ABSTRACT
Predation of starter lactic acid bacteria such as Streptococcus thermophilus by bacteriophages is a persistent and costly problem in the dairy industry. CRISPR-mediated bacteriophage insensitive mutants (BIMs), while straightforward to generate and verify, can quickly be overcome by mutant phages. The aim of this study was to develop a tool allowing the generation of derivatives of commercial S. thermophilus strains which are resistant to phage attack through a non-CRISPR-mediated mechanism, with the objective of generating BIMs exhibiting stable resistance against a range of isolated lytic S. thermophilus phages. To achieve this, standard BIM generation was complemented by the use of the wild-type (WT) strain which had been transformed with an antisense mRNA-generating plasmid (targeting a crucial CRISPR-associated [cas] gene) in order to facilitate the generation of non-CRISPR-mediated BIMs. Phage sensitivity assays suggest that non-CRISPR-mediated BIMs exhibit some advantages compared to CRISPR-mediated BIMs derived from the same strain.
IMPORTANCE The outlined approach reveals the presence of a powerful host-imposed barrier for phage infection in S. thermophilus. Considering the detrimental economic consequences of phage infection in the dairy processing environment, the developed methodology has widespread applications, particularly where other methods may not be practical or effective in obtaining robust, phage-tolerant S. thermophilus starter strains.
KEYWORDS: bacteriophage, resistance, insensitive, mutant, mRNA, adsorption, receptor, sedimentation
INTRODUCTION
Streptococcus thermophilus is a widely employed bacterium used for milk acidification and subsequent product texturizing, which are required for the manufacture of cheeses and yoghurts in the dairy industry (1). This process, however, can be disrupted, retarded, or halted by contaminant (bacterio)phages, which present a persistent problem in dairy fermentations (2; reviewed in reference 3). Several approaches may be utilized in an attempt to mitigate this costly problem. These include (i) starter rotational schemes (i.e., the rotation of strain sets with nonoverlapping phage sensitivities), (ii) heat or chemical treatments of dairy plant equipment in order to reduce the phage burden (4), and (iii) generation of bacteriophage insensitive mutants (BIMs) of S. thermophilus starters for incorporation into the rotational schemes mentioned above. BIMs may be generated by one of several methods, including insertional mutagenesis (5), immunoselection combined with cell sorting (6), construction of mutant libraries followed by phage exposure (7), the secondary culture method (8), serial passaging in the presence of high titer phages (9, 10), and chemical mutagenesis (11–13).
Generated BIMs should, ideally, (i) be robust, i.e., insensitive to a high level of and/or repeatedly introduced phages, (ii) be resistant to a wide variety of genetically distinct phages, (iii) possess technological properties identical to those of the parent strain from which they were derived, (iv) be derived from parent S. thermophilus strains at a high frequency, and (v) be easy to characterize both genetically and phenotypically. In the context of robustness, defined here as the ability of a BIM to survive high-titer phage exposure, repeated phage challenges (such as the Heap-Lawrence test [14]) and/or challenges using genetically distinct phages, the mechanism conferring resistance to the BIM may be relevant. Several S. thermophilus phage resistance systems have been documented. Restriction/modification (R/M) systems are thought to be widespread in this species and are, for the most part, chromosomally encoded (15–19), with one described example of a complete, plasmid-encoded R/M system (20). A lactococcus-derived R/M system was also shown to function in S. thermophilus when introduced using a compatible cloning vector (21). A lactococcal abortive infection (Abi) system was shown to function in S. thermophilus (22), and indeed, evidence of a native Abi system has been detected (23), although these systems are not well characterized in S. thermophilus. Superinfection exclusion (Sie) has also been described in prophages of S. thermophilus (24), although, considering the apparent rarity of resident prophages in the species (24, 25), prophage-encoded systems of this type probably are not widespread.
