ABSTRACT
Gut microbiota dysbiosis has been observed in a number of autoimmune diseases. However, the role of the gut microbiota in systemic lupus erythematosus (SLE), a prototypical autoimmune disease characterized by persistent inflammation in multiple organs of the body, remains elusive. Here we report the dynamics of the gut microbiota in a murine lupus model, NZB/W F1, as well as intestinal dysbiosis in a small group of SLE patients with active disease. The composition of the gut microbiota changed markedly before and after the onset of lupus disease in NZB/W F1 mice, with greater diversity and increased representation of several bacterial species as lupus progressed from the predisease stage to the diseased stage. However, we did not control for age and the cage effect. Using dexamethasone as an intervention to treat SLE-like signs, we also found that a greater abundance of a group of lactobacilli (for which a species assignment could not be made) in the gut microbiota might be correlated with more severe disease in NZB/W F1 mice. Results of the human study suggest that, compared to control subjects without immune-mediated diseases, SLE patients with active lupus disease possessed an altered gut microbiota that differed in several particular bacterial species (within the genera Odoribacter and Blautia and an unnamed genus in the family Rikenellaceae) and was less diverse, with increased representation of Gram-negative bacteria. The Firmicutes/Bacteroidetes ratios did not differ between the SLE microbiota and the non-SLE microbiota in our human cohort.
IMPORTANCE SLE is a complex autoimmune disease with no known cure. Dysbiosis of the gut microbiota has been reported for both mice and humans with SLE. In this emerging field, however, more studies are required to delineate the roles of the gut microbiota in different lupus-prone mouse models and people with diverse manifestations of SLE. Here, we report changes in the gut microbiota in NZB/W F1 lupus-prone mice and a group of SLE patients with active disease.
KEYWORDS: lupus, microbiota
INTRODUCTION
Systemic lupus erythematosus (SLE) is a prototypical autoimmune disease that affects almost all organs of the body. It exhibits diverse manifestations, which represents a challenge to clinicians. The etiology of SLE is unclear, but evidence has shown that it is influenced by genetic, environmental, hormonal, and epigenetic factors (1). Together, these factors act on the immune system and cause abnormalities, including generation of autoantibody-producing B cells and autoreactive T cells and abnormal production of proinflammatory cytokines. The disease is characterized by severe inflammation in target organs, leading to tissue damage. The products of damaged cells are picked up by autoantibodies to create immune complexes that activate antigen-presenting cells, which in turn induce more inflammatory T cells and proinflammatory cytokines, creating a vicious cycle of inflammation (2). Current standard-of-care treatments for SLE are mainly nonselective immunosuppressants (3). Not all patients respond to these treatments, and systemic immunosuppression is a major cause of concern. Higher incidences of infection and more-severe infections have been observed among patients receiving long-term nonselective immunosuppressant therapy (4). There is a critical need for better understanding of the pathogenesis of SLE, from which new treatment strategies may be developed.
Environmental factors, including diet, modern medicine, and environmental microbes, are involved in the etiology of SLE. This is supported by observations in both humans and mice. The incidence of SLE is significantly higher in African Americans than West Africans, although the two populations are genetically very similar (5). In lupus-prone mice, the involvement of environmental factors is evidenced by the progressive loss of the SLE-like phenotype observed in the classic lupus-prone strain MRL/Mp-Faslpr (MRL/lpr) over several years at The Jackson Laboratory under specific pathogen-free conditions (stock no. 006825), while no deviation from the original strain was detected at the genetic or epigenetic level. Because diet, medicine, and environmental microbes can influence the composition of the host microbiota, efforts have been made to understand the dynamics of the microbiota (largely commensal bacteria living in the gut) in the pathogenesis of SLE. Oral antibiotics are known to trigger lupus flares (6–8), suggesting a role for commensal bacteria in SLE. Our research team recently described changes in the gut microbiota in lupus-prone mice versus healthy controls (9); a decrease in Lactobacillaceae and an increase in Lachnospiraceae were observed in the MRL/lpr mouse model (stock no. 000485; The Jackson Laboratory). Interestingly, a treatment that improved lupus-like symptoms also restored lactobacilli (9). Based on these observations, we administered Lactobacillus spp. to MRL/lpr mice and showed a striking effect of the probiotics in ameliorating lupus nephritis (10). This suggests that lupus disease could be controlled by changes in the gut microbiota. In another lupus-prone mouse model, SNF1, greater abundance of the Rikenellaceae family of commensal bacteria was found to be associated with more severe SLE-like disease (11). Moreover, increased bacterial diversity was observed in both the MRL/lpr and SNF1 mouse models (9, 11). In human SLE, a cross-sectional study showed that the fecal microbiota of SLE patients with inactive disease had a significantly lower Firmicutes/Bacteroidetes ratio than did that of healthy controls (12). A lower Firmicutes/Bacteroidetes ratio is also evident in other autoimmune diseases (13, 14). The fecal microbiota of SLE patients was a stronger inducer of Th17 differentiation than was the healthy control fecal microbiota (15), and the Th17 response is considered a primary driver of autoimmunity in SLE (16–20). Here, we report the dynamics of the gut microbiota in another murine lupus model, NZB/W F1, as well as an altered gut microbiota in a small group of SLE patients.
