controls (146 samples).
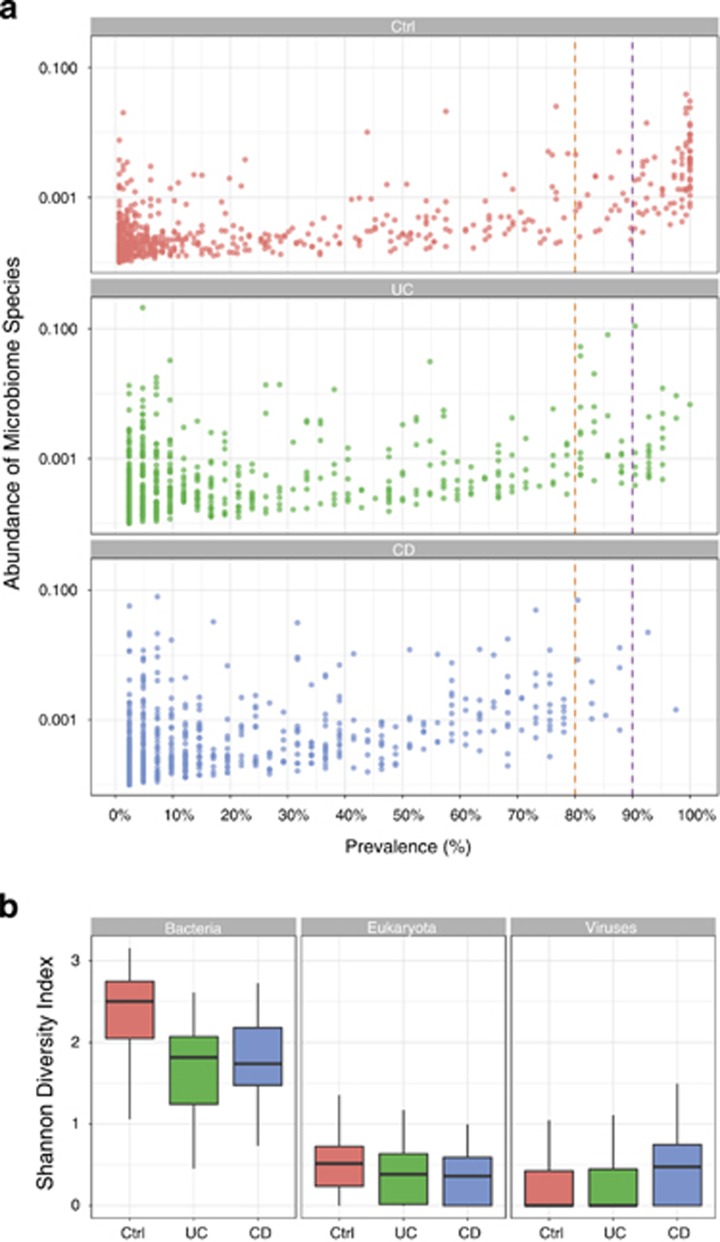
Metagenomic sequencing of IBD patients revealed hallmarks of microbiome dysbiosis with a general reduction in diversity of the microbial community compared to healthy controls (Figure 4a). Strikingly, although 29 bacterial species were shared among 100% of the healthy controls, not a single species was universally shared among IBD patients. Furthermore, among species prevalent in 90% of subjects, 78 bacterial species were shared among the healthy controls, but only 4 bacterial species were shared among the IBD patients. The common bacterial species present in the large majority of healthy controls (Supplementary Table. S3) may represent an important common core of bacteria that are central to maintaining homeostasis and health.
Figure 4.
Microbiome dysbiosis in IBD. (a). Prevalence of microbial species per cohort. Each dot represents a microbial species. Prevalence is estimated as the number of individuals with the corresponding species. Abundance is defined as the total length of mapped reads divided by the reference genome length. (b). Microbial diversity per super kingdom in UC and CD patients and healthy controls is shown using Shannon diversity index.
Consistent with prior studies describing dysbiosis in IBD patients, bacterial diversity was significantly reduced in IBD patients compared to the healthy controls, as measured by the Simpson diversity index, p-value=8.3e-05 (Figure 4b). Sensitivity analysis excluding early onset IBD, use of antibiotics, and J pouch demonstrated a consistent effect on diversity (Supplementary Figures). Overall, these data reflect a destruction of a core and diversified microbiome in the setting of IBD.
