Abstract
Purpose
The first aim of this study was to develop a novel inactivated porcine epidemic diarrhea virus (PEDV) vaccine using the recently isolated Korean PEDV QIAP1401 strain and to evaluate its protective efficacy in growing pigs. The second was to determine the optimum adjuvant formulation of the inactivated PEDV vaccine that induces protection against viral challenge.
Materials and Methods
To generate high titers of infectious PEDV, the QIAP1401 isolate was passaged in Vero cells. The experimental vaccines were prepared from a binary ethyleneimine-inactivated QIAP1401 strain passaged sequentially 70 times (QIAP1401-p70), formulated with four commercial adjuvants, and administered twice intramuscularly to growing pigs. Challenge studies using a virulent homologous strain of PEDV QIAP1401-p11, which was passaged 11 times after isolation, were performed to assess protection against disease progression and viral shedding during the 15-day observation period. The vaccine-induced antibody responses were measured in serum samples collected at predetermined time points by indirect enzyme-linked immunosorbent assay and virus neutralization test.
Results
The QIAP1401-p70 strain had 42 amino acid (aa) mutations, including a 25 aa deletion, and was selected as the inactivated PEDV vaccine candidate. Although none of the pigs that received the experimental vaccines were completely protected against subsequent viral challenge, they exhibited a significantly higher immune response than did non-vaccinated control pigs. Among the vaccine groups, the highest antibody responses were observed in the pigs that received an oil-based multiphasic water/oil/water (W/O/W) emulsion adjuvanted vaccine, which delayed the onset of clinical symptoms and viral shedding.
Conclusion
A novel inactivated PEDV vaccine formulated with a W/O/W emulsion adjuvant was both immunogenic and protective against viral challenge.
Keywords: Porcine epidemic diarrhea virus, Vaccines, Adjuvant, Growing pigs
Introduction
Porcine epidemic diarrhea (PED), caused by the PED virus (PEDV), is highly contagious and lethal in piglets. The disease is characterized by clinical signs such as diarrhea, vomiting, anorexia, and dehydration, with high mortality in neonatal piglets [1,2,3]. PED was first reported in England in 1971 [4], after which the virus was identified as coronavirus-like strain CV777 in Belgium in 1978 [5]. Subsequently, PED has occurred in many swine-producing countries in Europe and Asia and has had a substantial detrimental effect on the swine industry over the past three decades. Since the late 1990s, several PEDV vaccines have been commercialized in Asian countries as part of a comprehensive approach to disease prevention, because the clinical impact of PEDV is more acute and severe in Asia than in Europe [6]. Although the use of commercial vaccines may have led to a decline in the prevalence of the disease, the vaccines do not induce complete protection against the current epidemic PEDV strains in newborn piglets [7].
PEDV, a member of the genus Alphacoronavirus in the family Coronaviridae, is a large, enveloped virus that contains a 28 kb single-stranded positive-sense RNA genome encoding three non-structural proteins (replicases ORF1a and 1b, and ORF3) and four structural proteins (the spike [S], envelope, membrane and nucleocapsid [N] proteins) [8,9]. Among the structural proteins, the S protein is a type I glycoprotein present on the surface of virions that plays an important role in virus attachment to cell receptors, and it is a target for induction of neutralizing antibodies, as it harbors virus-neutralizing epitopes [10,11,12]. In addition, the S protein is associated with adaptation of viruses to growth in cell culture and attenuation of PEDV virulence in the natural host [13]. Therefore, the S protein of PEDV has been the subject of sequence diversity investigations aiming to understand the genetic and phylogenetic relationships of PEDV isolates and the epidemic status of PEDV in the field.
Since 2010, severe PED epidemics have occurred in many swine-producing countries, causing substantial financial losses to their domestic swine industries [14]. In Korea, severe acute diarrhea outbreaks associated with a high mortality rate (approximately 100%) occurred in neonatal piglets in the beginning of November 2013 [15]. Subsequently, the 2014 PED epidemics affected over 33,600 piglets at 169 pig farms (https://www.kahis.go.kr/), despite implementation of a national PEDV vaccination program to prevent the disease on pig farms. The field isolates responsible for this PEDV epidemic are genetically and antigenically different from the PEDV vaccine strains [14,16]. Therefore, the recent severe PED epidemics have emphasized the importance of establishing effective disease prevention nationwide, which requires development of next-generation vaccines against PEDV.