RESULTS
Dynamics of the gut microbiota in NZB/W F1 mice.
Like in MRL/lpr mice, the disease phenotype in NZB/W F1 mice resembles human SLE and is characterized by high levels of antinuclear antibodies, hemolytic anemia, proteinuria, and progressive immune complex glomerulonephritis. These mice have been used as a model for human SLE since the early 1960s, as these mice and humans with this multifactorial disease have similarly complex genomic landscapes. Similar to human SLE, which has a strong female bias, the disease is most pronounced in female NZB/W F1 mice. The average life span for females is 8 months, with disease onset at approximately 5 months (20 weeks) of age. Thus, disease progression is much slower in NZB/W F1 mice than in MRL/lpr mice.
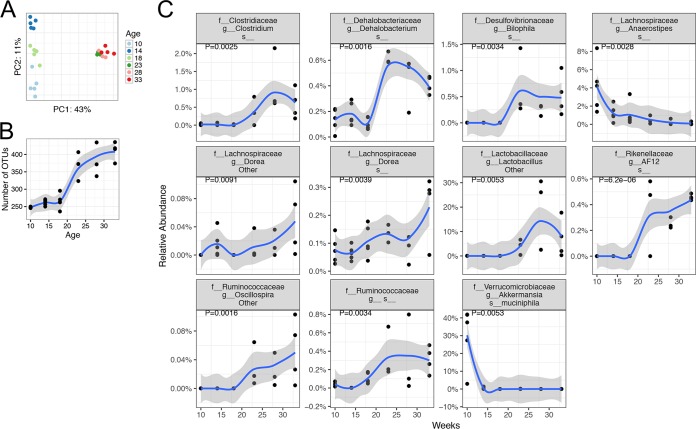
To determine the dynamics of the gut microbiota during lupus progression in female NZB/W F1 mice, we analyzed fecal pellets collected at three predisease time points (10, 14, and 18 weeks of age) and three post-disease-onset, or diseased, time points (23, 28, and 33 weeks of age). The structure and diversity of the lupus-associated microbiotas changed continuously over time (Fig. 1). The unweighted UniFrac distance-based principal-coordinate analysis (PCoA) showed that the gut microbiotas were distinct at three predisease time points but clustered together at the diseased time points (Fig. 1A; also see Fig. S1 in the supplemental material). Importantly, the UniFrac distance between predisease and diseased time points was greater than the distance observed among the three predisease time points. In addition, the gut microbiotas split into two groups along the PC1 axis (P < 0.01, permutational multivariate analysis of variance [PerMANOVA]), and the split happened at ∼20 weeks of age, suggesting a dramatic change in the gut microbiotas upon the onset of SLE-like symptoms. It is worth noting that we cannot rule out the possibility that cage effects are driving the differences in Fig. 1A, and we do not have a wild-type control for the effects of aging on the microbiota.
FIG 1.
Dynamics of the gut microbiota in NZB/W F1 mice. Fecal pellets were collected at 10, 14, and 18 weeks of age (predisease time points; n = 5 per time point) and 23, 28, and 33 weeks of age (post-disease-onset time points; n = 3 or 4 per time point) and were subjected to 16S rRNA sequencing analysis. The disease onset in NZB/W F1 mice is ∼20 weeks of age. (A) PCoA plot, showing alterations of overall community structures (P < 0.01). (B) Bacterial diversity, as indicated by the number of OTUs. The increase of OTUs from the pre-disease-onset stage to the post-disease-onset stages was significant (P < 0.01). (C) Time-dependent changes in the relative abundance of different bacterial species. Smoothing was performed with the ggplot2 package, using locally weighted regression. In all figures, P values were corrected for multiple comparisons by controlling the FDRs.
We next determined the change in microbiota diversity during lupus progression in NZB/W F1 mice. The number of operational taxonomic units (OTUs) in these mice increased significantly from the predisease stage to the diseased stage (P < 0.001) (Fig. 1B), suggesting increased bacterial diversity as the disease progressed. This is consistent with other lupus-prone mouse models, in which lupus-associated increases in microbiota diversity were also observed (9, 11). Individual bacterial species fluctuated over the time period tested, and the species with significant changes are shown in Fig. 1C. Specifically, significant increases from the predisease stage to the diseased stage were observed for several bacterial species in the genera Clostridium, Dehalobacterium, Lactobacillus, Oscillospira, Dorea (family Lachnospiraceae), Bilophila (family Desulfovibrionaceae), and AF12 (family Rikenellaceae) and an unnamed genus within the family Ruminococcaceae (P < 0.01 in all cases). Akkermansia muciniphila and a species within the genus Anaerostipes (family Lachnospiraceae), however, significantly decreased from the predisease stage to the diseased stage (P < 0.01). These results suggest that the composition of the gut microbiota changed markedly from before to after the onset of lupus disease in NZB/W F1 mice, with greater diversity and increased representation of several bacterial species as lupus progressed from the predisease stage to the diseased stage. It is worth noting that these changes may also be caused by the maturation of bacterial communities as the mice aged.