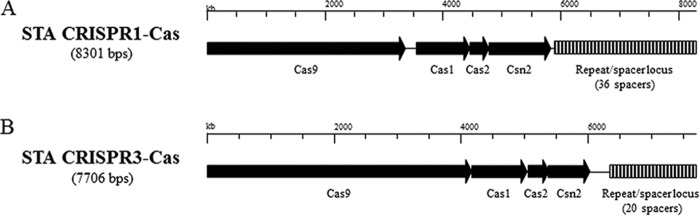
The most widely characterized phage resistance system in S. thermophilus is the clustered regularly interspaced short palindromic repeats (CRISPR) and their CRISPR-associated, or cas, genes, together referred to as the CRISPR-Cas system (26–29), first proven to be a phage resistance mechanism by Barrangou and colleagues (30). Four distinct CRISPR-Cas systems have been identified in S. thermophilus (31), two of which are known to actively provide phage immunity to this species (26, 27, and this study), namely, CRISPR1-Cas and CRISPR3-Cas. Both of these systems are classified as type II-A systems, of which the cas components comprise cas9, cas1, cas2, and csn2, each performing a specific function in the context of the system (32).
These systems fundamentally act to digest incoming DNA using the product of the cas9 gene, the signature gene of the type II-A group (33, 34), which is essential for CRISPR-based immunity (30, 35, 36). Segments (approximately 30 bp) of the foreign DNA (known as spacers) are incorporated into the chromosome at specific repeat-spacer loci in a mechanism which is believed to involve the product of the csn2 gene (30, 37), potentially in complex with Cas9, Cas1, and Cas2 (38). The Cas-encoding genes, as well as the repeat-spacer loci, are constitutively transcribed (18, 39), and the resultant crRNA acts as a guide to particular Cas proteins to recognize and target subsequent incoming DNA. This process is reliant on the presence of an approximately 30-nucleotide DNA fragment on the phage genome termed the protospacer (i.e., the specific sequence on the phage genome that is targeted by the CRISPR-Cas system [40]) as well as a short sequence flanking this target sequence, termed the protospacer adjacent motif, or PAM (26, 40, 41).
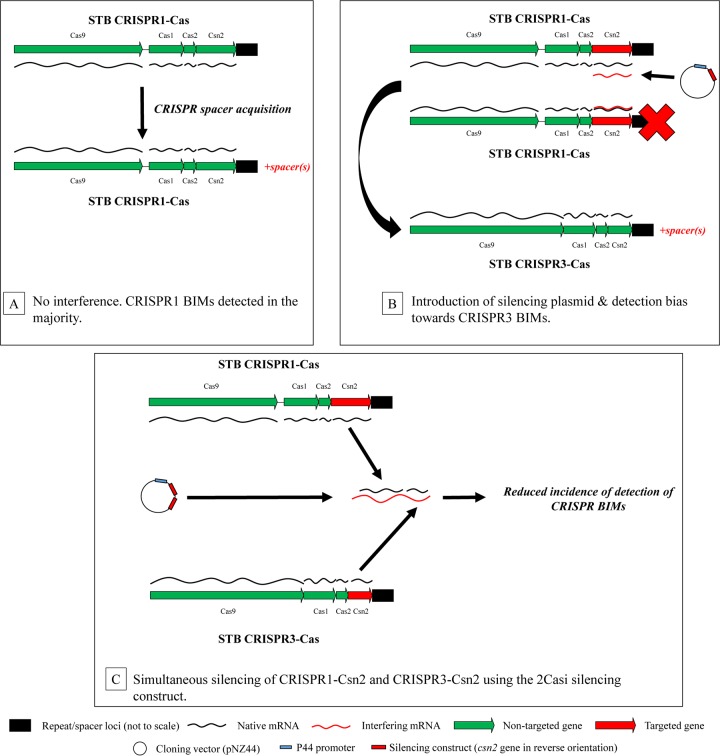
The presence of up to four distinct CRISPR-Cas systems in characterized S. thermophilus strains (31) suggests that this system has been instrumental in shaping the dynamics of the phage-host relationship on an evolutionary scale. The efficiency and specificity of this immune system are matched only by the phages' adaptive response to it. Deveau and colleagues (40) showed that phages can rapidly overcome CRISPR-mediated resistance through mutations in either the target protospacer or adjacent PAM. Similar results were reported more recently by Sun and colleagues (42). Thus, in order to develop robust BIMs, it may be prudent to maximize the number of systems targeting the incoming phage, an approach which has recently been shown to significantly increase phage resistance by combining the protective effect of a CRISPR-Cas system with that of a functional R/M system (43). In order to facilitate the selection of non-CRISPR-mediated BIMs, we endeavored to inactivate (by mRNA silencing) purportedly crucial genes in the CRISPR1-Cas and CRISPR3-Cas systems, namely, cas9 and csn2.