Recently, the highly virulent PEDV strain QIAP1401 was isolated from pooled intestinal homogenates and sequentially subjected to 70 passages in Vero cells in our laboratory. Phylogenetic analysis based on the full-length PEDV S gene revealed that the QIAP1401 strain belongs to the G2b genogroup and has a close phylogenetic relationship with the U.S. PEDV strains that emerged in 2014 [17]. In the present study, we developed an inactivated Korean PEDV vaccine and assessed its efficacy against homologous PEDV challenge in growing pigs. Furthermore, various adjuvants were evaluated to determine the optimum vaccine-adjuvant mixture for inducing humoral immunity in pigs.
Materials and Methods
Virus propagation
A Korean PEDV strain, QIAP1401, isolated in our laboratory was propagated on African green monkey kidney (Vero) cells (ATCC CCL-81, Manassas, VA, USA), as described previously [17], and was used after 70 passages (QIAP1401-p70). In brief, for viral infection, confluent monolayers of Vero cells were washed with phosphate-buffered saline (PBS) and inoculated with the PEDV isolate at a 0.1 multiplicity of infection in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 1 µg/mL crystallized trypsin (Sigma, St Louis, MO, USA). After incubation for 1 hour at 37℃ in an atmosphere of 5% CO2, the culture medium was replaced to remove extracellular virions. The PEDV-infected cells were maintained at 37℃ in 5% CO2 and monitored daily for a cytopathic effect (CPE) using a microscope. Upon observation of a CPE, the culture flask was subjected to three rounds of freezing and thawing, and clarified viral fluid was collected by centrifugation at 1,000×g for 10 minutes. Serial passaging of the virus was routinely conducted in six-well plates using the sequential limit dilution culture method. After each passage, an aliquot of the virus was stored at −70℃ until use. PEDV QIAP1401-p11, which was passaged 11 times after isolation, was used as the challenge strain.
Sequencing analysis
Viral RNA was extracted from 200 µL aliquots of QIAP1401-p70 using the RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The complete genome of the virus was sequenced at an external facility (Macrogen Inc., Seoul, Korea) by a next-generation sequencing technique. The whole genome sequence was compared with those of the following PEDV reference strains isolated in the United States: USA/Iowa303/2014 (GenBank No. KR265827), USA/Minnesota250/2014 (KR265776), and OH1414 (KJ408801). Multiple-sequence alignments were generated using Clustal X 2.0 [18], and the results were visualized using ESPript 3.0 [19].
Preparation of the vaccines
PEDV QIAP1401-p70 was chemically inactivated using binary ethyleneimine (BEI). In brief, 107.0 TCID50/mL virus were obtained from Vero cells cultured in roller bottles. After three rounds of freezing and thawing, the collected viral fluid was clarified and then inactivated by addition of 1 M BEI to a final concentration of 2 mM at 37℃ overnight. The remaining BEI was neutralized by addition of 20% sodium thiosulfate. Virus inactivation was verified by the absence of viral growth in Vero cell cultures.
Experimental vaccines were prepared by mixing BEI-inactivated PEDV QIAP1401-p70 with a commercial adjuvant according to the manufacturers' recommendations. The adjuvants used were Montanide IMS 1313 N VG PR (IMS 1313, Seppic, Paris, France), Montanide GEL (IMS gel), Montanide ISA 201 VG (ISA 201), and Montanide ISA 206 VG (ISA 206). ISA201 and ISA206 are oil-based multiphasic-emulsion adjuvants, and IMS1313 (nanoparticle-based) and IMSgel (synthetic polymer-based) are aqueous adjuvants. Vaccine formulations using IMS1313 and IMSgel were prepared by adding the adjuvants to BEI-inactivated viral fluid to final adjuvant concentrations of 30% and 10% (v/v), followed by vortexing at room temperature for 15 minutes. Vaccine formulations using ISA201 and ISA206 were prepared by emulsifying inactivated viral fluid with an equal volume of adjuvant using a homogenizer with a stirrer-propeller blade at 500 rpm for 10 minutes at 32℃ in a water bath. After cooling at 20℃ for 1 hour, formation of water/oil/water (W/O/W) multiple emulsions was confirmed by microscopy. All adjuvanted vaccines contained 0.01% of thimerosal to prevent growth of microorganisms, and were aseptically transferred to sterile vaccine vials and stored at 4℃ before use.