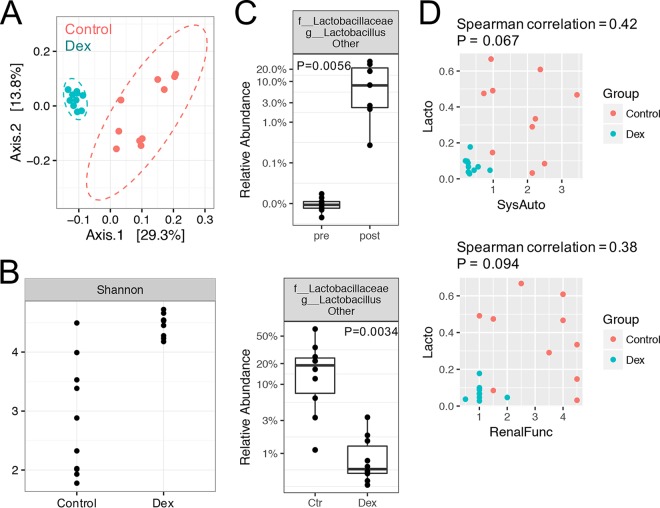
To determine whether a common treatment for SLE could reverse the changes in the gut microbiota, we treated NZB/W F1 mice with 2 mg/kg body weight dexamethasone (Dex) from 20 to 34 weeks of age (14 weeks of treatment). Prior studies showed that Dex suppresses the development of disease in NZB/W F1 lupus-prone mice (21). Treatment with Dex altered the microbiota as the animals aged, compared to the vehicle-treated controls (Fig. 2). The overall microbiota structure with Dex treatment was distinct from that with vehicle treatment (control), as shown in the unweighted UniFrac-based PCoA plot (P < 0.01) (Fig. 2A). Interestingly, instead of decreasing the bacterial diversity, which was already high in diseased mice (Fig. 1B), Dex appeared to further increase the diversity, with a significantly higher Shannon index and more observed OTUs (P < 0.001 for both cases) (Fig. 2B; also see Fig. S2). Notably, the community variability decreased (Fig. 2A) while diversity increased (Fig. 2B) in Dex-treated mice, compared to control mice. We suggest that high diversity in the Dex-treated group may lead to a more stable community and thus lower observed variability than for the control group.
FIG 2.
Changes in the gut microbiota in NZB/W F1 mice in response to immunosuppressant (Dex) treatment. NZB/W F1 mice were treated with 2 mg/kg body weight Dex (intraperitoneal injections five times per week) from 20 to 34 weeks of age. Fecal pellets were collected at 34 weeks of age and subjected to 16S rRNA sequencing analysis. (A) PCoA plot, showing the separation of overall bacterial structures (P < 0.01). (B) Bacterial diversity, as indicated by the Shannon index (P < 0.001). (C) Identification of a bacterial species for which there was a significant change from the pre-disease-onset stage (pre) to the post-disease-onset stage (post) and Dex treatment significantly reversed the change (P < 0.01 in both cases). Ctr, control. (D) Spearman correlation analysis between the abundance (shown as a proportion, where, e.g., 0.2 represents 20%) of the bacterial species (Lacto [Lactobacillaceae, Lactobacillus, other]) identified in panel C and two measurements of the SLE disease state, i.e., renal function (RenalFunc) and systemic autoimmunity (SysAuto). See Materials and Methods for calculation of these two measurements.
We next identified the bacterial species with significant changes from the pre-disease-onset stage to the post-disease-onset stage, and we investigated whether Dex treatment significantly reversed such changes. Only one bacterial species, i.e., “Lactobacillaceae, Lactobacillus, other,” fulfilled these criteria (Fig. 2C). The relative abundance of this species increased significantly, from 0.01% at the pre-disease-onset stage to 10% at the post-disease-onset stage, and Dex was able to significantly decrease the abundance to 1% (P < 0.01 in both cases). BLAST analysis showed that the OTU sequences within Lactobacillaceae, Lactobacillus, other, were at least 98% identical to those of Lactobacillus murinus, Lactobacillus kimchicus, and Lactobacillus senmaizukei; among the three, only Lactobacillus murinus was isolated from mice (the other two were isolated from kimchi and Japanese pickle, respectively). However, there could be unsequenced isolates that would be represented by the sequence obtained from the 16S analysis; therefore, the identity of the isolate(s) is unknown, but the isolate(s) could be one or more of the three detected species. We further studied Spearman association coefficients for this bacterial species and two measurements of SLE disease state, namely, renal function and systemic autoimmunity (Fig. 2D). The renal function was calculated as a composite score reflecting the level of proteinuria and the histopathological score, whereas systemic autoimmunity was calculated as a composite score reflecting the level of autoantibodies against double-stranded DNA (dsDNA) and the weight of spleen (see Materials and Methods for details). Correlation analysis showed that Lactobacillaceae, Lactobacillus, other, was positively associated with renal function (correlation efficient of 0.38; P = 0.094) and systemic autoimmunity (correlation efficient of 0.42; P = 0.067), although the associations were not statistically significant. These results suggest that a greater abundance of a group of lactobacilli (for which a species assignment could not be made) in the gut microbiota may be associated with more severe clinical signs in female NZB/W F1 mice. This is distinct from findings observed for female MRL/lpr mice, in which a greater abundance of Lactobacillus reuteri was found to be associated with disease attenuation (9, 10).
Gut microbiota dysbiosis in human SLE patients.
