ABSTRACT
Endoxylanases are important enzymes in bioenergy research because they specifically hydrolyze xylan, the predominant polysaccharide in the hemicellulose fraction of lignocellulosic biomass. For effective biomass utilization, it is important to understand the mechanism of substrate recognition by these enzymes. Recent studies have shown that the substrate specificities of bacterial and fungal endoxylanases classified into glycoside hydrolase family 30 (GH30) were quite different. While the functional differences have been described, the mechanism of substrate recognition is still unknown. Therefore, a gene encoding a putative GH30 endoxylanase was cloned from Streptomyces turgidiscabies C56, and the recombinant enzyme was purified and characterized. GH30 glucuronoxylan-specific xylanase A of Streptomyces turgidiscabies (StXyn30A) showed hydrolytic activity with xylans containing both glucuronic acid and the more common 4-O-methyl-glucuronic acid side-chain substitutions but not on linear xylooligosaccharides, suggesting that this enzyme requires the recognition of glucuronic acid side chains for hydrolysis. The StXyn30A limit product structure was analyzed following a secondary β-xylosidase treatment by thin-layer chromatography and mass spectrometry analysis. The hydrolysis products from both glucuronoxylan and 4-O-methylglucuronoxylan by StXyn30A have these main-chain substitutions on the second xylopyranosyl residue from the reducing end. Because previous structural studies of bacterial GH30 enzymes and molecular modeling of StXyn30A suggested that a conserved arginine residue (Arg296) interacts with the glucuronic acid side-chain carboxyl group, we focused on this residue, which is conserved at subsite −2 of bacterial but not fungal GH30 endoxylanases. To help gain an understanding of the mechanism of how StXyn30A recognizes glucuronic acid substitutions, Arg296 mutant enzymes were studied. The glucuronoxylan hydrolytic activities of Arg296 mutants were significantly reduced in comparison to those of the wild-type enzyme. Furthermore, limit products other than aldotriouronic acid were observed for these Arg296 mutants upon secondary β-xylosidase treatment. These results indicate that a disruption of the highly conserved Arg296 interaction leads to a decrease of functional specificity in StXyn30A, as indicated by the detection of alternative hydrolysis products. Our studies allow a better understanding of the mechanism of glucuronoxylan recognition and enzyme specificity by bacterial GH30 endoxylanases and provide further definition of these unique enzymes for their potential application in industry.
IMPORTANCE Hemicellulases are important enzymes that hydrolyze hemicellulosic polysaccharides to smaller sugars for eventual microbial assimilation and metabolism. These hemicellulases include endoxylanases that cleave the β-1,4-xylose main chain of xylan, the predominant form of hemicellulose in lignocellulosic biomass. Endoxylanases play an important role in the utilization of plant biomass because in addition to their general utility in xylan degradation, they can also be used to create defined compositions of xylooligosaccharides. For this, it is important to understand the mechanism of substrate recognition. Recent studies have shown that the substrate specificities of bacterial and fungal endoxylanases that are classified into glycoside hydrolase family 30 (GH30) were distinct, but the difference in the mechanisms of substrate recognition is still unknown. We performed characterization and mutagenesis analyses of a new bacterial GH30 endoxylanase for comparison with previously reported fungal GH30 endoxylanases. Our study results in a better understanding of the mechanism of substrate specificity and recognition for bacterial GH30 endoxylanases. The experimental approach and resulting data support the conclusions and provide further definition of the structure and function of GH30 endoxylanases for their application in bioenergy research.
KEYWORDS: glycoside hydrolase family 30, glucuronoxylan, Streptomyces turgidiscabies, xylanase
INTRODUCTION
Lignocellulosic biomass is a nonfood renewable resource currently receiving increased attention for efficient bioconversion to value-added chemicals and fuels. The second most abundant polysaccharide in lignocellulosic biomass is hemicellulose, which constitutes between 20 and 30% (wt/wt) of the total mass. Due to its chemical complexity and interactions within the woody lignocellulose composite, efficient utilization of hemicellulose is not highly developed. Hemicellulose is a heteropolysaccharide, and its carbohydrate composition varies depending on the plant origin and stage of tissue development (1, 2). In a typical bioconversion process, hemicellulose is chemically degraded to monosaccharides by sulfuric acid or water at high temperatures to extract it from the biomass. Fermentation inhibitors produced under these severe hydrolysis conditions make it difficult to utilize the resulting hemicellulose hydrolysate (3, 4, 5). For more efficient utilization, hemicellulases, the enzymes that selectively degrade hemicellulose, may be developed to target the production of specific sugar products from hemicellulose. These enzymes have great potential to increase the efficiency of biomass utilization because it is possible to regulate the structure and size of products through an understanding of the substrate specificity of the endoxylanases employed.
Collectively, hemicellulose represents several types of noncellulose, biomass-derived, polymeric sugars. Of these polysaccharides, xylan is the most abundant form of hemicellulose found in all land plants and represents the second largest biomass resource next to cellulose. Xylans consist of a backbone of β-1,4-linked xylopyranose units, which may be substituted with acetyl (Ac), arabinofuranosyl, and glucuronosyl side chains (1, 2). Xylans substituted with 4-O-methyl α-1,2-glucuronic acid (GlcA) groups are known as glucuronoxylans and are the primary hemicellulose type found in hardwoods and crop residues. Substitution types are specific for the plant, and substitution characteristics depend on plant age and the tissue source. Endoxylanases (EC 3.2.1.8), which randomly hydrolyze the β-1,4-xylan backbone, are primarily classified according to the CAZy (Carbohydrate-Active Enzymes) database (http://www.cazy.org/) (6, 7) as glycoside hydrolase family 10 (GH10) or GH11 endoxylanases. Recently, new endoxylanases specific for glucuronoxylan have been reported (8). These glucuronoxylan xylanohydrolases are classified into the GH30 family, recognize GlcA side chains of glucuronoxylan, and hydrolyze the glycosidic β-1,4 linkages of the xylan backbone in an endo-specific manner, as defined by the GlcA position. The bacterial GH30 endoxylanases XynC from Bacillus subtilis (9), XynA from Erwinia chrysanthemi (10), Xyn5B from Bacillus sp. strain BP-7 (11), Xyn30D from Paenibacillus barcinonensis (12), and Xyn30A from Clostridium thermocellum (CtXyn30A) (13, 14) have been characterized. Interpretation of crystal structure data from XynC and XynA indicates that the interaction with a GlcA side chain at the −2 subsite of the catalytic cleft is the source of specific molecular contacts, which discriminate the structure of glucuronoxylan (15, 16). These bacterial, glucuronoxylan-specific GH30 endoxylanases belong to subfamily 8 of GH30 (GH30-8) (17). In contrast, GH30 endoxylanases from subfamily 7 (GH30-7) are derived primarily from fungi and display a variety of β-1,4-xylanase-related functional properties. XYN IV from Trichoderma reesei is a GH30-7 endoxylanase that shows both endo- and exoxylanase activities, not having any detected specificity for GlcA appendages as observed for bacterial GH30-8 endoxylanases. This unique xylanase can hydrolyze glucuronoxylan, arabinoxylan, and linear xylooligosaccharide but has its highest observed rate of hydrolysis on rhodymenan an algal β-1,3-β-1,4-xylan. Interestingly, the dual exo/endoxylanase functionality results in the ultimate accumulation of xylose (X1) (18). T. reesei is also the source of a second novel GH30-7 endoxylanase (19). XYN VI, recently characterized by Biely et al. (19), preferentially degrades glucuronoxylan with a dependence upon the GlcA appendage. For this GlcA-dependent endoxylanase, the specificity for glucuronoxylan was shown to be nearly identical to that of the bacterial GH30-8 glucuronoxylanases. However, the substrate specificity of XYN VI is somewhat different from that of bacterial GH30-8 endoxylanases, as it displayed a low level of activity for hydrolyzing linear neutral xylooligosaccharides and arabinoxylan. Both of the unique T. reesei xylanases belong to subfamily 7 of GH30 (17). The difference in the mechanisms of substrate recognition between bacterial GH30 subfamily 8 and fungal GH30 subfamily 7 remains unknown.