Pig vaccination-challenge experiment
Twenty growing pigs (6 weeks old) were purchased from a PEDV-free pig farm and housed at the laboratory animal facility of the Animal and Plant Quarantine Agency in accordance with Institutional Animal Care and Use Committee protocols (ethics approval number: 2017-573). Before the start of the experiment, all pigs were verified as serologically negative for PEDV. After 1 week of acclimatization, pigs were randomly divided into four vaccination groups of four pigs each and comingled in two separate rooms for the duration of the experiment. Pigs in the vaccination groups were immunized twice intramuscularly at a 2-week interval with 2 mL of the experimental PEDV vaccine. The four remaining contact pigs were not vaccinated and were used as controls. At 42 days after the first vaccination, all pigs were challenged orally with PEDV QIAP1401-11p (1 mL 104.0 TCID50/mL). Clinical signs of PEDV infection were observed daily for 15 days, and the clinical score was determined based on diarrhea severity: 0, no clinical signs, healthy; 1, moderate diarrhea; and 2, severe diarrhea, vomiting and dehydration. Fecal samples were collected daily for the duration of the challenge to monitor viral shedding in feces. Serum samples for immunoserological analysis were collected at 0, 14, and 28 days post-vaccination (dpv) and 15 days post-challenge (dpc), i.e., 43 dpv.
Immunoserological analyses
Serum-neutralizing antibody titers were determined by virus neutralization (VN) test in 96-well cell culture plates. Briefly, serum samples collected from the pigs after vaccination and challenge were diluted twofold (1:4 to 1:512) in serum-free DMEM containing 1 µg/mL trypsin and mixed with an equal volume of a virus suspension containing 100–200 TCID50/0.1 mL PEDV. After incubation for 1 hour at 37℃, 200 µL of the virus-serum mixture were added to confluent Vero cell monolayers and incubated for 1 hour at 37℃ in 5% CO2. Thereafter, the cultures were washed three times with PBS containing 1 µg/mL trypsin and replenished with viral growth medium. After incubation for 48 hours, VN titers were established as reciprocal values of the highest serum dilution that inhibited PEDV-specific CPE.
Serum PEDV-specific antibody levels were measured by indirect enzyme-linked immunosorbent assay (I-ELISA). The commercial PEDV N protein-based ID Screen PEDV Indirect Screening test (IDvet, Montpellier, France) was used, and IELISA was conducted according to the manufacturer's instructions. Optical densities at 450 nm were measured using an enzyme-linked immunosorbent assay (ELISA) plate reader.
Real-time reverse transcription polymerase chain reaction
Viral RNA was extracted from fecal samples using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. Following extraction, 5 µL viral RNA template were added to 20 µL real-time reverse transcription polymerase chain reaction (rRT-PCR) master mix from the PowerChek PEDV Real-Time RT-PCR kit (Kogenbiotech, Seoul, Korea). The reactions were run in presence/absence mode on the Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Middlesex, MA, USA) at 50℃ for 30 minutes, 95℃ for 10 minutes, and then 40 cycles of 95℃ for 15 seconds and 60℃ for 1 minute. A FAM-labeled probe was used to detect the PEDV, and a VIC-labeled probe was used to detect the internal control. The results were analyzed by the StepOne Plus v2.3 software (Applied Biosystems).
Statistics
The results, except for the clinical data, are expressed as mean±standard deviation. Statistical analysis was performed using SPSS statistics ver. 21.0 (IBM Corp., Armonk, NY, USA). In the ELISA and VN test, differences between the experimental groups were analyzed by unpaired Student's t test and one-way ANOVA, followed by Tukey's post hoc test. A chi-square or Fisher exact test was employed to compare viral shedding in feces between experimental groups at each time point. A p-value of <0.05 was considered to indicate statistical significance.