Microbiome taxonomic profile in IBD
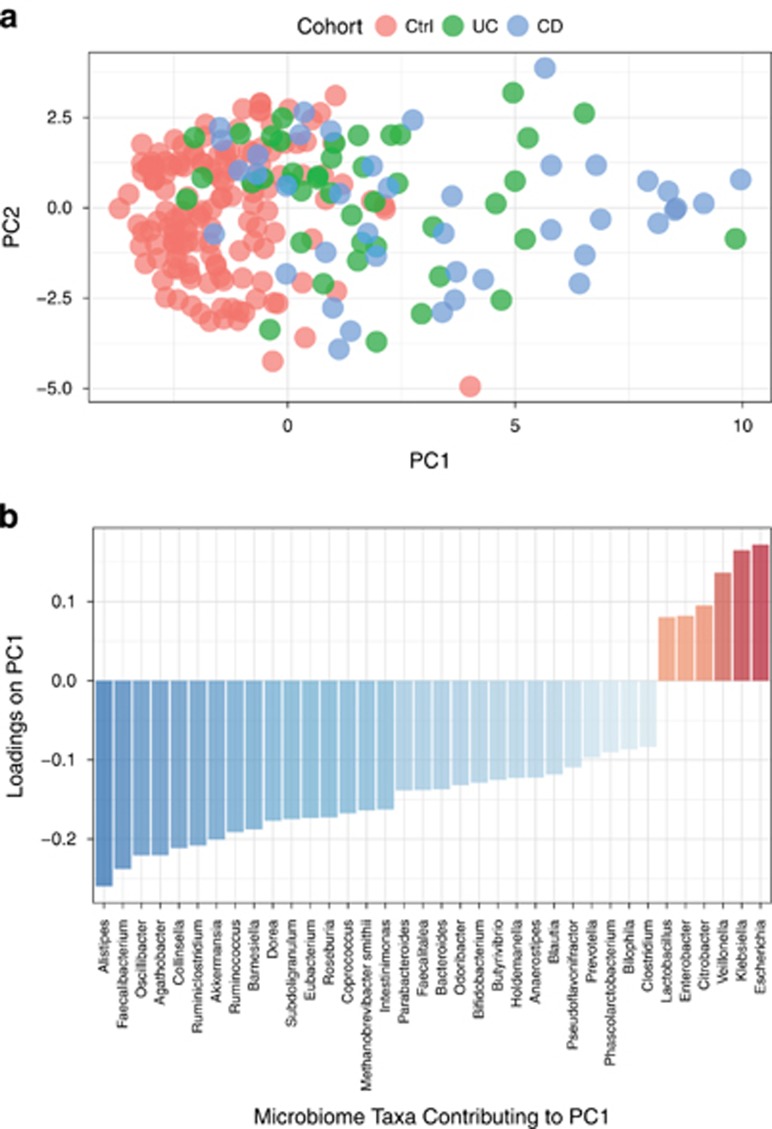
To characterize the composition of the microbiome in IBD, we first conducted a principal component analysis of the microbial abundances. PC1 indicated a significant separation between healthy controls and the IBD study population, p-value=3.6e-7 (Figure 5a).
Figure 5.
Analysis of microbiome taxonomic abundance. Principal component analysis was used to characterize the microbial populations across controls, UC and CD. (a). PC1 on the x-axis explains 26% of the variance and PC2 on the y-axis explains 10% of the variance. (b). Major taxa contributing to PC1. Major contributing taxa were identified by loadings≥standard deviation±median.
Analysis of the loadings of the first principal component identified the most enriched taxa in IBD patients as compared to healthy controls. The enriched taxa were primarily Proteobacteria (e.g., Escherichia and Klebsiella). In contrast, the most depleted taxa in IBD patients as compared with controls were Bacteroidetes (e.g., Alistipes and Barnesiella) and Firmicutes (e.g., Faecalibacterium, Oscillibacter, Agathobacter, Ruminococcus) (Figure 5b). Microbiome taxa that are statistically different in IBD patients and healthy controls are presented in Supplementary Figure S2. We also examined the relationship between genetic risk and microbiome composition in IBD and healthy control individuals. We observed a weak correlation between genetic risk and the microbiome taxonomic composition (Supplementary Figure S3).