In this study, to help understand the mechanism by which bacterial GH30-8 glucuronoxylanases recognize GlcA substitutions for endohydrolysis of the xylan chain and to establish the difference in substrate specificities between bacterial GH30-8 and fungal GH30-7 endoxylanases, we cloned a GH30-8 endoxylanase from Streptomyces turgidiscabies C56 (StXyn30A) and characterized the properties of the enzyme in the hydrolysis of 4-O-methylglucuronoxylan (MeGXn), a 4-O-methylglucuronoacetylxylan, and cotton-derived glucuronoxylan, which lacks the common 4-O-methyl modification on the GlcA substitution. Mutations of Arg296 were utilized to establish the importance of this amino acid as a portion of the overall GlcA binding motif of GH30-8 endoxylanases. The functional specificities of StXyn30A and the Arg296 mutant forms on diverse xylan substrates were characterized by utilizing thin-layer chromatography (TLC) and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS). A secondary enzyme treatment was utilized to simultaneously simplify and confirm our findings. The results show that a disruption of the dual-salt bridge/hydrogen bond interaction predicted to occur in bacterial GH30-8 endoxylanases involving the conserved arginine residue equivalent to Arg296 in StXyn30A results in decreased activity and specificity, underscoring the importance of this specific interaction for GH30-8 functional specificity.
RESULTS
Cloning, expression, and purification of the GH30 glucuronoxylan-specific xylanase from S. turgidiscabies C56.
The gene Stxyn30A from S. turgidiscabies C56 is 1,254 bp and encodes 418 amino acids. Nascent StXyn30A consists of a 21-amino-acid signal sequence at the N terminus and a 397-amino-acid catalytic domain belonging to GH30 subfamily 8. Comparison of amino acid sequences of StXyn30A with those of previously studied xylanases such as XynC from B. subtilis (9) and XynA from E. chrysanthemi (10) showed that StXyn30A is similar to XynC and XynA in overall length and had amino acid sequence identities of 61% and 36% with these GH30-8 endoxylanase, respectively (Fig. 1). The acid/base catalyst and catalytic nucleophile glutamates, which are conserved in GH30 enzymes, are also conserved in StXy30A (E165 and E254, respectively), and amino acid side chains involved in GlcA coordination by GH30-8 endoxylanases were also well conserved in StXyn30A (Fig. 1).
FIG 1.
Comparison of amino acid sequences of GH30 xylanases from S. turgidiscabies C56, E. chrysanthemi, B. subtilis, T. reesei XYN VI, T. reesei XYN IV, Bispora sp. strain MEY-1, and Leptosphaeria maculans. Amino acid sequences were aligned by using ClustalW software (37). Identical amino acids are shown in black and gray boxes. ●, catalytic residue; *, conserved arginine in bacterial GH30 enzymes. Amino acid residues located at cleft region conserved in bacterial GH30 enzymes are boxed.
The N-terminal signal sequence was removed by PCR amplification of the target StXyn30A sequence and, following sequence verification, was cloned into pET30, fusing the C-terminal reading frame to include the plasmid-encoded His tag and stop codon. This construct was expressed in isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Escherichia coli Tuner(DE3) cells. The recombinant enzyme was purified by affinity chromatography using the His tag encoded at the C terminus of the protein expression product. The purified protein showed a single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, with the expected molecular mass of 42 kDa (Fig. 2).
FIG 2.
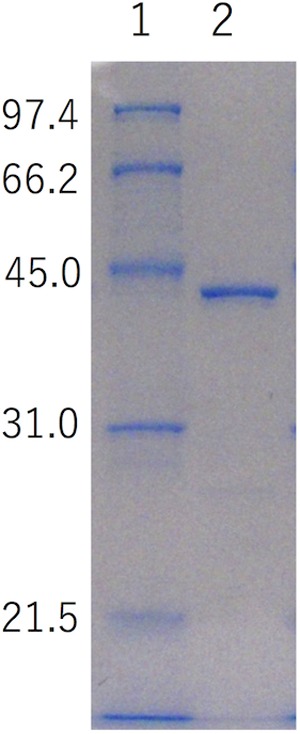
SDS-PAGE analysis of recombinant StXyn30A and enzymatic properties of StXyn30A. The molecular mass for each band in the standard is shown to the left (in kDa). Lane 1, molecular mass marker (1 μg each band); lane 2, purified StXyn30A (1 μg). SDS-PAGE was carried out with a 12% polyacrylamide gel according to the method of Laemmli (29).
The effects of pH and temperature on StXyn30A activity and stability were determined by using soluble birchwood xylan as the substrate. Maximal enzyme activity was detected at pH 6.5 and at 55°C for a 10-min reaction time. StXyn30A was stable between pH 4.0 and 10.0 at 55°C for 1 h and was also stable at temperatures of up to 40°C during 1 h of incubation at pH 6.5 (data not shown).
Substrate specificity and analysis of hydrolysis products.