Results
Characterization of the complete genome of PEDV QIAP1401-p70
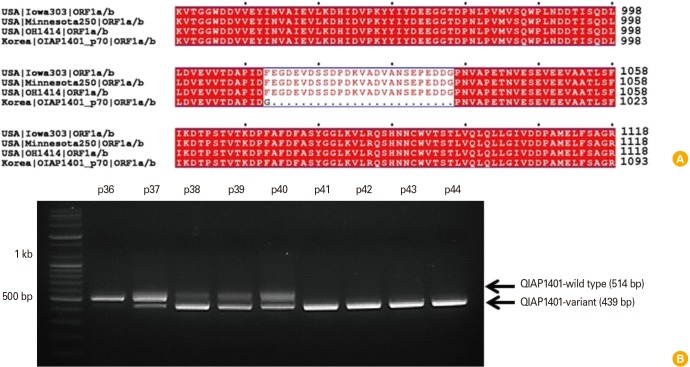
The complete genome of the QIAP1401-p70 isolate, comprising 27,920 nucleotides, was determined by next-generation sequencing. The whole genome of the PEDV USA/Minnesota250/2014 strain was used as a reference for sequencing because of its close genetic relationship to QIAP1401 isolates. To assess whether the PEDV QIAP1401 variant had emerged after sequential passaging, the genome sequence was compared with that of U.S. PEDV strains. The PEDV QIAP1401-p70 isolate had > 99.4% sequence homology to the USA/Iowa303/2014, USA/Minnesota250/2014 and OH1414 U.S. PEDV strains. However, we identified 42 amino acid (aa) mutations, including a 25 aa deletion, in the ORF1a gene of the QIAP1401-p70 isolate (Fig. 1A). To determine at which passage the deletion arose, viral RNA isolated from each passage was screened by reverse transcription polymerase chain reaction using ORF1a-specific PCR primers. The results showed that the deletion mutant emerged after 41 passages (Fig. 1B). The deletion mutation detected at passage 37 was present during the following three passages. Moreover, viral stock at passages 37 to 40 contained both parental and mutant viruses.
Fig. 1. (A) Genetic characterization of QIAP1401-p70. A cell-culture-adapted QIAP1401 variant was generated by passaging 70 times using the sequential limit dilution culture method. The whole genome sequence of QIAP1401-p70 was determined by next-generation sequencing technology and compared with the reference sequences of genogroup G2: USA/Iowa303/2014 (KR265827), USA/Minnesota250/2014 (KR265776), and OH1414 (KJ408801). QIAP1401-p70 had 42 aa variations, of which a 25 aa deletion in ORF1a was notable. (B) QIAP1401-p70 emerged after 41 sequential passages.
Clinical assessments
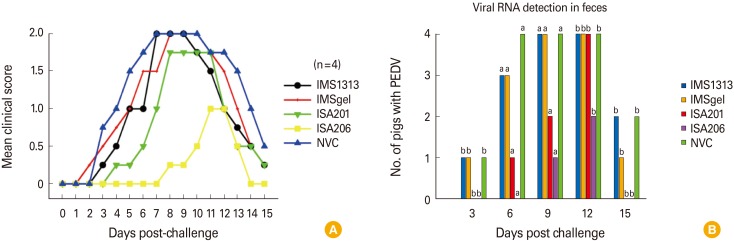
All growing pigs were apparently healthy and had no clinical symptoms before oral challenge. The protective efficacy of the experimental vaccines against challenge with virulent homologous virus (104.0 TCID50/mL PEDV QIAP1401-p11) was determined by the absence of clinical signs and decreased viral shedding during the 15-day observation period. Viral shedding in feces was determined using a commercial rRT-PCR kit. All pigs in the non-vaccinated control (NVC) group exhibited early signs of disease, typically mild diarrhea and loss of appetite (mean clinical score ≤1.0) after 2 dpc and experienced severe watery diarrhea with vomiting thereafter (mean clinical score ≤1.75) (Fig. 2A). The IMS1313p<and IMSgel-adjuvanted vaccine groups exhibited a similar clinical presentation to that of the NVC group. In contrast, the ISA206-adjuvanted vaccine group exhibited weaker and delayed clinical signs, mainly mild diarrhea (mean clinical score ≤1.0). In the ISA201 vaccine group, clinical disease progression was slow, and clinical severity was relatively weak, compared with the NVC group, but watery diarrhea eventually developed.