Metagenomics has expanded the ability to detect not only microbial communities, but also to provide insight into functional effects of the microbial composition in IBD. In particular, metagenomics allows the investigation of the distribution of virulence factors across the microbiome which has not been systematically studied or previously reported in IBD. We observed a greater prevalence of virulence factors from E. coli in IBD patients. Virulence factors from Clostridium perfringens were only present in patients with IBD and undetectable in healthy controls. Overall, virulence factors were identified in 51% of the CD patients and 26% of the UC patients (~39% of the IBD patients) as compared to 14% of the healthy controls. The most prevalent virulence factors that were identified among the IBD cohort are Enterotoxin (senB) (22%), Haemoglobin protease (vat) (17%), Hemolysin A (hlyA) (11%), Hemolysin B (hlyB) (10%), Hemolysin C (hlyC) (10%), Hemolysin D (hlyD) (10%), Cytotoxic necrotizing factor 1 (cnf1) (10%), invasion protein IbeA (ibeA) (9%), Secreted autotransporter toxin (sat) (8%), and Tir domain containing protein TcpC (tcpC) (8%). Thus, expression of virulence factors in a dysbiotic microbiome may contribute to pathogenesis in IBD (Figure 6).
Figure 6.
Prevalence of virulence factors in IBD patients vs. controls. Prevalence of is estimated as the number of individuals with one virulence factor or more relative to the total number of individuals per the group.
Discussion
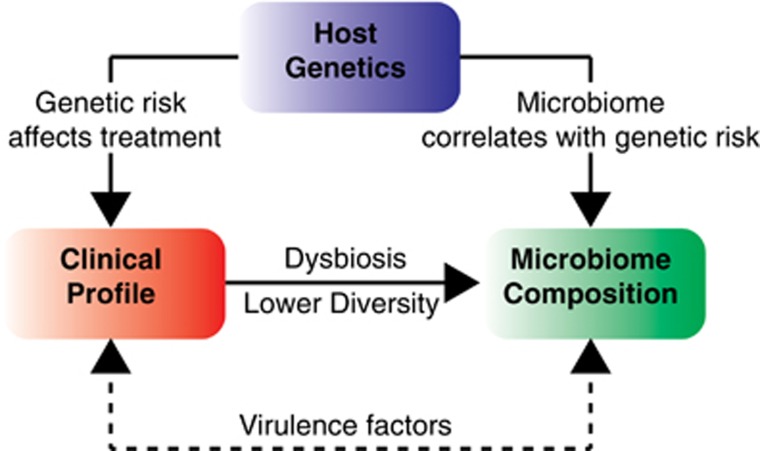
Recent advances in sequencing and data analysis have transformed our understanding of the human genome and microbiome, and the complex interaction with clinical phenotype is slowly being unraveled (Figure 7). High throughput of metagenomes is also proving to be more valuable than 16S rRNA sequencing which has become widely accepted in the human microbiome field. Using metagenomics data from well-characterized cohort of IBD patients and healthy controls, we confirm the presence of a striking dysbiosis in IBD based on diversity indices for bacteria. The dysbiosis in IBD was characterized by enrichment of bacteria that are not commonly present in healthy controls and depletion of taxa and species that are typically present in most healthy controls. In particular, we observed the depletion of a core microbiome an expansion of virulence factors in IBD patients that may facilitate overgrowth of potentially pathogenic bacteria, although we cannot exclude the possibility that the therapies used to treat IBD may have contributed to these observations.
Figure 7.
Integration of clinical, genetic and microbiome features of IBD.
Our results confirm prior studies identifying depletions in specific phylum and bacterial species that have been shown to play beneficial roles in maintaining homeostasis in the colon. Specifically, our data confirmed a reduction in the firmicutes phylum in IBD,12, 27, 28 and also support prior observations that butyrate-producing species are depleted in IBD patients as compared to healthy controls. Faecalibacterium, an anaerobe which has been associated with ameliorating colitis in mouse models, was depleted in our cohort, as has been consistently shown in multiple recent studies.8, 29, 30 In addition, Roseburia, a butyrate-producer, was also depleted in our cohort, consistent with prior studies.9, 29 Alistipes, a bile tolerant organism that is enriched in individuals on animal-based diets,31 was among the most significantly depleted bacteria in our IBD cohort. Alistipes has been shown to be depleted in elderly patients hospitalized with C. difficile in colitis.32 Overall, these findings, in conjunction with prior studies, underscore the importance of butyrate-producing bacteria that help provide energy and maintain homeostasis in colonic epithelial cells.