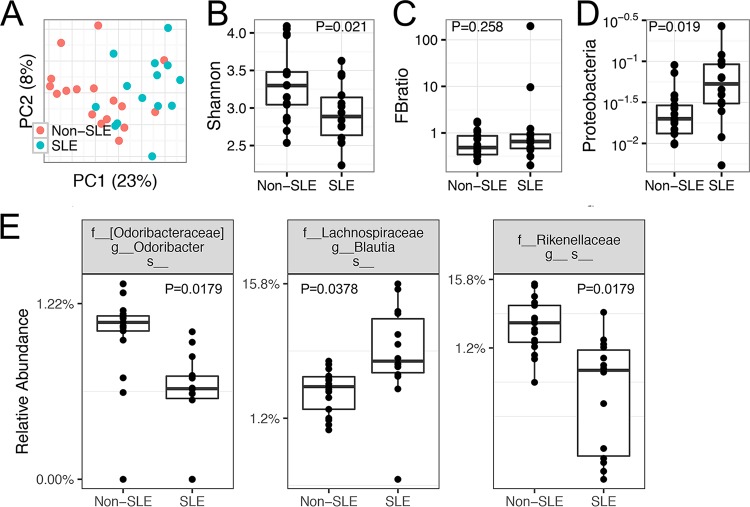
To study the differences in the gut microbiota of patients with SLE, compared to patients without immune-mediated diseases, we enrolled 14 patients with active SLE and 17 non-SLE controls. Although the overall community structures could not be separated by unweighted UniFrac-based PCoA analysis (Fig. 3A), SLE subjects had significantly lower diversity, measured with the Shannon index (P < 0.05, Mann-Whitney test) (Fig. 3B). Unlike the published comparison between healthy controls and SLE patients, in which the Firmicutes/Bacteroidetes ratio was significantly lower in SLE patients in remission (12), the ratios were not significantly different for our cohort of SLE patients versus non-SLE controls (P > 0.05) (Fig. 3C). In addition, the abundance of the bacterial phylum Proteobacteria (a representation of facultative anaerobic Gram-negative bacteria) was significantly greater in the gut microbiota of SLE patients (P < 0.05) (Fig. 3D). This is consistent with the increased serum levels of lipopolysaccharide (LPS) endotoxin in SLE patients that were observed by others (22). Three bacterial species appeared to be differentially represented in SLE patients, compared to non-SLE controls (P < 0.05, based on both nonparametric Mann-Whitney tests and DESeq2 analysis) (Fig. 3E); the species were within the genera Odoribacter and Blautia (family Lachnospiraceae) and an unnamed genus (family Rikenellaceae). BLAST analyses of the OTU sequences were performed, and the identified species are presented in Table 1. These results suggest that, compared to controls without immune-mediated diseases, SLE patients with active lupus disease possessed an altered gut microbiota that differed in several bacterial species and was less diverse, with increased representation of Gram-negative bacteria. Medication may influence the composition and diversity of the gut microbiota (Table 2) but, due to the small sample size, we did not attempt to analyze the effects of medication.
FIG 3.
Gut microbiota dysbiosis in SLE patients with active disease, compared to non-SLE controls. The inclusion and exclusion criteria are listed in Materials and Methods. A fecal sample was collected from each patient or control subject at the time of the first visit, and the samples were subjected to 16S rRNA sequencing analysis. (A) PCoA plot, showing no separation of overall bacterial structures (P > 0.05). (B) Significant difference in bacterial diversity levels, as indicated by the Shannon index (P < 0.05). (C) No significant difference in Firmicutes/Bacteroidetes (FB) ratios (P > 0.05). (D) Significant difference in the relative abundance of the phylum Proteobacteria, a representation of facultative anaerobic Gram-negative bacteria (P < 0.05). (E) Significant changes in several bacterial species, with the P values indicated (nonparametric Mann-Whitney test). DESeq2 analysis was also performed, and the DESeq2 P values were 0.037, 0.007, and 0.007, respectively. P values were corrected for multiple comparisons by controlling the FDRs.
TABLE 1.
Differentially represented bacterial species between SLE and non-SLE individuals
| Bacterial species | Strain name | OTU no. | Sequence identity (%) | SLE vs non-SLE |
|---|---|---|---|---|
| Blautia wexlerae | AUH-JLD17 | 7 | 100 | Increased |
| Blautia wexlerae | AUH-JLD56 | 7 | 100 | Increased |
| Blautia sp. strain Marseille | P3602 | 7 | 100 | Increased |
| Blautia sp. strain GD8 | 23 | 100 | Increased | |
| Blautia faecis | M25 | 25 | 100 | Increased |
| Blautia caecimuris | SJ18 | 492 | 99 | Increased |
| Blautia sp. strain AUH-JLD3 | 585 | 98 | Increased | |
| Blautia sp. strain Marseille | P3387 | 585 | 98 | Increased |
| Odoribacter laneus | JCM | 63 | 100 | Decreased |
| Odoribacter laneus | YIT | 63 | 100 | Decreased |
| Odoribacter laneus | JCM | 63 | 100 | Decreased |
| Odoribacter splanchnicus | 67 | 100 | Decreased | |
| Alistipes onderdonkii | 13 | 100 | Decreased | |
| Alistipes sp. | LS-J | 13 | 100 | Decreased |
| Alistipes sp. | LS-M | 13 | 100 | Decreased |
| Alistipes shahii | WAL 8301 | 164 | 100 | Decreased |
| Alistipes obesi | ph8 | 164 | 100 | Decreased |
| Alistipes ihumii | AP11 | 188 | 100 | Decreased |
| Alistipes sp. strain cv1 | 188/210 | 100 | Decreased | |
| Alistipes indistinctus | JCM | 188/252 | 100 | Decreased |
| Alistipes indistinctus | YIT | 188/252 | 100 | Decreased |
| Alistipes sp. strain S216 | 252 | 100 | Decreased | |
| Alistipes finegoldii | DSM 17242 | 398 | 99 | Decreased |
| Alistipes finegoldii | JCM 16770 | 398 | 99 | Decreased |
| Alistipes finegoldii | CIP 107999 | 398 | 99 | Decreased |
| Alistipes finegoldii | AHN 2437 | 398 | 99 | Decreased |
TABLE 2.