Fig. 2. Clinical score and viral shedding in pigs after challenge with virulent homologous virus (porcine epidemic diarrhea virus [PEDV] QIAP1401-p11). Diarrhea severity was scored (A), and viral shedding in feces was monitored by real-time reverse transcription polymerase chain reaction (B). The letter “a” above the bars indicates a significant difference among the experimental groups (p<0.05, Fisher exact test), whereas “b” indicates no significant difference. NVC, non-vaccinated control.
Viral shedding was defined as the presence of PEDV RNA as detected by rRT-PCR. PEDV shedding in feces in all groups was generally accompanied by clinical signs of disease (Fig. 2B). PEDV RNA was first detected in one of four pigs in the IMS1313, IMSgel, and NVC groups at 3 dpc. The duration of viral shedding differed among the groups. In the ISA206 and ISA201 groups, viral shedding in feces continued for 3 and 6 days, respectively. In contrast, in the other vaccinated groups and in the NVC group, viral shedding in feces was observed until the end of the challenge experiment. The viral shedding rates differed significantly among the groups (number of pigs with PEDV/total number of fecal samples) at both 6 and 9 dpc (p<0.05).
Serum antibody response
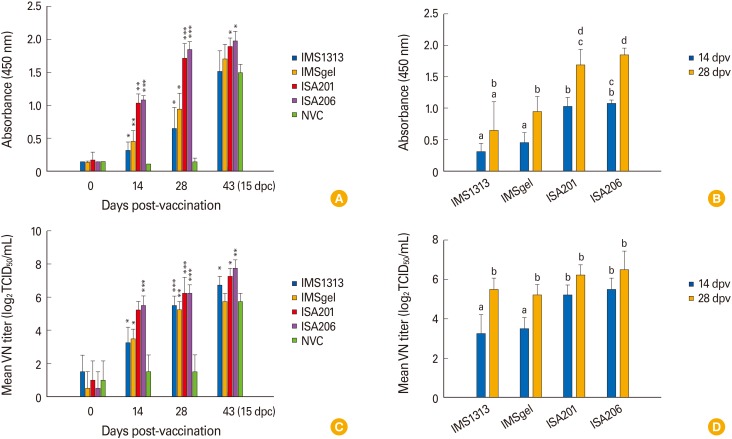
The serum antibody response induced by the experimental vaccines was assessed by I-ELISA and VN. All of the experimental vaccines induced a significant PEDV-specific serum IgG response to PEDV. The mean IgG levels in all vaccine groups, but not the NVC group, increased continuously after the first and second vaccinations. At 14 and 28 dpv, the mean IgG levels in all vaccine groups were significantly higher than those in the NVC group (p<0.01, unpaired Student's t test) (Fig. 3A). The mean IgG levels were significantly higher in the ISA201- and ISA206-adjuvanted vaccine groups than in the IMS1313- and IMSgel-adjuvanted vaccine groups at 14 and 28 dpv (p<0.05, Tukey's post hoc test) (Fig. 3B), indicating that the ISA201 and ISA206 adjuvants induced the strongest PEDV-specific IgG response in growing pigs.
Fig. 3. Serum immune response of pigs in the IMS1313, IMSgel, ISA201, and ISA206 groups. Pigs were vaccinated at 2-week intervals and then challenged with virulent homologous virus (porcine epidemic diarrhea virus [PEDV] OIAP1401-p11). Serum samples collected at 0, 14, 28, and 48 days post-vaccination (dpv) (15 days post-challenge [dpc]) were subjected to enzyme-linked immunosorbent assay (ELISA) (A and B) and virus neutralization (VN) test (C and D) of PEDV-specific IgG and virus-neutralizing antibody titers, respectively. Bars represents the means standard deviation of five independent samples. *, **, and *** indicate significant differences from the non-vaccinated control (NVC) group (p<0.05, p<0.01, and p<0.001, respectively, unpaired Student's t test). Different lower-case letters above the bars indicate significant differences among the groups (p<0.05, Tukey's post hoc test).