Here, we present a method whereby BIMs of S. thermophilus, whose phage resistance is mediated by a CRISPR-independent mechanism, may be selected for. This method relies on antisense mRNA-producing plasmids, a method previously used to confer phage resistance to Lactococcus lactis and S. thermophilus by targeting intracellular phage replication (44–47). Here, the principle of antisense-mediated CRISPR-Cas silencing was validated by targeting the cas9-1 (CRISPR1-associated) or cas9-3 (CRISPR3-associated) gene of three BIMs, thereby inhibiting phage resistance conferred by a previously acquired spacer. In order to generate BIMs of a given S. thermophilus strain that are resistant to phages by the action of a non-CRISPR-mediated mechanism, an antisense plasmid construct simultaneously silencing both the csn2 gene of the CRISPR1-Cas system and its analogue in the CRISPR3-Cas system was employed.
(A patent application describing elements of this work has been submitted under reference number WO2016012552.)
RESULTS AND DISCUSSION
BIMs of S. thermophilus STA. (i) Generation of CRISPR- and non-CRISPR-mediated BIMs of STA.
Considering that the two bacterial strains used in this study were observed to behave differently when subjected to phage attack, distinct cas silencing approaches were adopted. For this reason, S. thermophilus strains ST47795 (here referred to as strain STA) and ST64985 (strain STB) will be discussed separately. BIMs of S. thermophilus STA (Table 1) were generated by phage exposure and validated by phage sensitivity assays and CRISPR locus sequencing (Table 2; see also Table S1 in the supplemental material). BIMs of this strain were readily generated in this manner (occurring at a frequency of approximately 1 × 10−5, estimated by dividing the CFU per milliliter of visible BIMs after overnight incubation by the CFU per milliliter of the overnight culture used in the plaque assay), and two BIMs (designated STA BIM1 and STA BIM2; Table 1), both generated against phage 7951, were selected for further characterization. The relative efficiencies of plaquing (EOP) of each assessed phage on a given BIM are shown in Table 2. While STA BIM1 was found to be resistant only to the phage used in the challenge (7951), STA BIM2 was shown to be resistant to all four, genetically distinct (49), phages available against the parent strain (phages 7951, 7952, 7953, and 7954; here referred to as phages 7951-4; Table 1). In addition, while presumed escape mutant phages were detected on STA BIM1 using the plaque assay method (as indicated by single, well-formed plaques occurring in the bacterial lawn upon challenge using undiluted phage lysate), this was not the case for STA BIM2, for which no escape mutants could be isolated (Table 2). Determination of the sequences of the CRISPR1, CRISPR2, and CRISPR3 loci of the two BIMs (CRISPR4 is not detected by PCR and presumed to be absent) and comparison to those of the parent strain from which they had been derived showed that STA BIM1 had acquired a single spacer in the CRISPR1 locus (schematic representations of the CRISPR1-Cas and CRISPR3-Cas loci are given in Fig. 1) (see Tables S1 and S7). The sequence of this added spacer displayed 100% identity to a region on the genome of phage 7951 (partly located within ORF357951, which encodes a predicted RecT recombinase [49]), consistent with protospacer targeting (30). In contrast, the sequences of the CRISPR1, CRISPR2, and CRISPR3 loci of STA BIM2 were identical to the parent strain (Table S1). Taken together, these data indicate that the mechanisms by which STA BIM1 and STA BIM2 had become resistant to phage 7951 were distinct.
TABLE 1.