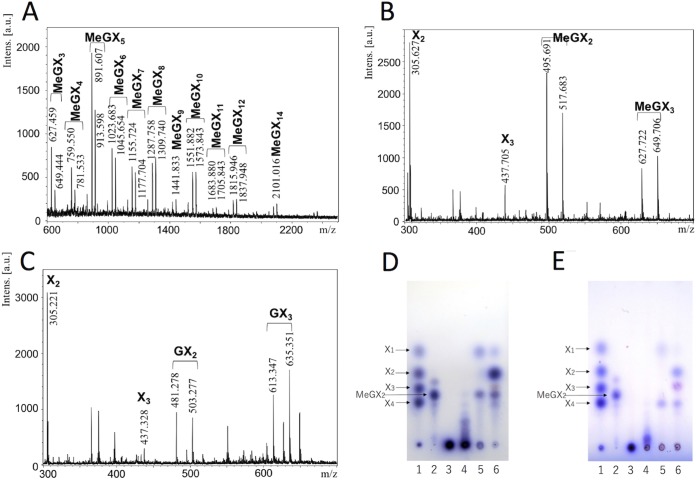
The activities of StXyn30A on various polysaccharides and oligosaccharides were examined. StXyn30A showed hydrolytic activity on beechwood, birchwood, oat spelt, and cotton seed xylans, which have 4-O-methyl-glucuronic acid or glucuronic acid side chains, but showed no activity on wheat arabinoxylan and β-1,3-xylan (Table 1). No hydrolysis products were detected when StXyn30A was incubated with xylooligosaccharides (degree of polymerization [DP], 2 to 6) (data not shown). The hydrolysis products of StXyn30A from birchwood 4-O-methylglucuronoxylan were analyzed by MALDI-TOF MS (Fig. 3A). A series of oligosaccharides forming ion clusters was observed. Each cluster is formed from sodium adduct ions, [M + Na]+ and [M + 2Na − H]+. The difference of m/z values between clusters was 132, which indicated the presence of xylooligosaccharides consisting of 3 to 15 xylopyranosyl residues containing a single 4-O-methyl-glucuronic acid (4-O-methyl glucuronoxylan; MeGX3~MeGX15) in each residue. While MALDI-TOF MS is not quantitative, it is interesting that the most intense peak is that of the aldouronate MeGX5, indicating characteristics of the GlcA substitution pattern in birchwood glucuronoxylan.
TABLE 1.
Substrate specificity toward polysaccharidesa
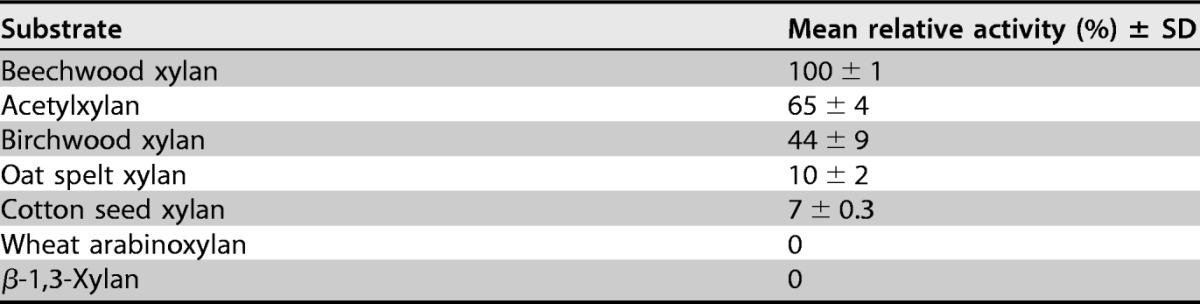
The reactions were performed with 50 mM acetate buffer (pH 6.0) containing 0.5% (wt/vol) substrate, 0.1% (wt/vol) BSA, and 1 μM enzyme at 37°C for 16 h. The reaction was stopped by heating the solutions at 100°C for 20 min. The hydrolytic activity was determined by the amounts of reducing sugars by the Somogyi-Nelson method (31). The assay was performed by using samples in triplicate.
FIG 3.
Analysis of the xylan hydrolysate of StXyn30A and SoXyn10A or β-xylosidase. (A to C) MALDI-TOF MS analysis of the xylan hydrolysate following a 2-day digestion with StXyn30A or with StXyn30A and SoXyn10A. (A) Soluble birchwood xylan hydrolysate with StXyn30A. (B) Soluble birchwood xylan hydrolysate with StXyn30A and SoXyn10A. (C) Cotton seed xylan hydrolysate with StXyn30A and SoXyn10A. Intens., intensity; a.u., arbitrary units. (D and E) Thin-layer chromatography analysis of the xylan hydrolysate following a 2-day digestion of StXyn30A with β-xylosidase or SoXyn10A. (D) Hydrolysis of 4-O-methylglucuronoxylan (birchwood xylan); (E) hydrolysis of glucuronoxylan (cotton seed xylan). Lane 1, xylose standard of xylose (X1), xylobiose (X2), xylotriose (X3), and xylotetraose (X4); lane 2, aldotriouronic acid (MeGX2); lane 3, birchwood xylan; lane 4, hydrolysis product with StXyn30A; lane 5, hydrolysis product with StXyn30A and β-xylosidase; lane 6, hydrolysis product with StXyn30A and SoXyn10A. StXyn30A was mixed with 0.5% (wt/vol) soluble birchwood xylan, acetyl xylan, or 2.5% (wt/vol) cotton seed xylan in 50 mM acetate buffer (pH 6.0) containing 0.1% (wt/vol) BSA. After incubation at 37°C for 20 h, GH3 β-xylosidase or SoXyn10A (20) was added to the reaction mixture, and the mixture was incubated at 37°C for 2 days.
Next, in order to identify the location of GlcA substitutions on the generated aldouronates, the StXyn30A hydrolysate was treated with a β-xylosidase of GH3 or an endoxylanase of GH10, both of which have known functions, and these hydrolysates were subsequently analyzed by TLC and MALDI-TOF MS. The β-xylosidase is known to hydrolyze the individual xylose residues from the nonreducing terminus up until a substituted xylose, and the GH10 endoxylanase is known to accommodate a GlcA substitution linked to a xylose positioned in its −3 and +1 subsites. The products of birchwood 4-O-methylglucuronoxylan hydrolysis by StXyn30A and β-xylosidase were xylose and MeGX2 (Fig. 3D, lane 5, and see Fig. 7BI). MeGX2 was confirmed by MALDI-TOF MS analysis. This result indicated that the birchwood xylan hydrolysate of StXyn30A having an aldouronate mixture of MeGX3∼MeGX15 as detected by MALDI-TOF MS has a GlcA substitution at the second xylopyranosyl residue from the reducing terminus. Similarly, two hydrolysis products, xylose and glucuronosyl-xylobiose (GX2), were detected when the hydrolysate of cotton seed xylan was hydrolyzed by the β-xylosidase (Fig. 3E, lane 5). These results indicate that glucuronic acid substitutions in cotton seed xylan were not methylated and that the 4-O-methyl group of glucuronic acid does not affect the function of StXyn30A.
FIG 7.