VN titers also increased continuously in all vaccine groups after the first and second vaccinations and were significantly higher than those in the NVC group (p<0.01, unpaired Student's t test) (Fig. 3C). Although the maximum VN titers in all vaccine groups were detected at 28 dpv, there was no significant difference among the values (p<0.05, Tukey's post hoc test). In contrast, at 14 dpv, the mean VN titer of the ISA201 group (5.0 log2 VN titer) was significantly higher than those of the IMS1313 (3.25 log2 VN titer) and IMS gel (3.5 log2 VN titer) groups. The ISA206 group also exhibited a higher mean VN titer (5.5 log2 VN titer) than those of the IMS1313 and IMSgel groups, while the mean VN titers of the ISA206 and ISA201 groups were comparable (p<0.05, Tukey's post hoc test) (Fig. 3D). These findings indicated that the ISA201 and ISA206 adjuvants induce a virus-neutralizing antibody response more rapidly than do the IMS1313 and IMSgel adjuvants.
Discussion
Recent outbreaks and global re-emergences of PED pose a substantial threat to the swine industry in Asia and the United States. Indeed, the efficacies of the commercially available inactivated PEDV vaccines based on the SM98, DR-13, and CV-777 strains used in Asian swine-producing countries are uncertain [20]. Most PEDV strains associated with recent severe PED outbreaks were of the G2b genogroup and are; thus, they are antigenically and genetically distinct from the PEDV vaccine strains [14]. This may explain the low protective efficacy of PEDV vaccines against outbreak strains, and suggests that development of an efficacious vaccine is urgently required.
A virulent Korean PEDV strain, QIAP1401, was recently isolated and passaged in our laboratory. This isolate is almost genetically identical to the PEDV strains responsible for the severe PED epidemics in Asia and the United States since 2013 (G2b genogroup) [17]. Therefore, QIAP1401 is a suitable candidate for development of an inactivated vaccine against current epidemic PEDV strains.
Adaptation of viruses to cell culture passaging has been used to alter the mechanism of viral replication [21]. For instance, wild-type hepatitis A virus (HAV) produces a low yield of progeny virus due to its low replication capacity, but a cell-culture-adapted HAV variant generated by repeated passaging produces higher titers of infectious viral particles [22]. A comparative study in which growing pigs were vaccinated with 106.0 to 108.0 TCID50/mL inactivated PEDV vaccine reported that the humoral immune response increased with an increase in the viral titer inoculated [23]. Therefore, cell culture-adapted viral variants that yield higher titers should be used in the development of inactivated viral vaccines.
To this end, we passaged the QIAP1401 isolate 70 times in Vero cells at 3-day intervals. A growth kinetics analysis showed that the infectious titer of QIAP1401 increased significantly to 107.0 TCID50/mL after 70 passages [17]. This indicated that the virus had acquired adaptive mutations that enhanced production of infectious virus. To investigate the underlying mechanism, we analyzed the whole genome of QIAP1401-p70 by next-generation sequencing. Compared with reference G2b genogroup PEDVs, the adapted QIAP1401-p70 genome harbored mutations in genes encoding structural and nonstructural proteins. Among these mutations, the most remarkable was a 25 aa deletion in the ORF1a region of the nonstructural protein. We hypothesized that this 25 aa deletion reduced the duration of the replication cycle of QIAP1401-p70. This finding is supported by previous reports that the 5' two-thirds of the coronavirus genome contains ORF1a and ORF 1b, which encode the viral RNA replicase [24,25].
Pregnant sows are suitable models for evaluating the effectiveness of inactivated PEDV vaccines in target populations, such as neonatal piglets, because transfer of maternal antibodies to piglets via colostrum may play an important role in the development of protective immunity against PEDV in sucking piglets [26]. However, considerations of size of litter, housing, and reproduction have limited their use in vaccination experiment. And the correspondingly higher cost associated with housing and experimentation is needed. Therefore, in the present study, we used growing pigs to evaluate the protective efficacy of our experimental vaccines, because pigs of all ages are susceptible to PEDV infection [8].