Demographic, immunological, and clinical features of SLE patients
| Subject no. | Age (yr) | Sexa | Raceb | Immunological featuresc | Clinical symptomsd | SLEDAI score | BMI (kg/m2)e | Medication(s)f |
|---|---|---|---|---|---|---|---|---|
| 01 | 39 | F | AA | ANA, dsDNA, C3, C4, hem | CL, PS | 0 | 36.6 | HCQ, belimumab |
| 02 | 21 | F | Caucasian | ANA, dsDNA, sm, C3, C4, hem | Alopecia, LN, PS, Neuro | 8 | 36.4 | HCQ, MMF, belimumab |
| 03 | 40 | F | Caucasian | ANA, C3, C4, hem | Alopecia, SE | 3 | 21.5 | HCQ, MMF, belimumab |
| 04 | 64 | F | AA | ANA, C3, C4 | Alopecia, MR, PS | 6 | 31.4 | HCQ, MMF, belimumab |
| 05 | 65 | F | Caucasian | ANA, dsDNA, C3, C4 | LN | 13 | 26.2 | HCQ, MMF |
| 06 | 41 | M | Caucasian | ANA, dsDNA, C3 | LN, PS, IA | 2 | 43.5 | MTX |
| 07 | 27 | M | AA | ANA, dsDNA, sm, ACL, C3, C4 | MR, LN | 6 | 31.6 | HCQ, MMF |
| 09 | 56 | F | Caucasian | ANA, dsDNA, hem | PS | 1 | 36.5 | HCQ, MMF, belimumab |
| 10 | 25 | F | Caucasian | ANA, dsDNA, ACL, B2G, C3, C4 | MR, LN, PS | 8 | 39.1 | HCQ, MMF, rituximab |
| 11 | 73 | F | Caucasian | ANA, sm | SE | 0 | 18.4 | HCQ, MMF |
| 17 | 23 | M | Caucasian | ANA, dsDNA, hem | LN | 2 | 21.0 | HCQ, AZA |
| 19 | 36 | M | Caucasian | ANA, dsDNA, sm, C4, hem | MR, SE | 0 | 37.2 | HCQ, MMF |
| 20 | 29 | F | AA | ANA, dsDNA, sm, C3, C4 | MR, IA | 2 | 33.9 | HCQ |
| 23 | 66 | F | Caucasian | ANA, dsDNA, hem | None | 0 | 33.2 | HCQ, AZA, tacrolimus |
F, female; M, male.
Race categories were African American (not Caribbean) (AA) and Caucasian, non-Hispanic.
Immunological features included antinuclear antibodies (ANA), anti-dsDNA antibodies (dsDNA), anti-Smith antibodies (sm), anticardiolipin antibodies (ACL), β2-glycoprotein (B2G), lupus anticoagulant (LAC), complement C3 (C3), complement C4 (C4), and hematological manifestation (hem).
Clinical symptoms included alopecia, cutaneous lupus (CL), malar rash (MR), lupus nephritis (LN), photosensitivity (PS), inflammatory arthritis (IA), serositis (SE), and neurolupus (Neuro).
BMI, body mass index.
Medications included hydroxychloroquine (HCQ), mycophenolate mofetil (MMF), methotrexate (MTX), azathioprine (AZA), belimumab, rituximab, and tacrolimus.
DISCUSSION
In our studies, we analyzed the gut microbiota of NZB/W F1 mice and SLE patients with active disease by using 16S rRNA sequencing. In NZB/W F1 mice, significant differences in the gut microbiota were observed between predisease and diseased mice and between untreated mice and mice treated with the immunosuppressive drug Dex. The microbiota tended to be more diverse as the disease progressed and after Dex treatment. In addition, greater relative abundance of a group of lactobacilli (for which a species assignment could not be made) in the gut microbiota may be associated with progressing disease in NZB/W F1 mice. Fewer differences were found in the comparison between non-SLE subjects and SLE patients with active disease. The fecal microbiota was less diverse in SLE patients, whereas the Firmicutes/Bacteroidetes ratios were not different between the two groups. Interestingly, the relative abundance of Proteobacteria, representing LPS-containing Gram-negative bacteria, was significantly increased in the SLE microbiota.