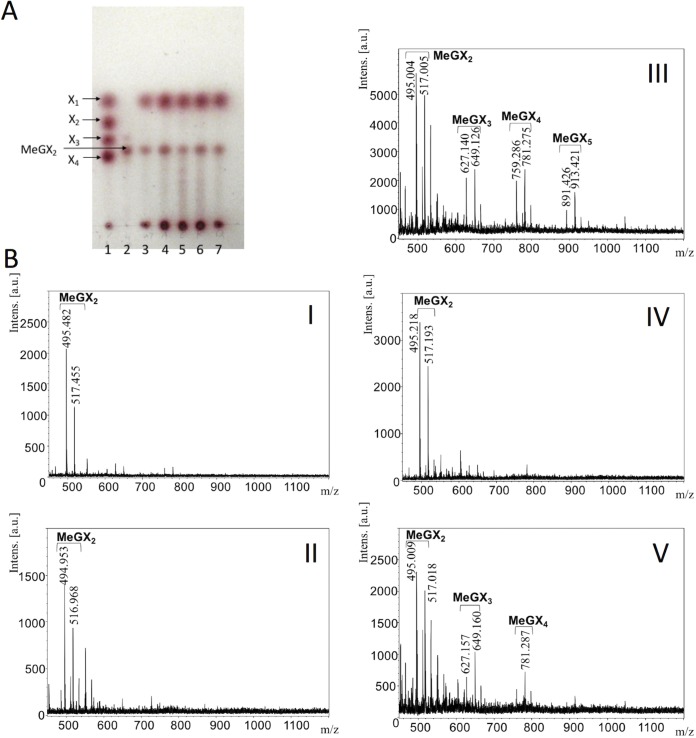
Analysis of birchwood xylan hydrolysates from StXyn30A mutants and β-xylosidase. (A) Thin-layer chromatography analysis. Sugars were developed on a silica gel 60 TLC plate with an n-butanol–acetic acid–water (2:1:1 [vol/vol/vol]) solvent and detected by using N-(1-naphthyl)ethylenediamine dihydrochloride (34, 35). Lane 1, xylose standard of xylose (X1), xylobiose (X2), xylotriose (X3), and xylotetraose (X4); lane 2, aldotriuronic acid (MeGX2); lane 3, hydrolysis products of StXyn30A and β-xylosidase; lane 4, hydrolysis products of the R296A mutant and β-xylosidase; lane 5, hydrolysis products of the R296E mutant and β-xylosidase; lane 6, hydrolysis products of the R296K mutant and β-xylosidase; lane 7, hydrolysis products of the R296Q mutant and β-xylosidase. (B) MALDI-TOF MS analysis. Shown are the hydrolysis products of StXyn30A and β-xylosidase (I), the R296A mutant and β-xylosidase (II), the R296E mutant and β-xylosidase (III), the R296K mutant and β-xylosidase (IV), and the R296Q mutant and β-xylosidase (V).
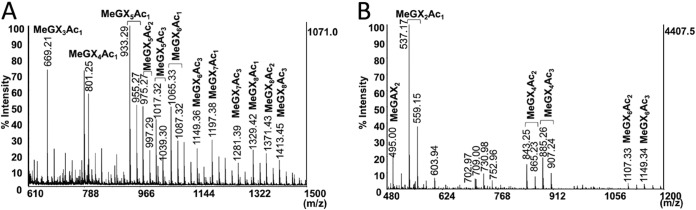
Next, the StXyn30A hydrolysate was hydrolyzed by a Streptomyces olivaceoviridis GH10 xylanase (SoXyn10A). The GH10 endoxylanase is known to accommodate a GlcA substitution linked to a xylose positioned in its −3 and +1 subsites (20). When xylans were digested by StXyn30A followed by SoXyn10A, the products X1, xylobiose (X2), xylotriose (X3), MeGX2, and MeGX3 (Fig. 3B and D, lane 6) and the products X1, X2, X3, GX2, and GX3 (Fig. 3C and E, lane 6) were detected from birchwood xylan and cotton seed xylan, respectively. Additionally, we determined whether StXyn30A was able to hydrolyze acetylated MeGXn (MeGXxAcy, where the subscript x and y represent the relevant X and Ac numbers) (Fig. 4A and B). As the results show, StXyn30A is able to hydrolyze acetylated MeGXn, and mass peaks corresponding to MeGX3Ac1∼MeGX8Ac1, MeGX5Ac2, MeGX8Ac2, and MeGX5Ac3∼MeGX8Ac3 were observed at m/z 669 (MeGX3Ac1-Na adduct), m/z 801 (MeGX4Ac1-Na adduct), m/z 933 (MeGX5Ac1-Na adduct), m/z 975 (MeGX5Ac2-Na adduct), m/z 1,017 (MeGX5Ac3-Na adduct), m/z 1,065 (MeGX6Ac1-Na adduct), m/z 1,149 (MeGX6Ac3-Na adduct), m/z 1,197 (MeGX7Ac1-Na adduct), m/z 1,239 (MeGX7Ac2-Na adduct), m/z 1,281 (MeGX7Ac3-Na adduct), m/z 1,329 (MeGX8Ac1-Na adduct), m/z 1,371 (MeGX8Ac2-Na adduct), and m/z 1,413 (MeGX8Ac3-Na adduct) (Fig. 4A). These peaks correspond to sodium-additive ions of a few acetylated aldouronic acids. Peaks corresponding to nonsubstituted linear acetylated xylooligosaccharides were not observed, and the peaks corresponding to MeGX2, MeGX2Ac1, MeGX4Ac3, MeGX6Ac2, and MeGX6Ac3 were observed as the hydrolysis products of StXyn30A and β-xylosidase.
FIG 4.
MALDI-TOF mass spectra of hydrolysis products of acetylated xylan. (A) Acetylated glucuronoxylan hydrolysate of StXyn30A. (B) Acetylated glucuronoxylan hydrolysate of StXyn30A and GH3 β-xylosidase. Acetylxylan (0.5% [wt/vol]) was incubated with StXyn30A in 50 mM acetate buffer (pH 6.0) containing 0.1% (wt/vol) BSA at 37°C for 20 h. GH3 β-xylosidase was then added to the reaction mixture, and the mixture was incubated at 37°C for 2 days.
Mutagenesis study of Arg296 in StXyn30A.
Figure 5 shows a substrate binding model of StXyn30A. For XynC from B. subtilis (9) and XynA from E. chrysanthemi (10), a conserved GlcA coordination motif has been defined, which involves numerous amino acid residues located in the substrate binding cleft. All these amino acid residues were conserved in StXyn30A, including Trp52, Trp110, Tyr168, Trp172, Tyr229, Tyr256, Trp292, Tyr293, Arg296, and Tyr298 (Fig. 1 and 5). Importantly, the arginine equivalent to Arg296 in StXyn30A is required for the recognition of the glucuronic acid, as it establishes a hydrogen bond and a salt bridge with the C-6 carboxylate of GlcA. Interestingly, this arginine residue is not conserved in fungal GH30-7 xylanases shown in Fig. 1 (glutamine [Q] in Leptosphaeria, glutamine in Trichoderma XYN IV, glutamic acid [E] in Trichoderma XYN VI, and glutamic acid in Bispora). We thought that the presence or absence of the positively charged arginine residue would affect the substrate discrimination at subsite −2 and may explain the difference in substrate specificities between bacterial GH30 xylanases and fungal GH30 xylanases. Therefore, we constructed Arg296 mutants of StXyn30A, including R296A, R296E, R296K, and R296Q, and their activities and hydrolysis products were compared with those of the wild-type enzyme. As shown in Fig. 6, the hydrolysis activities of soluble beechwood xylan by mutants were significantly reduced compared to those of wild-type StXyn30A. The activity of the R296E mutant was barely detectable over a 60-min time course (Fig. 6), most likely due to electrostatic repulsion between the C-6 carboxylate of GlcA and the newly occurring glutamate amino acid side chain. To examine the structure of hydrolysis products, soluble birchwood xylan was hydrolyzed completely by StXyn30A mutants, and the products were digested by a β-xylosidase. These hydrolysis products were then analyzed by TLC and MALDI-TOF MS (Fig. 7). Xylose and MeGX2 were detected as the main products in all mutant hydrolysates by TLC analysis (Fig. 7A), and the other products were also observed in the R296E mutant hydrolysate (Fig. 7A, lane 5). To investigate the size of hydrolysis products in more detail, we performed MALDI-TOF MS analysis of the hydrolysis products of the mutants (Fig. 7B). The peaks of low intensity of several 4-O-methylglucuronoxylooligosaccharides were detected in the R296E (Fig. 7BIII) and R296Q (Fig. 7BV) mutants, together with xylose and MeGX2, which were the products of wild-type StXyn30 following β-xylosidase treatment. MeGX3, MeGX4, and MeGX5 were observed as products of the R296E mutant, and MeGX3 and MeGX4 were observed as products of the R296Q mutant.