Bacterial strains and bacteriophages applied during this study
| Strain/phage/plasmid | Description | Source or reference |
|---|---|---|
| S. thermophilus strains | ||
| STA | Industrial starter parent strain | DSM, Delft, The Netherlands |
| STB | Industrial starter parent strain | DSM, Delft, The Netherlands |
| STA BIM1 | CRISPR-BIM of STA generated against phage 7951 | UCC, Cork, Ireland |
| STA BIM2 | Non-CRISPR BIM of STA generated against phage 7951 | UCC, Cork, Ireland |
| STB BIM1 | CRISPR-BIM of STB generated against phage 9854 | UCC, Cork, Ireland |
| STB BIM2 | CRISPR-BIM derivative of STB generated against phage 9851 | UCC, Cork, Ireland |
| STB BIM3 | Non-CRISPR BIM of STB generated against phage 9854 | UCC, Cork, Ireland |
| L. lactis strain | ||
| NZ9000 | Transformation host | 60 |
| Phages | ||
| 7951 | Lytic pac-containing phage infecting STA | DSM, Delft, The Netherlands |
| 7952 | Lytic pac-containing phage infecting STA | DSM, Delft, The Netherlands |
| 7953 | Lytic pac-containing phage infecting STA | DSM, Delft, The Netherlands |
| 7954 | Lytic pac-containing phage infecting STA | DSM, Delft, The Netherlands |
| 9851 | Lytic cos-containing phage infecting STB | DSM, Delft, The Netherlands |
| 9853 | Lytic pac-containing phage infecting STB | DSM, Delft, The Netherlands |
| 9854 | Lytic cos-containing phage infecting STB | DSM, Delft, The Netherlands |
| Plasmid constructs | ||
| pNZ44 | Transformation vector | |
| pNZ44+Cas9-1i | STA CRISPR1-Cas9 silencing vector | This study |
| pNZ44+Cas9-1i | STB CRISPR1-Cas9 silencing vector | This study |
| pNZ44+Cas9-3i | STB CRISPR3-Cas9 silencing vector | This study |
| pNZ44+Csn2-1i | STB CRISPR1-Csn2 silencing vector | This study |
| pNZ44+2Csni | STB CRISPR1-Csn2 and CRISPR3-Csn2 silencing vector | This study |
TABLE 2.
Relative EOPs of four phages on STA, its derived BIMs, and their CRISPR-Cas-silenced derivatives
| Strain | EOP for phagea: |
|||
|---|---|---|---|---|
| 7951 | 7952 | 7953 | 7954 | |
| STA WT | 1 | 1 | 1 | 1 |
| STA BIM1 | (8.33 ± 0.12) × 10−8 | 0.55 ± 0.16 | 0.75 ± 0.28 | 0.84 ± 0.49 |
| STA BIM1::pNZ44 | (4.87 ± 0.61) × 10−6 | 4.95 ± 1.10 | 6.02 ± 0.68 | 6.33 ± 1.90 |
| STA BIM1::pNZ44+Cas9-1i | 1.03 ± 0.07 | 1.08 ± 0.27 | 0.93 ± 0.07 | 1.43 ± 0.30 |
| STA BIM1::pNZ44+Cas9-1i (cured) | (2.21 ± 3.00) × 10−6 | 4.08 ± 3.50 | 4.04 ± 2.76 | 3.89 ± 3.41 |
| STA BIM2 | ≤3 × 10−7 | ≤1 × 10−8 | ≤4 × 10−8 | ≤9 × 10−7 |
| STA BIM2::pNZ44+Cas9-1i | ≤3 × 10−7 | ≤4 × 10−7 | (1.85 ± 2.62) × 10−7 | ≤4 × 10−7 |
Values preceded by a less-than-or-equal-to sign indicate the limit of detection of phage titer.
FIG 1.
Schematic representation of the CRISPR1-Cas and CRISPR3-Cas systems of S. thermophilus STA.