The IBD microbiome appears to be enriched in Enterobacteriaceae; specifically, enrichment in E. coli is consistent with findings in the RISK cohort of pediatric patients with early Crohn’s disease.8 Enterobacteriaceae may contribute to the enrichment in virulence factors in the IBD cohort that we observed in the present study. To our knowledge, this finding has not been previously reported, due to inability of 16S rDNA sequencing to capture information on virulence factors. Thus, our findings suggest the intriguing possibility that virulence factors may play a significant role in modifying and exacerbating the effect of dysbiosis in patients with IBD.
While the increased genetic risk based on known SNPs and HLA alleles associated with IBD cohort is not novel, our finding of a correlation between genetic risk and need for biologics as well as escalation to second-line biologics in patients with IBD offers new potential insight into strategies for risk stratification of patients with IBD. This observation will require further validation in prospective studies, particularly given that similar results have not emerged from large genetic consortium studies. Individuals with higher genetic risk were more likely to fail conventional therapies and required use of biologics with clinical success. Despite the limited number of individuals in the study, we also observed an association between genetic risk and microbiome composition. Our findings suggest that genetic risk scores may provide early risk stratification to identify those patients who may benefit from early escalation of management to modify their disease course.
A limitation of the study is the small number of patients compared to prominent reports from recent genetic consortia.25, 26 To maintain our sample size, we included a range of patients with IBD, including pediatric onset and those with prior colonic resections, with the goal of identifying distinct subtypes of IBD in a descriptive manner through the visualization of clinical metadata. Notably, Cleynen et al25 included nearly 30 000 patients with inflammatory bowel disease and identified differences between subtypes that could not be detected in our study. Lee JC et al26 examined extreme phenotypes in over 2,500 Crohn’s disease patients with genotype arrays. Poor-prognosis was defined as individuals who had frequent flares, treatment refractory disease based on the need for≥2 immune modulators, or multiple bowel surgeries. With this approach, they identified genetic susceptibility loci but not loci associated with prognosis. While our study was not powered to refute the findings of prior studies, we notice that not only genetic risk, but also HLA class II coincide in the association with need for biologics.
Our analysis of the fecal microbiome, rather than that of the mucosal-associated microbiome, may be a second limitation of our study. Gevers et al8 has shown that the fecal microbiome may be less sensitive in measuring dysbiosis especially early in the course of IBD. Though our cohort had well-established IBD, the stool microbiome and metagenomics may miss more subtle and relevant changes in the mucosal-associated microbiome which may play a critical role in the pathophysiology of IBD.33, 34, 35 Additional studies using intestinal biopsies will be needed to understand the mucosal-associated changes in the microbiome and metagenome.
Thus, using host whole genome sequencing and shotgun metagenomic sequencing of the microbiome in a cohort of IBD patients, we were able to confirm prior observations about genetic risk as well as dysbiosis in the stool microbiome of patients with IBD. In addition, we show that genetic risk may have a role in an early stratification strategy in the care of IBD patients and that expression of virulence factors in a dysbiotic microbiome may contribute to pathogenesis.
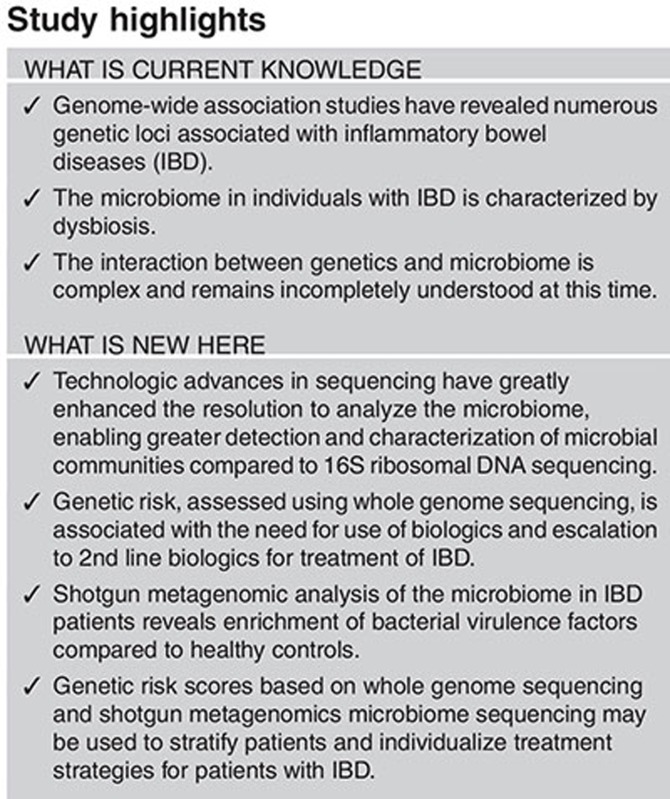
Study Highlights
Acknowledgments
Current affiliation of AM is The American University, Cairo, Egypt. Current affiliation of SY is University of Central Florida, Orlando, FL 32816. Current affiliation of MBJ is Regeneron Pharmaceuticals, Inc., Tarrytown, NY 10591. Current affiliation of AT is The Scripps Research Institute, La Jolla, CA 92037.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Publisher’s note
Springer Nature remains neutral with regard to jurisditional claims in published maps and institutional affiliations.