FIG 5.
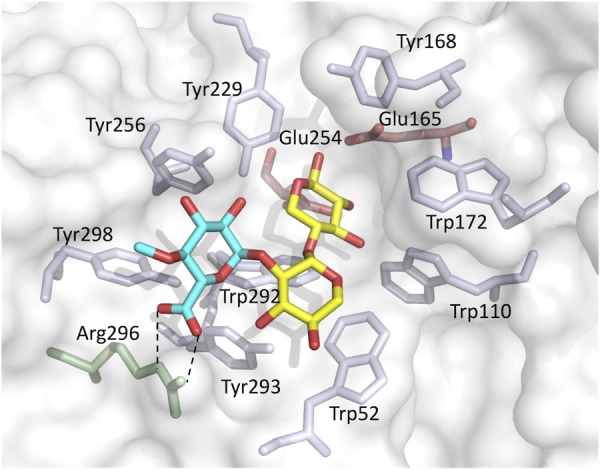
Model of the active site of StXyn30A with MeGX2. Blue, 4-O-methyl-glucuronic acid; yellow, xylose; transparent red, Glu165 and Glu254, catalytic residues. Trp52, Trp110, Tyr168, Trp172, Tyr229, Tyr256, Trp292, Tyr293, Arg296, and Tyr298 are conserved in bacterial GH30 enzymes. Two dashed lines indicate the salt bridge between the Arg296 side chain and 4-O-methyl-glucuronic acid of MeGX2. The homology model structure of StXyn30A with the substrate was modeled on the basis of the crystal structure of XynC from B. subtilis (9) (PDB accession number 3KL5) using the software Modeller (https://salilab.org/modeller/) (36).
FIG 6.
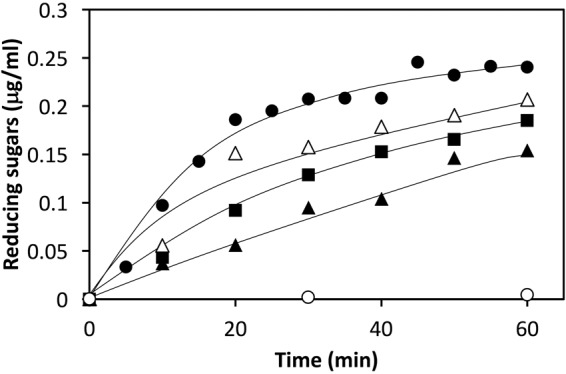
Activities of StXyn30A mutants. A 1% soluble beechwood solution in 50 mM acetate buffer (pH 6.0) was incubated with StXyn30A (closed circles) and the R296A (closed squares), R296E (open circles), R296K (open triangles), and R296Q (closed triangles) mutants at 37°C. Hydrolytic activity was determined by the amounts of reducing sugars according to the Somogyi-Nelson method (31).
DISCUSSION
It is known that the substrate specificities of canonical bacterial GH30-8 enzymes and fungal GH30-7 enzymes are significantly different, although these enzymes belong to the same family of glycoside hydrolases. However, the difference in the mechanisms of substrate specificity between these enzymes has been unclear. In this study, we performed a detailed analysis of mutant enzymes and evaluated the mechanism of the substrate specificity of StXyn30A.
We cloned and expressed the bacterial GH30 xylanase from S. turgidiscabies C56 (StXyn30A). The optimum pH for StXyn30A (pH 6.5) was similar to that for Xyn30D from P. barcinonensis (12). The optimum temperature for StXyn30A (55°C) was lower than those for XynC (65°C) and CtXynGH30 (70°C) and higher than those for Xyn30D (50°C) and XynA (35°C) (data not shown). StXyn30A exhibits hydrolysis activity toward 4-O-methylglucuronoxylans such as beechwood and birchwood xylans but had no activity toward wheat arabinoxylan or linear xylooligosaccharides, thereby confirming that StXyn30A is a glucuronoxylan-specific endoxylanase. In a previous study, Xyn30D from P. barcinonensis did not hydrolyze oat spelt xylan, but StXyn30A was shown to have activity with this substrate (Table 1). The reason for the difference in activity toward oat spelt xylan is not clear, but oat spelt xylan is a 4-O-methylglucuronoarabinoxylan (21, 22), so our result is reasonable. In addition, StXyn30A was able to hydrolyze cotton seed xylan. This type of xylan has been reported to contain both 4-O-methyl-glucuronic acid and glucuronic acid side chains (23). However, from our results, 4-O-methyl-glucuronic acid was not detected in our prepared cotton seed xylan (Fig. 3C). The analytical results for the hydrolysates of birchwood xylan and cotton seed xylan (Fig. 3B to E) indicate that StXyn30A recognizes the glucuronic acid with or without methylation at the C-4 hydroxyl position and can therefore hydrolyze glucuronoxylans not containing the 4-O-methyl-GlcA derivative.
In order to examine the oligosaccharide composition of the StXyn30A hydrolysate, a β-xylosidase or endoxylanase (SoXyn10A) with known specificity was used. The addition of a β-xylosidase or SoXyn10A to the StXyn30A hydrolysate simplified the oligomeric structure of the hydrolysate. By analysis of the resulting hydrolysate, we confirmed that the hydrolysis products generated by StXyn30A have a structure with the glucuronic acid side chain located on the second xylopyranosyl residue from the reducing end. This result indicated that StXyn30A had the same catalytic properties as those of XynC from B. subtilis (9) and XynA from E. chrysanthemi (10).
In this paper, we also performed an analysis of the acetylated glucuronoxylan hydrolysate of StXyn30A. As a result of MS analysis, acetylated MeGXn was digested by StXyn30A, and peaks corresponding to MeGX3Ac1∼MeGX8Ac1, MeGX5Ac2, MeGX8Ac2, and MeGX5Ac3∼MeGX8Ac3 were observed in the mass spectrum (Fig. 4A). These peaks correspond to sodium-additive ions of a few acetylated aldouronic acids. In addition, after digestion by GH3 β-xylosidase, MeGX2Ac1 (at m/z 537, suggested to be [M + Na]+) was mainly observed. MeGX2 at m/z 495 ([M + Na]+), MeGX4Ac2 at m/z 843 ([M + Na]+), MeGX4Ac3 at m/z 885 ([M + Na]+), MeGX6Ac2 at m/z 1,107, and MeGX6Ac3 at m/z 1,149 were slightly observed (Fig. 4B). Our finding that no linear acetylated xylooligosaccharides are detected during the hydrolysis of glucuronoacetylxylan by StXyn30A is in contrast to the unexpected results recently reported by Busse-Wicher et al. showing that an acetylated xylan lacking glucuronic acid was digested by the GH30-8 endoxylanase XynA from E. chrysanthemi (24).