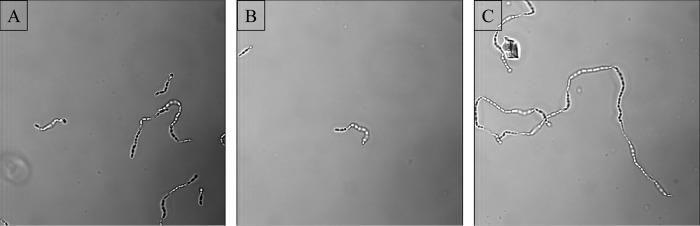
To further characterize the derived BIMs, STA BIM1 and STA BIM2 were subjected to adsorption (Table 3), sedimentation (Fig. 2), and morphological assays (Fig. 3, Table 4), as described in Materials and Methods. Adsorption efficiency of phage 7951 (which was used in the challenge) on STA BIM1 was not significantly different from that of the parent strain. Similarly, compared to the parent strain, this BIM did not show a significantly different sedimentation profile and cell chain length properties, as measured by counting the cells within continuous cell chains in a wet-mount preparation of the strain (cells per chain [CPC]). In contrast to this presumed CRISPR-mediated BIM, strain STA BIM2 was shown to exhibit a significant reduction in its ability to adsorb phage 7951, indicating that a phage receptor modification is responsible for the observed phage insensitivity of this BIM. Interestingly, this BIM also showed a distinct sedimentation profile relative to that of the STA wild type (WT) (and to STA BIM1), in that cells of STA BIM2 were shown to sediment to the bottom of the tube used for standard overnight growth, as visualized in Fig. 2. Indeed, this phenomenon has previously been observed in phage adsorption-deficient derivatives of Lactococcus lactis (50) and is indicative of a bacterial cell surface alteration. Lastly, a significant increase in cell chain length was observed in STA BIM2 compared to the parent strain and STA BIM1 (Fig. 3, Table 4), indicative of a cell envelope alteration and consistent with the observed sedimentation phenotype (51, 52). The nature of these mutations has been characterized and will be published elsewhere (B. McDonnell, L. Hanemaaijer, F. Bottacini, P. Kelleher, K. Lavelle, I. Sadovskaya, E. Vinogradov, E. Ver-Loren-Van-Themaat, T. Kouwen, J. Mahony, and D. van Sinderen, unpublished data).
TABLE 3.
Adsorption analysis of S. thermophilus parent strains and derived BIMs
| Strain/BIM | Adsorption of phage 7951 (%) | P value |
|---|---|---|
| STA WT | 79.9 ± 13.6 | |
| STA BIM1 | 83.2 ± 1.8 | 0.76 |
| STA BIM2 | 10.2 ± 8.2 | 0.0034 |
FIG 2.
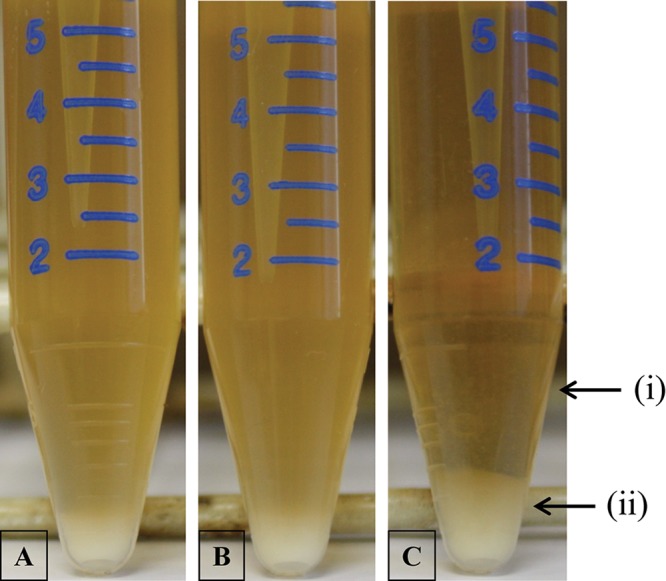
Observed sedimentation of S. thermophilus STA parent strain and derived BIMs. STA WT (A), STA BIM1 (CRISPR BIM) (B), STA BIM2 (non-CRISPR BIM) (C) are shown, where (i) indicates a partial clearing of the supernatant broth and (ii) indicates a visible increase in pellet size.
FIG 3.
Representative images of cell chains visualized using confocal laser scanning microscopy. (A) STA WT; (B) STA BIM1 (CRISPR BIM); (C) STA BIM2 (non-CRISPR BIM).
TABLE 4.
Relative CPC of S. thermophilus parent strains and derived BIMs
| Strain | CPC (total no. of cells) | % CPC (avg vs parent) | P value |
|---|---|---|---|
| STA (parent) | 10.84 ± 8.64 (n = 28) | NAa | NA |
| STA BIM1 | 7.4 ± 8.30 (n = 32) | 68.3 | 0.12 |
| STA BIM2 | 25.9 ± 13.0 (n = 51) | 238.9 | 3.9 × 10−5 |
NA, not applicable.