We found that human SLE patients had an altered microbiome in active disease, compared to non-SLE controls. The studies expand those of Hevia et al. (12), who reported that SLE patients with inactive lupus disease had significantly lower Firmicutes/Bacteroidetes ratios than did healthy control individuals. However, we did not observe a similar change of the ratios in our cohort. This is possibly due to differences in the study designs, i.e., (i) there may be differences in the gut microbiotas of patients with active versus inactive disease; (ii) we included both female and male SLE patients of different ethnicities and with various levels of lupus disease manifestations, whereas Hevia et al. (12) included only female Caucasian patients with SLE disease activity index (SLEDAI) scores of <8; and (iii) we chose to include SLE patients in need of immunosuppressive therapies, whereas Hevia et al. excluded those who had received steroid or immunological treatments. Our inclusion/exclusion criteria selected subjects who may better represent the SLE patient population, because SLE disproportionately affects African Americans and does occur in men as well as women.
The use of nonselective immunosuppressive therapies, such as Dex and azathioprine, for lupus patients may have broad effects on the gut microbiota. A study by Huang et al. showed that Dex treatment could alter the gut microbiota in healthy wild-type C57BL/6 mice by increasing the prevalence of Firmicutes, Actinobacteria, and Bifidobacterium and, in an inflammatory bowel disease model, the microbiota from Dex-treated donor mice could reduce colonic inflammation through reduced interleukin 12p40 (IL-12p40) and IL-17 production, compared to control donor mice (23). This evidence suggests that, even as immunosuppressive medications are acting directly to suppress inflammation from the patient's cells, these medications may be contributing to the formation of a microbiome that promotes the regulation of inflammation locally in the gastrointestinal tract and potentially at distant sites in the body. All human fecal samples were from patients taking immunosuppressive medications. The characterization of the gut microbiotas of SLE patients who have not yet started therapy, compared to their respective microbiotas following treatment, would allow for a better understanding of how these broad-acting medications are able to modulate microbial dysbiosis and to affect the resultant autoimmune disease phenotype.
In our previous studies, we compared the fecal microbiota of MRL/lpr mice to that of MRL control mice (9). In this study, however, we did not compare the fecal microbiota of NZB/W F1 mice to that of a control mouse strain. People have used NZB or NZW mice as controls for NZB/W F1 mice, but both of these strains can develop anti-DNA autoantibodies and glomerulonephritis (according to descriptions on The Jackson Laboratory website) like NZB/W F1. C57BL/6 mice represent another possible control strain. Without a control mouse strain, our findings on the changes in the microbiota over time (Fig. 1) could be interpreted as either disease related or age related. Either way, our results are informative because they provide new information on the longitudinal changes in the gut microbiota in one additional lupus-prone mouse strain. In addition, we tried to control for the cage effect (see Fig. S1 in the supplemental material). For the 10-, 14-, and 18-week time points, we sampled the same cage of animals, depending on the age, but the microbiotas were distinct. For the 23-, 28-, and 33-week time points, while the microbiotas clustered together, these samples were collected from three different cages, with each cage representing a time point. While our intention had been to control for the cage effect, this design was not adequate. In particular, our results might have been confounded by the time of sample collection. A better study design would involve both the same cage of animals sampled at different times and different ages of animals sampled at the same time. It also would be interesting to cohouse mice of different ages and to determine whether the microbiotas were still distinct.
We attempted to identify consensus in the changes in the gut microbiota between mice (NZB/W F1, MRL/lpr, and SNF1) and our cohort of SLE patients. While it was consistent among different lupus-prone mouse models that the gut microbiotas were more diverse as the disease progressed, the microbial diversity was significantly lower in SLE patients with active disease, compared with non-SLE controls. The only consensus between mice and humans was the relative abundance of Lachnospiraceae, which was significantly greater in both MRL/lpr mice (9) and SLE patients. In humans, however, the increase was limited to a species in the genus Blautia; when the entire Lachnospiraceae family was examined, the difference was no longer evident. In future investigations, we will try to enroll a larger population of SLE patients, to divide them based on disease manifestations, and to analyze the gut microbiotas separately. In this way, consensus between mice and humans may be achieved, enabling us to continue using mice to model the human disease with respect to the roles of the microbiome in lupus.
MATERIALS AND METHODS
Mice.
Female NZB/W F1 mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and maintained in a specific-pathogen-free facility according to the requirements of the Institutional Animal Care and Use Committee (IACUC) at Virginia Tech. For the time course, fecal pellets were collected from untreated mice at 10, 14, 18, 23, 28, and 33 weeks of age. For the 10-, 14-, and 18-week time points, the same cage of mice (n = 5) was sampled according to age. For the 23-, 28-, and 33-week time points, three different cages of mice (each representing a time point; n = 3 or 4 per cage) were sampled. For the experiment involving Dex, age-matched mice treated with either Dex or vehicle control were euthanized at 34 weeks of age. Hydroxypropyl methylcellulose (HPMC) (Sigma, St. Louis, MO) was diluted in sterile deionized water at a concentration of 0.05%, autoclaved, and then used as the vehicle for the drug solutions. Ten mice (n = 5 per cage per group) were included in each of the two groups, treated with vehicle control (HPMC) or 2 mg/kg Dex. Intraperitoneal injections were performed 5 times per week, with a 50-μl volume of the respective treatments beginning at 20 weeks of age, and treatments were continued until euthanasia. Proteinuria (measured with Chemstrips) and body weight were measured biweekly before treatment and then weekly after treatment began.