Whereas the bacterial GH30-8 endoxylanases specifically degrade glucuronoxylan, fungal GH30-7 xylanases are not glucuronoxylan specific and degrade a variety of xylan substrates (18, 19). T. reesei XYN IV belongs to GH30-7 and exhibits its highest rate of hydrolysis toward rhodymenan, a linear, soluble β-1,3-β-1,4-xylan, and displays extremely low activity toward xylan containing glucuronoxylan. This enzyme has exo- and endoxylanase activities and degrades glucuronoxylans, arabinoxylans, and also xylooligosaccharides, producing xylose as its primary hydrolysis product. Although not considered for its role in the hydrolysis of rhodymenan, the GH30-7 endoxylanase from Bispora has been reported (25). While that research was not as detailed as the studies of T. reesei XYN IV, the finding that it works on all tested xylan substrates and releases xylose as its major limit product suggests that this enzyme is similar to T. reesei XYN IV. Distinct from the other GH30-7 endoxylanases, XYN VI from T. reesei exhibits catalytic properties similar to those of the bacterial GH30-8 GlcA-dependent endoxylanases (19). However, this enzyme also exhibits low hydrolysis activity toward arabinoxylan and rhodymenan. The catalytic features of fungal GH30 endoxylanases are strikingly different from those of bacterial GH30 endoxylanases.
Urbániková et al. (16) noted that the interaction of the Erwinia GH30 enzyme with the glucuronic acid side chain is stronger than that with the xylopyranosyl residues in the −2 subsite, and ionic interaction with arginine residue and glucuronic acid is important for the first contact of the enzyme and substrates. In bacterial GH30-8 endoxylanases, this arginine residue is conserved, but it is not conserved in fungal GH30-7 endoxylanases (Fig. 1). Not having this conserved arginine residue may be one of the reasons why fungal GH30-7 endoxylanases are not glucuronoxylan specific. To understand this further, we performed a mutagenesis study of StXyn30A by creating four unique Arg296 mutants (R296A, R296E, R296K, and R296Q). When Arg296 of StXyn30A was replaced with a negatively charge glutamic acid (R296E), the hydrolysis activity toward glucuronoxylan was remarkably reduced (Fig. 6, open circles). The results indicated that the interaction of Arg296 with the carboxyl group of the glucuronic acid side chain is very important for the strict substrate recognition of StXyn30A and suggested that substrate binding and discrimination were changed in the R296E mutant by the replacement of the positively charged arginine residue with the negatively charged glutamic acid residue. It was considered that glucuronoxylan may not fit well into the substrate binding cleft of the R296E mutant due to charge-charge repulsion. Actually, xylose, MeGX2, MeGX3, MeGX4, and MeGX5 were observed when β-xylosidase was used to treat the hydrolysis product of the R296E mutant (Fig. 7), indicating that the R296E mutant was not able to cleave xylan in the manner expected for the wild-type enzyme form.
Furthermore, we examined whether mutants have activity toward other xylan types. When xylooligosaccharides, arabinoxylan, and β-1,3-xylan were used as the substrates for StXyn30A mutants, enzyme activity such as that seen in T. reesei XYN IV was not observed (data not shown). This result suggested that the topologies of the substrate binding cleft are basically different between fungal and bacterial enzymes.
In conclusion, we characterized the StXyn30A xylanase from S. turgidiscabies C56, and we showed that the ionic interaction between the glucuronic acid substitution and the Arg296 residue, which is conserved in canonical bacterial GH30-8 endoxylanases, is very important for molecular interactions that control substrate specificity. Previous studies using synthesized glucoxylans also revealed a large decrease in activity, indicating a disruption of the ionic interaction between the C-6 carboxylate of GlcA and the conserved arginine (26, 27). However, it was difficult to explain the difference in the substrate recognition mechanisms between bacterial GH30 enzymes (subfamily 8) and fungal GH30 enzymes (subfamily 7) by the absence or presence of an arginine residue. Whereas the overall structures of bacterial and fungal GH30 endoxylanases are similar, these enzymatic properties are completely different. In order to clarify the difference in the substrate recognition mechanisms between bacterial and fungal enzymes, further mutagenesis studies are needed.
MATERIALS AND METHODS
Cloning of StXyn30A and construction of StXyn30A mutants.
The gene encoding a putative glucuronoxylan-specific xylanase (Stxyn30A) was cloned from the genome of S. turgidiscabies C56. The Stxyn30A gene was amplified by PCR with KOD-Plus-Neo (Toyobo, Osaka, Japan), using the following primer pair: C56-F (5′-CATATGGCTCCGGCGACGGCCGCCGCC-3′) and C56-R (5′-GCGGCCGCGGTCGTCACGAACGTCGTCAC-3′) (the underlined sequences represent restriction sites). Before insertion of the PCR products into the expression vector, the amplified fragment was subcloned by Target Clone-Plus (Toyobo, Osaka, Japan) and was confirmed by sequencing with a 3130 genetic analyzer (Applied Biosystems, Tokyo, Japan). The Stxyn30A gene was digested from the subcloned plasmid with NdeI and NotI and cloned between the NdeI and NotI sites of pET30a (Novagen, Darmstadt, Germany). The resulting pET30-stxyn30a recombinant plasmid was transformed into Escherichia coli Tuner(DE3) (Merck KGaA, Darmstadt, Germany). The transformants were grown in Luria-Bertani medium (28) at 37°C with shaking. IPTG was added to the culture at a final concentration of 0.1 mM when the optical density at 600 nm (OD600) reached 0.2. After the addition of IPTG, the cultures were grown at 25°C for 20 h. The cells were collected and resuspended in 50 mM phosphate buffer (pH 7.2), followed by sonication for 5 min. The StXyn30A protein was purified by immobilized metal affinity chromatography using the vector-derived C-terminal 6× histidine tag. This was subsequently dialyzed against distilled H2O. The purity of the enzyme was determined by using SDS-PAGE with 12% cross-linking according to the method of Laemmli (29). The protein was stained with Coomassie brilliant blue R-250 and then destained with 10% acetic acid in 30% methanol. The molecular weight of the enzyme was determined by SDS-PAGE using the SDS-PAGE Standard Low molecular weight marker (Bio-Rad, WA, USA).
Mutants were constructed by site-directed mutagenesis using the pET30-stxyn30a expression vector as a template DNA and the following primer pairs: R296A-F (5′-TACATCCGGGCGAGTTACGGTCCG-3′) and R296A-R (5′-GACCGTAACTCGCCCGGATGTACCA-3′), R296E-F (5′-TACATCCGGGAAAGTTACGGTCCG-3′) and R296E-R (5′-GACCGTAACTTTCCCGGATGTACCA-3′), R296K-F (5′-TACATCCGGAAAAGTTACGGTCCG-3′) and R296K-R (5′-GACCGTAACTTTTCCGGATGTACCA-3′), and R296Q-F (5′-TACATCCGGCAAAGTTACGGTCCG-3′) and R296Q-R (5′-GACCGTAACTTTGCCGGATGTACCA-3′). Each mutant was purified as described above for wild-type StXyn30A.