(ii) CRISPR-Cas silencing restores the sensitivity of CRISPR-mediated BIMs to phages.
An antisense mRNA silencing approach for CRISPR inactivation in S. thermophilus was performed to confirm that the observed phage resistance in STA BIM1 was conferred by the CRISPR-Cas system, in contrast to the phage resistance observed for STA BIM2. To this end, STA BIM1 and STA BIM2 were transformed using a cas9-interfering (Cas9i) antisense plasmid construct as described in Materials and Methods. The constitutive p44 promoter combined with the cloning, in reverse orientation, of the native STA cas9 gene was employed to produce cas9-antisense mRNA (partly) complementary to the cas9-encompassing mRNA. It was reasoned that if this cas9-antisense RNA hybridized with its sense counterpart it would interfere with translation of cas9 mRNA and, thus, with its associated CRISPR-mediated phage resistance. Table 2 shows the relative EOPs of phages on these CRISPR-silenced derivatives. It is clear that when STA BIM1 contains the cas9-antisense-producing plasmid (pNZ44+Cas9-1i), phage sensitivity is restored approximately to WT levels, thus demonstrating that this plasmid is capable of effective CRISPR silencing by presumed antisense mRNA production. STA BIM1::pNZ44+Cas9-1i then was subjected to plasmid curing as outlined above. Table 2 shows that, when the strain was cured of the silencing plasmid, the respective phage resistance profile of STA BIM1 was restored to that observed prior to the introduction of plasmid pNZ44+Cas9-1i. Interestingly, the introduction of plasmid pNZ44 without an insert (as a control) seemed to partially increase the sensitivity of STA BIM1 to all four phages (Table 2), although this increased sensitivity did not approach the level of restored sensitivity of STA BIM1::pNZ44+Cas9-1i to phage 7951 using the cas silencing approach (Table 2). The reason for this phenomenon is unknown.
To confirm that the insensitivity of S. thermophilus STA BIM2 was conferred by a resistance mechanism other than CRISPR1, plasmid pNZ44+Cas9-1i was introduced into this BIM, and the resulting strain then was subjected to the phage assays outlined above. It is clear from Table 2 that silencing the CRISPR1-Cas system had no significant effect on the ability of phages 7951-4 to produce plaques on this strain, supporting the notion that 7951-4 insensitivity in this BIM is not mediated by a CRISPR-Cas system.
BIMs of S. thermophilus STB. (i) STB predominantly produces CRISPR-mediated BIMs.
BIMs of S. thermophilus strain STB were generated and validated by CRISPR locus sequencing (Tables S2, S6, S8, and S9). Phages used in each challenge are listed in Table 1, and the relative EOP values of each phage on each BIM are given in Table 5. While BIMs with altered and unaltered CRISPR spacer content were readily derived from strain STA (as described above), all analyzed BIMs derived from STB (generally arising at an approximate frequency of 10−7) were shown to contain spacer alterations in either the CRISPR1 or CRISPR3 locus (Fig. 4 and Tables S3 and S9). This phenomenon is illustrated in Tables S3 and S9, which show that 100% of analyzed BIMs derived from WT STB contained an additional spacer in either locus.
TABLE 5.
EOPs of phages 9851, 9853, and 9854 on STB, its derived BIMs, and their CRISPR-Cas-silenced derivatives
| Strain | Value for phagea: |
||
|---|---|---|---|
| 9851 | 9853 | 9854 | |
| STB WT | 1 | 1 | 1 |
| STB BIM1 | 0.22 ± 0.09 | 0.26 ± 0.06 | (2.04 ± 0.79) × 10−6 |
| STB BIM1::pNZ44 | 0.27 ± 0.06 | 0.30 ± 0.09 | (4.01 ± 0.79) × 10−6 |
| STB BIM1::pNZ44+Cas9-3i | 2.35 ± 0.33 | 1.15 ± 0.24 | 0.22 ± 0.08 |
| STB BIM2 | (9.80 ± 5.93) × 10−7 | 0.13 ± 0.05 | ≤1 × 10−9 |
| STB BIM2::pNZ44 | (2.59 ± 1.04) × 10−6 | 0.23 ± 0.08 | ≤1 × 10−9 |
| STB BIM2::pNZ44+985Cas9-1i | 0.002 ± 0.0002 | 0.56 ± 0.17 | ≤1 × 10−9 |
Values preceded by a less-than-or-equal-to sign indicate the limit of detection of phage titer.