Evaluation of SLE-like disease in mice.
Renal function was assessed and calculated as described previously (9). The serum levels of anti-dsDNA autoantibodies were measured as described previously (24). Systemic autoimmunity was calculated as follows: (serum level of anti-dsDNA + [500 × spleen/body weight ratio])/2, where the coefficient 500 equalized the averages of the two variables.
Human subjects.
The patients were enrolled at the lupus clinic in a single center in southwest Roanoke, Virginia. All patients met the 2009 Systemic Lupus International Collaborating Clinic (SLICC) revised American College of Rheumatology (ACR) diagnostic criteria for SLE. At the time of enrollment, the medication list was collected and a SLEDAI score was determined. The inclusion criteria were as follows: age of 18 to 75 years, fulfilling the SLICC revised ACR criteria for SLE (2009), and receiving or eligible for immunosuppressive therapy. The exclusion criteria were as follows: age of <18 years or >75 years, not meeting the SLICC revised ACR criteria for SLE, disease not requiring immunosuppressive therapy, pregnancy, the presence of other immune diseases (such as inflammatory bowel disease), and current or recent use of antibiotics; vulnerable patients, including prisoners, non-English speakers, and critically ill patients, also were excluded.
Microbiota sampling, 16S rRNA analysis, and metagenome prediction.
Mouse fecal microbiotas were obtained by taking individual mice out of their cages and collecting a fecal pellet. Colonic microbiota samples were collected within 20 min after euthanasia. To avoid cross-contamination, each microbiota sample was collected by removing the colon and extruding the fecal material into a sterile 1.5-ml Eppendorf tube by using a new pair of sterile forceps. Human fecal samples were collected from patients at Carilion Medical Center (Roanoke, VA) between 2015 and 2016. For stool samples, patients were provided with a collection “pot” and requested to bring in a specimen collected the day before a routine visit. All procedures were approved by the institutional review board (IRB) of Virginia Tech Carilion School of Medicine. All microbiota samples were stored at −80°C until they were processed at the same time. Sample homogenization, cell lysis, DNA extraction, and PCR of the 16S V4 region were performed with the same methods as described in a previous report (9). Purified amplicons were sequenced bidirectionally (paired ends of 150 bp) on an Illumina MiSeq system at Argonne National Laboratory. Sequence read merging, quality filtering, dereplication, chimera removal, and OTU clustering were performed with the UPARSE pipeline implemented in the USEARCH program (version 8.1/1831) (25). Bacterial taxonomy was assigned by using the UclustConsensusTaxonAssigner function implemented in QIIME (26), with the Greengenes reference database (27, 28), and results were summarized at all taxonomic levels (29). Alpha and beta diversity metrics were computed with QIIME. Alpha diversity included the Shannon diversity index and observed OTUs, and beta diversity included unweighted UniFrac distance metrics (30, 31).
Statistics.
Two-sample comparisons were performed with Student's t test or a nonparametric Mann-Whitney rank-based test. For >2 groups, one-way analysis of variance (ANOVA) or the nonparametric Kruskal-Wallis test was performed. Because the Kruskal-Wallis test assumes identical distributions in each group, we also used the oneway.test function in R to compare means without assuming equal variances. Correlation analysis was performed using Spearman rank-based methods. PCoA results were tested for significance by a method described previously (32). When studying age as a fixed effect and mouse subject as a random effect, we used the linear mixed model implemented in the NLME package in R. P values were corrected for multiple comparisons by controlling the false discovery rates (FDRs). All statistical computations were performed in R (version 3.3.1).
Accession number(s).
Sequencing data have been deposited in the NCBI Sequence Read Archive (accession no. SRP092445), and the OTU results for all experiments can be found in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sarah Henderson for coordinating the collection of patient samples, Sarah Owens and Saikumar Karyala for assistance in Illumina MiSeq sequencing, Husen Zhang for microbiome analysis, and Yingxing Wu for assistance in statistical analysis.
We declare no conflicts of interest (as defined by American Society for Microbiology policies).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02288-17.