Various types of adjuvants are used in inactivated viral vaccines to induce a protective immune response. Therefore, determination of the optimum adjuvant for a particular vaccine is important. In this study, we compared the ability of experimental vaccines formulated with four adjuvants to induce protective immunity against PEDV. Although none of the vaccinated pigs were completely protected against subsequent viral challenge, they developed a significantly stronger immune response than that of NVC pigs. The strongest antibody responses were detected in pigs that received the vaccines containing oil-based W/O/W emulsion adjuvants, and the ISA206-based vaccine protected against both disease progression and viral shedding. Similarly, a foot and mouth disease vaccine formulated with ISA201 and ISA206 elicited the strongest antibody response and induced protection against the disease [27,28,29]. Such formulations also elicit protective immunity against viral and bacterial pathogens in various mammalian species [30,31].
Although emulsified adjuvants are frequently used in veterinary vaccines, instead of aluminum hydroxide, because they induce long-term immunity [32], improved adjuvants such as those based on nanoparticles or polymers are required. IMS1313 and IMSgel are aqueous adjuvants composed of nanoparticles and an immunostimulatory compound and synthetic polymer, respectively (https://www.seppic.com). According to the manufacturer, both adjuvants significantly enhance the strength and duration of the immune response and are suitable for a wide range of infectious agents. Indeed, the ability of both adjuvants to induce specific mucosal immunity against various pathogens in target animals has been assessed [33,34,35]. In this study, pigs that received the vaccines containing IMS1313 and IMSgel exhibited a strong serum PEDV-specific antibody response.
In summary, a novel inactivated PEDV vaccine formulated with a W/O/W emulsion adjuvant was immunogenic and effective in growing pigs. Our findings support use of growing pigs as an alternative experimental model for assessment of the protective efficacy of inactivated PEDV vaccines in pregnant sows and neonatal piglets. Therefore, our results may be used to reduce the risk of failure in future clinical trials of PEDV vaccines. Maternally derived lactogenic immunity (IgG and IgA) plays an important role in protection of neonatal piglets against PEDV infection [20]. Thus, future studies should assess the effect of vaccinating sows with our novel inactivated PEDV vaccine on neonatal piglets.
Footnotes
No potential conflict of interest relevant to this article was reported.
This study was supported financially by a grant (B-1543083-2015-17-01) from the Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs (MAFRA), Republic of Korea.
References
- 1.Debouck P, Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am J Vet Res. 1980;41:219–223. [PubMed] [Google Scholar]
- 2.Pensaert MB, Yeo SG. Porcine epidemic diarrhea. In: Straw BE, Zimmerman JJ, D'Allaire S, Taylor DJ, editors. Disease of swine. 9th ed. Ames, IW: Blackwell Publishing; 2006. pp. 367–372. [Google Scholar]
- 3.Pijpers A, van Nieuwstadt AP, Terpstra C, Verheijden JH. Porcine epidemic diarrhoea virus as a cause of persistent diarrhoea in a herd of breeding and finishing pigs. Vet Rec. 1993;132:129–131. doi: 10.1136/vr.132.6.129. [DOI] [PubMed] [Google Scholar]
- 4.Oldham J. Letter to the editor. Pig Farming. 1972;10:72–73. [Google Scholar]
- 5.Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song DS, Oh JS, Kang BK, et al. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res Vet Sci. 2007;82:134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saif LJ, Pensaert MP, Sestak K, Yeo SG, Jung K. Coronaviruses. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Diseases of swine. 10th ed. Ames, IW: Wiley-Blackwell; 2012. pp. 501–524. [Google Scholar]
- 9.Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang SH, Bae JL, Kang TJ, et al. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- 12.Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato T, Takeyama N, Katsumata A, Tuchiya K, Kodama T, Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes. 2011;43:72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S, Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg Infect Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park BK, Song D. Recent outbreaks and emergence of mutants of porcine epidemic diarrhea viruses (PEDV) in Korea. Jpn J Vet Res. 2016;64(Suppl 1):S25–S32. [Google Scholar]