Substrates.
4-O-Methylglucuronoxylan from beechwood and birchwood and 4-O-methylglucuronoarabinoxylan from oat spelt were obtained from Sigma Chemical Company (St. Louis, MO, USA). These xylans were suspended in an appropriate amount of distilled water and incubated at 60°C for 1 day. Each xylan solution was centrifuged, and the supernatant was recovered. The supernatant was lyophilized, dissolved in distilled water, and used in this experiment as a soluble xylan. Xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5), xyloheptaose (X6), and wheat arabinoxylan (low viscosity; 2 centistokes [cSt]) were obtained from Megazyme International (Wicklow, Ireland). Cotton seed glucuronoxylan was prepared as described previously (23). β-1,3-Xylan was prepared from the siphonous green alga Bryopsis maxima according to the method of Iriki et al. (30), and acetylglucuronoxylan (acetylxylan) was obtained from the Forest Products Laboratory (FPL) Collection.
Enzyme activity and substrate specificity.
To evaluate the substrate specificity of StXyn30A for polysaccharides, we selected wheat arabinoxylan, soluble oat spelt xylan, soluble birchwood xylan, soluble beechwood xylan, acetylxylan, and β-1,3-xylan as the substrates. The reactions were performed in 50 mM acetate buffer (pH 6.0) containing 0.5% (wt/vol) substrate, 0.1% (wt/vol) bovine serum albumin (BSA), and 1 μM enzyme at 37°C for 16 h. The reaction was stopped by heating the solutions at 100°C for 20 min. Hydrolytic activity was determined by the amounts of reducing sugars according to the Somogyi-Nelson method (31). The effects of pH and temperature on enzyme activity and stability were investigated as described previously (32). The assay was performed by using samples in triplicate.
Modes of action of xylans and xylooligosaccharides.
StXyn30A was incubated with 0.5% (wt/vol) xylooligosaccharide (X2, X3, X4, X5, or X6) in 50 mM acetate buffer (pH 6.0) at 37°C for 20 h. The hydrolysis products were analyzed by high-performance anion-exchange chromatography with a pulsed amperometric detection (HPAEC-PAD) system and a CarboPac PA1 column (4 by 250 mm) (Dionex Corp., Sunnyvale, CA), as described previously (33).
StXyn30A was mixed with 0.5% (wt/vol) soluble birchwood xylan, acetyl xylan, or 2.5% (wt/vol) cotton seed xylan in 50 mM acetate buffer (pH 6.0) containing 0.1% (wt/vol) BSA. After incubation at 37°C for 20 h, GH3 β-xylosidase or SoXyn10A (20) was added to the reaction mixture, and the mixture was incubated at 37°C for 2 days. The reactions were terminated by heating the mixtures at 100°C for 20 min, and the mixtures were treated with Amberlite 200 and then filtered through a 0.22-μm filter. β-Xylosidase was prepared from cellulase powder of Penicillium funiculosum (Sigma) by the following method. Cellulase powder (2 g) was suspended in 100 ml of 50 mM phosphate buffer (pH 6.0) and agitated overnight at 4°C. After centrifugation to remove insoluble materials, the supernatant was loaded onto a Q-Sepharose column (16 by 100 mm) equilibrated with 50 mM phosphate buffer (pH 6.0) at flow rates of 5 ml/min and eluted with a linear gradient of 50 mM phosphate buffer containing 250 mM NaCl (pH 6.0). The recovered protein was loaded onto a 1-ml CM-Sepharose column equilibrated with 10 mM acetate buffer (pH 4.0) and eluted with a linear gradient of 10 mM acetate buffer containing 500 mM NaCl (pH 4.0) at flow rates of 1.0 ml/min. The eluted proteins were verified by SDS-PAGE.
Hydrolysis products were analyzed by TLC on silica gel 60 plates (Darmstadt, Germany, Merck). Sugars were developed with an n-butanol–acetic acid–water (2:1:1 [vol/vol/vol]) solvent and detected by using N-(1-naphthyl)ethylenediamine dihydrochloride (34, 35). Hydrolysis products were also analyzed by MALDI-TOF MS on a Reflex II instrument (Bruker Daltonics) or a 4800 MALDI-TOF/TOF analyzer (Applied Biosystems) in the positive-ion mode. The samples were diluted 10-fold with TA buffer (0.1% trifluoroacetic acid–acetonitrile [2:1]). One microliter of the diluted sample and 1 μl of 10 mg/ml 2,5-dihydroxybenzoic acid (DHB) in 30% (vol/vol) ethanol were mixed and spotted onto the target plate. The samples/matrix spots were analyzed by MALDI-TOF MS.
Molecular modeling.
The homology model structure of StXyn30A with the substrate was modeled on the basis of the crystal structure of XynC from B. subtilis (9) (PDB accession number 3KL5), using the software Modeller (https://salilab.org/modeller/) (36), and the bound aldouronate MeGX2 was docked according to the structure of the B. subtilis XynC-MeGX2 complex (PDB accession number 3KL5).