REFERENCES
- 1.Tsokos GC. 2011. Systemic lupus erythematosus. N Engl J Med 365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Ohl K, Tenbrock K. 2011. Inflammatory cytokines in systemic lupus erythematosus. J Biomed Biotechnol 2011:432595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lech M, Anders HJ. 2013. The pathogenesis of lupus nephritis. J Am Soc Nephrol 24:1357–1366. doi: 10.1681/ASN.2013010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tektonidou MG, Wang Z, Dasgupta A, Ward MM. 2015. Burden of serious infections in adults with systemic lupus erythematosus: a national population-based study, 1996–2011. Arthritis Care Res (Hoboken) 67:1078–1085. doi: 10.1002/acr.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symmons DP. 1995. Frequency of lupus in people of African origin. Lupus 4:176–178. doi: 10.1177/096120339500400303. [DOI] [PubMed] [Google Scholar]
- 6.Lawson TM, Amos N, Bulgen D, Williams BD. 2001. Minocycline-induced lupus: clinical features and response to rechallenge. Rheumatology (Oxford) 40:329–335. doi: 10.1093/rheumatology/40.3.329. [DOI] [PubMed] [Google Scholar]
- 7.Margolis DJ, Hoffstad O, Bilker W. 2007. Association or lack of association between tetracycline class antibiotics used for acne vulgaris and lupus erythematosus. Br J Dermatol 157:540–546. doi: 10.1111/j.1365-2133.2007.08056.x. [DOI] [PubMed] [Google Scholar]
- 8.Pope J, Jerome D, Fenlon D, Krizova A, Ouimet J. 2003. Frequency of adverse drug reactions in patients with systemic lupus erythematosus. J Rheumatol 30:480–484. [PubMed] [Google Scholar]
- 9.Zhang H, Liao X, Sparks JB, Luo XM. 2014. Dynamics of gut microbiota in autoimmune lupus. Appl Environ Microbiol 80:7551–7560. doi: 10.1128/AEM.02676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, Ahmed SA, Yuan R, Li L, Cecere TE, Branson DB, Kirby JL, Goswami P, Leeth CM, Read KA, Oestreich KJ, Vieson MD, Reilly CM, Luo XM. 2017. Control of lupus nephritis by changes of gut microbiota. Microbiome 5:73. doi: 10.1186/s40168-017-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson BM, Gaudreau MC, Al-Gadban MM, Gudi R, Vasu C. 2015. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin Exp Immunol 181:323–337. doi: 10.1111/cei.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hevia A, Milani C, Lopez P, Cuervo A, Arboleya S, Duranti S, Turroni F, Gonzalez S, Suarez A, Gueimonde M, Ventura M, Sanchez B, Margolles A. 2014. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 5:e01548-14. doi: 10.1128/mBio.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, Queipo-Ortuno MI. 2013. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med 11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Man SM, Kaakoush NO, Mitchell HM. 2011. The role of bacteria and pattern-recognition receptors in Crohn's disease. Nat Rev Gastroenterol Hepatol 8:152–168. doi: 10.1038/nrgastro.2011.3. [DOI] [PubMed] [Google Scholar]
- 15.Lopez P, de Paz B, Rodriguez-Carrio J, Hevia A, Sanchez B, Margolles A, Suarez A. 2016. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep 6:24072. doi: 10.1038/srep24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen XQ, Yu YC, Deng HH, Sun JZ, Dai Z, Wu YW, Yang M. 2010. Plasma IL-17A is increased in new-onset SLE patients and associated with disease activity. J Clin Immunol 30:221–225. doi: 10.1007/s10875-009-9365-x. [DOI] [PubMed] [Google Scholar]
- 17.Amarilyo G, Lourenco EV, Shi FD, La Cava A. 2014. IL-17 promotes murine lupus. J Immunol 193:540–543. doi: 10.4049/jimmunol.1400931. [DOI] [PubMed] [Google Scholar]
- 18.Crispin JC, Oukka M, Bayliss G, Cohen RA, Van Beek CA, Stillman IE, Kyttaris VC, Juang YT, Tsokos GC. 2008. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J Immunol 181:8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doreau A, Belot A, Bastid J, Riche B, Trescol-Biemont MC, Ranchin B, Fabien N, Cochat P, Pouteil-Noble C, Trolliet P, Durieu I, Tebib J, Kassai B, Ansieau S, Puisieux A, Eliaou JF, Bonnefoy-Berard N. 2009. Interleukin 17 acts in synergy with B cell-activating factor to influence B cell biology and the pathophysiology of systemic lupus erythematosus. Nat Immunol 10:778–785. doi: 10.1038/ni.1741. [DOI] [PubMed] [Google Scholar]
- 20.Crispin JC, Tsokos GC. 2010. Interleukin-17-producing T cells in lupus. Curr Opin Rheumatol 22:499–503. doi: 10.1097/BOR.0b013e32833c62b0. [DOI] [PubMed] [Google Scholar]
- 21.Macanovic M, Sinicropi D, Shak S, Baughman S, Thiru S, Lachmann PJ. 1996. The treatment of systemic lupus erythematosus (SLE) in NZB/W F1 hybrid mice: studies with recombinant murine DNase and with dexamethasone. Clin Exp Immunol 106:243–252. doi: 10.1046/j.1365-2249.1996.d01-839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi L, Zhang Z, Yu AM, Wang W, Wei Z, Akhter E, Maurer K, Costa Reis P, Song L, Petri M, Sullivan KE. 2014. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS One 9:e93846. doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang EY, Inoue T, Leone VA, Dalal S, Touw K, Wang Y, Musch MW, Theriault B, Higuchi K, Donovan S, Gilbert J, Chang EB. 2015. Using corticosteroids to reshape the gut microbiome: implications for inflammatory bowel diseases. Inflamm Bowel Dis 21:963–972. doi: 10.1097/MIB.0000000000000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao X, Ren J, Wei CH, Ross AC, Cecere TE, Jortner BS, Ahmed SA, Luo XM. 2015. Paradoxical effects of all-trans-retinoic acid on lupus-like disease in the MRL/lpr mouse model. PLoS One 10:e0118176. doi: 10.1371/journal.pone.0118176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.