- 16.Lee DK, Park CK, Kim SH, Lee C. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Res. 2010;149:175–182. doi: 10.1016/j.virusres.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang DK, Kim HH, Lee SH, Yoon SS, Park JW, Cho IS. Isolation and characterization of a new porcine epidemic diarrhea virus variant that occurred in Korea in 2014. J Vet Sci. 2017 Jul 10; doi: 10.4142/jvs.2018.19.1.71. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song D, Moon H, Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Xiao L, Nelson C, Hagedorn CH. A cell culture adapted HCV JFH1 variant that increases viral titers and permits the production of high titer infectious chimeric reporter viruses. PLoS One. 2012;7:e44965. doi: 10.1371/journal.pone.0044965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daemer RJ, Feinstone SM, Gust ID, Purcell RH. Propagation of human hepatitis A virus in African green monkey kidney cell culture: primary isolation and serial passage. Infect Immun. 1981;32:388–393. doi: 10.1128/iai.32.1.388-393.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collin EA, Anbalagan S, Okda F, Batman R, Nelson E, Hause BM. An inactivated vaccine made from a U.S. field isolate of porcine epidemic disease virus is immunogenic in pigs as demonstrated by a dose-titration. BMC Vet Res. 2015;11:62. doi: 10.1186/s12917-015-0357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiel V, Herold J, Schelle B, Siddell SG. Viral replicase gene products suffice for coronavirus discontinuous transcription. J Virol. 2001;75:6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing N, Guan X, An B, et al. Ultrasensitive detection of porcine epidemic diarrhea virus from fecal samples using functionalized nanoparticles. PLoS One. 2016;11:e0167325. doi: 10.1371/journal.pone.0167325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goede D, Murtaugh MP, Nerem J, Yeske P, Rossow K, Morrison R. Previous infection of sows with a “mild” strain of porcine epidemic diarrhea virus confers protection against infection with a “severe” strain. Vet Microbiol. 2015;176:161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Park ME, Lee SY, Kim RH, et al. Enhanced immune responses of foot-and-mouth disease vaccine using new oil/gel adjuvant mixtures in pigs and goats. Vaccine. 2014;32:5221–5227. doi: 10.1016/j.vaccine.2014.07.040. [DOI] [PubMed] [Google Scholar]
- 28.Comi G, Freedman MS, Kappos L, et al. Pooled safety and tolerability data from four placebo-controlled teriflunomide studies and extensions. Mult Scler Relat Disord. 2016;5:97–104. doi: 10.1016/j.msard.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Ibrahim Eel-S, Gamal WM, Hassan AI, Mahdy Sel D, Hegazy AZ, Abdel-Atty MM. Comparative study on the immunopotentiator effect of ISA 201, ISA 61, ISA 50, ISA 206 used in trivalent foot and mouth disease vaccine. Vet World. 2015;8:1189–1198. doi: 10.14202/vetworld.2015.1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouguyon E, Goncalves E, Shevtsov A, et al. A new adjuvant combined with inactivated influenza enhances specific CD8 T cell response in mice and decreases symptoms in swine upon challenge. Viral Immunol. 2015;28:524–531. doi: 10.1089/vim.2014.0149. [DOI] [PubMed] [Google Scholar]
- 31.Aziz-Boaron O, Gleser D, Yadin H, et al. The protective effectiveness of an inactivated bovine ephemeral fever virus vaccine. Vet Microbiol. 2014;173:1–8. doi: 10.1016/j.vetmic.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 32.Aucouturier J, Dupuis L, Ganne V. Adjuvants designed for veterinary and human vaccines. Vaccine. 2001;19:2666–2672. doi: 10.1016/s0264-410x(00)00498-9. [DOI] [PubMed] [Google Scholar]
- 33.Jang SI, Lillehoj HS, Lee SH, et al. Mucosal immunity against Eimeria acervulina infection in broiler chickens following oral immunization with profilin in Montanide adjuvants. Exp Parasitol. 2011;129:36–41. doi: 10.1016/j.exppara.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Walders B, Raschke A, Neugebauer M, et al. Blending of a conventional Mycoplasma hyopneumoniae vaccine with a positive marker: tracking of immunised pigs by peptide-specific antibodies raised to the marker component. Res Vet Sci. 2005;78:135–141. doi: 10.1016/j.rvsc.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Deville S, Arous JB, Bertrand F, Borisov V, Dupuis L. Efficacy of intranasal and spray delivery of adjuvanted live vaccine against infectious bronchitis virus in experimentally infected poultry. Procedia Vaccinol. 2012;6:85–92. doi: 10.1016/j.provac.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]