Guarantors of the article: Brigid S. Boland and Amalio Telenti.
Specific author contributions: Planning the study: KEN, WJS, JTC, AT, BSB. Collecting data: BSB. Generation and analysis of microbiome and genomic data: AM, WL, ELA, EHMW. Sequencing: WB. Interpreting data: AM, WL, AT, JTC, BSB. Drafting manuscript: AM, JTC, AT, BSB. Approval of final draft: AM, WL, ELA, EHMW, PSD, WJS, WB, SY, MBJ, JCV, KEN, JTC, AT, BSB.
Financial Support: P.S.D. was supported by US National Institute of Diabetes and Digestive and Kidney Diseases (5T32DK007202). J.T.C was supported by the US National Institutes of Health (DK0935007). B.S.B was supported by the Crohn’s and Colitis Foundation and UCSD KL2 (1KL2TR001444).
Potential competing interests: WL, ELA, WB, JCV, KEN are employees and stock holders of Human Longevity Inc. There is no intellectual property associated with this work. There are no other competing interests.
Data access: The metagenomic data were deposited at NCBI under Bioproject PRJNA423297, available from https://www.ncbi.nlm.nih.gov/bioproject/423297.
Supplementary Material
References
- Jostins L, Ripke S, Weersma RK et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015; 47: 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Jostins L, Spain SL et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016; 48: 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R, Russell RK, Reiff C et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol 2012; 107: 1913–1922. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KA, Bertha M, Hofmekler T et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med 2016; 8: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN St, Amand AL, Feldman RA et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007; 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014; 15: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010; 139: 1844–1854 e1. [DOI] [PubMed] [Google Scholar]
- Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology 2017; 152: 327–339 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Nishida A, Fujimoto T et al. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn's Disease. Digestion 2016; 93: 59–65. [DOI] [PubMed] [Google Scholar]
- Imhann F, Vich Vila A, Bonder MJ et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2016; 67: 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS, Zhu A, Benes V et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 2016; 352: 586–589. [DOI] [PubMed] [Google Scholar]
- Luo C, Knight R, Siljander H et al. ConStrains identifies microbial strains in metagenomic datasets. Nat Biotechnol 2015; 33: 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S et al. Genomic variation landscape of the human gut microbiome. Nature 2013; 493: 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stekhoven DJ, Buhlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics 2012; 28: 112–118. [DOI] [PubMed] [Google Scholar]
- Telenti A, Pierce LC, Biggs WH et al. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci U S A 2016; 113: 11901–11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MB, Highlander SK, Anderson EL et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proc Natl Acad Sci U S A 2015; 112: 14024–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Batzoglou S. What is the expectation maximization algorithm? Nat Biotechnol 2008; 26: 897–899. [DOI] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 2006; 34: 5623–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zheng D, Liu B et al. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 2016 44: D694–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Yeo ZX, Wong M et al. Fast and accurate HLA typing from short-read next-generation sequence data with xHLA. Proc Natl Acad Sci U S A 2017; 114: 8059–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg MS, Satsangi J, Ahmad T et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol 2005; 19 (Suppl A): 5A–36A. [DOI] [PubMed] [Google Scholar]
- Goyette P, Boucher G, Mallon D et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet 2015; 47: 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I, Boucher G, Jostins L et al. Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016; 387: 156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Biasci D, Roberts R et al. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn's disease. Nat Genet 2017; 49: 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006; 55: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Kabakchiev B, Waldron L et al. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol 2015; 16: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels K, Joossens M, Sabino J et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014; 63: 1275–1283. [DOI] [PubMed] [Google Scholar]
- Varela E, Manichanh C, Gallart M et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther 2013; 38: 151–161. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Ticinesi A, Gerritsen J et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep 2016; 6: 25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe 2015; 17: 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 2011; 140: 1720–1728. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 2014; 146: 1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.