REFERENCES
- 1.Aspinall GO. 1980. Chemistry of cell wall polysaccharides, p 473–500. In Preiss J. (ed) The biochemistry of plants (a comprehensive treatise), vol 3 Carbohydrates: structure and function. Academic Press, New York, NY. [Google Scholar]
- 2.Saha BC. 2003. Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30:279–291. doi: 10.1007/s10295-003-0049-x. [DOI] [PubMed] [Google Scholar]
- 3.Dunlop AP. 1948. Furfural formation and behavior. Ind Eng Chem 40:204–209. doi: 10.1021/ie50458a006. [DOI] [Google Scholar]
- 4.Larsson S, Palmqvist E, Hahn-Hagerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant N. 1999. The generation of inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb Technol 24:151–159. doi: 10.1016/S0141-0229(98)00101-X. [DOI] [Google Scholar]
- 5.Jönsson L, Alriksson B, Nilvebrant NO. 2013. Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol Biofuels 6:16. doi: 10.1186/1754-6834-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishitani K, Nevins DJ. 1991. Glucuronoxylan xylanohydrolase. A unique xylanase with the requirement for appendant glucuronosyl units. J Biol Chem 266:6539–6543. [PubMed] [Google Scholar]
- 9.St John FJ, Rice JD, Preston JF. 2006. Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J Bacteriol 188:8617–8626. doi: 10.1128/JB.01283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vršanská M, Kolenová K, Puchart V, Biely P. 2007. Mode of action of glycoside hydrolase family 5 glucuronoxylan xylanohydrolase from Erwinia chrysanthemi. FEBS J 274:1666–1677. doi: 10.1111/j.1742-4658.2007.05710.x. [DOI] [PubMed] [Google Scholar]
- 11.Gallardo O, Fernández-Fernández M, Valls C, Valenzuela SV, Roncero MB, Vidal T, Díaz P, Pastor FI. 2010. Characterization of a family GH5 xylanase with activity on neutral oligosaccharides and evaluation as a pulp bleaching aid. Appl Environ Microbiol 76:6290–6294. doi: 10.1128/AEM.00871-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valenzuela SV, Diaz P, Pastor FI. 2012. Modular glucuronoxylan-specific xylanase with a family CBM35 carbohydrate-binding module. Appl Environ Microbiol 78:3923–3931. doi: 10.1128/AEM.07932-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verma AK, Goyal A. 2016. A novel member of family 30 glycoside hydrolase subfamily 8 glucuronoxylan endo-β-1,4-xylanse (CtXynGH30) from Clostridium thermocellum orchestrates catalysis on arabinose decorated xylans. J Mol Catal B Enzym 129:6–14. doi: 10.1016/j.molcatb.2016.04.001. [DOI] [Google Scholar]
- 14.St John FJ, Crooks C, Dietrich D, Hurlbert J. 27 March 2017. Xylanase 30 A from Clostridium thermocellum functions as a glucuronoxylan xylanohydrolase. J Mol Catal B Enzym doi: 10.1016/j.molcatb.2017.03.008. [DOI] [Google Scholar]
- 15.St John FJ, Hurlbert JC, Rice JD, Preston JF, Pozharski E. 2011. Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase. J Mol Biol 407:92–109. doi: 10.1016/j.jmb.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Urbániková L, Vršanská M, Mørkeberg Krogh KBR, Hoff T, Biely P. 2011. Structural basis for substrate recognition by Erwinia chrysanthemi GH30 glucuronoxylanase. FEBS J. 278:2105–2116. doi: 10.1111/j.1742-4658.2011.08127.x. [DOI] [PubMed] [Google Scholar]
- 17.St. John FJ, González JM, Pozharski E. 2010. Consolidation of glycosyl hydrolase family 30: a dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett 584:4435–4441. doi: 10.1016/j.febslet.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 18.Tenkanen M, Vršanská M, Siika-aho M, Wong DW, Puchart V, Penttilä M, Saloheimo M, Biely P. 2012. Xylanase XYN IV from Trichoderma reesei showing exo- and endo-xylanase activity. FEBS J 280:285–301. doi: 10.1111/febs.12069. [DOI] [PubMed] [Google Scholar]
- 19.Biely P, Puchart V, Stringer MA, Mørkeberg Krogh KB. 2014. Trichoderma reesei XYN VI—a novel appendage-dependent eukaryotic glucuronoxylan hydrolase. FEBS J 281:3894–3903. doi: 10.1111/febs.12925. [DOI] [PubMed] [Google Scholar]
- 20.Fujimoto Z, Kaneko S, Kuno A, Kobayashi H, Kusakabe I, Mizuno H. 2004. Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86. J Biol Chem 279:9606–9614. doi: 10.1074/jbc.M312293200. [DOI] [PubMed] [Google Scholar]
- 21.Kormelink FJ, Voragen AG. 1993. Degradation of different [(glucurono)arabino] xylans by a combination of purified xylan-degrading enzymes. Appl Microbiol Biotechnol 38:688–695. doi: 10.1007/BF00182811. [DOI] [Google Scholar]
- 22.Sun HJ, Yoshida S, Park NH, Kusakabe I. 2002. Preparation of (1→4)-beta-d-xylooligosaccharides from an acid hydrolysate of cotton-seed xylan: suitability of cotton-seed xylan as a starting material for the preparation of (1→4)-beta-d-xylooligosaccharides. Carbohydr Res 337:657–661. doi: 10.1016/S0008-6215(02)00031-9. [DOI] [PubMed] [Google Scholar]
- 23.Matsuo N, Yoshida S, Kusakabe I, Murakami K. 1991. Chemical structure of xylan in cotton-seed cake. Agric Biol Chem 55:2905–2907. doi: 10.1271/bbb1961.55.2905. [DOI] [PubMed] [Google Scholar]
- 24.Busse-Wicher M, Gomes TC, Tryfona T, Nikolovski N, Stott K, Grantham NJ, Bolam DN, Skaf MS, Dupree P. 2014. The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J 79:492–506. doi: 10.1111/tpj.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo H, Yang J, Li J, Shi P, Huang H, Bai Y, Fan Y, Yao B. 2010. Molecular cloning and characterization of the novel acidic xylanase XYLD from Bispora sp. MEY-1 that is homologous to family 30 glycosyl hydrolases. Appl Microbiol Biotechnol 86:1829–1839. doi: 10.1007/s00253-009-2410-0. [DOI] [PubMed] [Google Scholar]
- 26.Hurlbert JC, Preston JF. 2001. Functional characterization of a novel xylanase from a corn strain of Erwinia chrysanthemi. J Bacteriol 183:2093–2100. doi: 10.1128/JB.183.6.2093-2100.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biely P, Malovíková A, Hirsch J, Morkeberg Krogh KB, Ebringerová A. 2015. The role of the glucuronoxylan carboxyl groups in the action of endoxylanases of three glycoside hydrolase families: a study with two substrate mutants. Biochim Biophys Acta 1850:2246–2255. doi: 10.1016/j.bbagen.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 29.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Iriki Y, Suzuki T, Nishizawa K, Miwa T. 1960. Xylan of siphonaceous green algae. Nature 187:82–83. doi: 10.1038/187082a0. [DOI] [PubMed] [Google Scholar]
- 31.Somogyi M. 1952. Notes on sugar determination. J Biol Chem 195:19–23. [PubMed] [Google Scholar]
- 32.Ichinose H, Yoshida M, Fujimoto Z, Kaneko S. 2008. Characterization of a modular enzyme of exo-1,5-α-l-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893. Appl Microbiol Biotechnol 80:399–408. doi: 10.1007/s00253-008-1551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichinose H, Araki Y, Michikawa M, Harazono K, Yaoi K, Karita S, Kaneko S. 2012. Characterization of an endo-processive-type xyloglucanase having a β-1,4-glucan-binding module and an endo-type xyloglucanase from Streptomyces avermitilis. Appl Environ Microbiol 78:7939–7945. doi: 10.1128/AEM.01762-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bounias M. 1980. N-(1-Naphthyl)ethylenediamine dihydrochloride as a new reagent for nanomole quantification of sugars on thin-layer plates by a mathematical calibration process. Anal Biochem 106:291–295. doi: 10.1016/0003-2697(80)90523-0. [DOI] [PubMed] [Google Scholar]
- 35.Biely P, Mastihubová M, Côté GL, Greene RV. 2003. Mode of action of acetylxylan esterase from Streptomyces lividans: a study with deoxy and deoxy-fluoro analogues of acetylated methyl beta-d-xylopyranoside. Biochim Biophys Acta 1622:82–88. doi: 10.1016/S0304-4165(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 36.Eswar N, Eramian D, Webb B, Shen MY, Sali A. 2008. Protein structure modeling with MODELLER. Methods Mol Biol 424:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]