FIG 4.
CRISPR spacer addition patterns in STB WT (A), STB::pNZ44+Csn2-1i (B), and STB::pNZ44+2Csni (C).
The observation that phage exposure invariably results in the addition of CRISPR spacers in STB was exploited to produce a BIM containing iterative additions of spacers acquired upon exposure to previously unencountered phages: STB BIM1 was used to isolate STB BIM2, having been exposed to phages 9854 and 9851 independently and respectively. CRISPR spacer content of each BIM relative to parent strain STB is shown in Tables S2 and S8. Successive spacer addition was shown to correspond to phage insensitivity, i.e., additional spacers conferred resistance to the phage against which the BIM had been challenged (Table 5 and as previously shown [40]). It is noteworthy that while presumed phage CRISPR escape mutants (CEMs) were detected in most cases, phage 9854 did not produce plaques at a detectable level on STB BIM2. This may be explained by the fact that the added spacer in the CRISPR1 locus of STB BIM2 (added subsequent to challenge with phage 9851) also shows 100% nucleotide identity to phage 9854 (already targeted by a spacer in CRISPR3). It is plausible that the lack of escape mutants is due to the effect of being targeted by two distinct CRISPR-Cas systems through the addition of these two spacers, a phenomenon which has been observed previously using a similar approach (40).
(ii) CRISPR-Cas silencing in CRISPR1- and CRISPR3-mediated STB BIMs.
To prove the broader feasibility of the silencing method (as outlined above for strain STA), this approach was adopted in STB derivatives STB BIM1 and STB BIM2, purported to be CRISPR-mediated BIMs. The EOP values given in Table 5 clearly show that in STB BIM1 harboring the Cas9-3i construct (or, in the case of STB BIM2, the Cas9-1i construct), sensitivity to phages used in the initial challenge is largely reverted, where this effect is not seen in STB BIM1 and STB BIM2 which harbor the control pNZ44 plasmid.
The specificity of this system is highlighted by restoration of the sensitivity of STB BIM2 to phage 9851 (and not 9854), indicating that only those phages which had previously been targeted by the now-silenced CRISPR-Cas system regain the ability to infect. In the case of STB BIM2::pNZ44+Cas9-1i, however, the restoration of sensitivity to phage 9851 is not complete (the EOP of the phage being approximately 10−3 relative to that of WT STB) (Table 5). It is possible that the silencing protocol was not operating at optimal efficiency, potentially due to the plasmid being targeted by the CRISPR1-Cas or (more likely, considering the CRISPR1-Cas9 silencing nature of the insert) the CRISPR3-Cas system also being present in this strain.
(iii) CRISPR-Cas silencing reduces the incidence of CRISPR spacer addition during BIM generation.
Considering the evident spacer addition in analyzed BIMs of STB (mentioned above), we endeavored to inhibit the generation of CRISPR-mediated BIMs in this strain. For this purpose, two silencing plasmid constructs were employed separately. The first (pNZ44+Csn2-1i) was used to target the csn2-1 gene, i.e., the CRISPR1-Cas system only. The second construct (pNZ44+2Csni) produced a single antisense transcript targeting csn2-1 and csn2-3, i.e., the action of both the CRISPR1-Cas and CRISPR3-Cas systems simultaneously. The encoded product of csn2-1 has previously been implicated in spacer acquisition in S. thermophilus (30) as well as other species (37), and it was hypothesized that csn2-3 encodes a protein of similar function in the CRISPR3-Cas system based on its analogous position in the CRISPR3-Cas locus (Fig. 1 and 4) (26, 53). The csn2 genes were employed for gene-silencing purposes in this case (in contrast to cas9 genes in previous experiments) due to their smaller size, potentially enhancing plasmid stability and enabling the construction of a tandem construct targeting both systems (for reasons discussed in Materials and Methods). These constructs were introduced separately into STB WT (Table S3), after which BIMs were generated against phage 9854.
BIMs derived from STB WT, as well as BIM derivatives from STB containing either pNZ44, pNZ44+Csn2-1i (targeting CRISPR1-Cas), or pNZ44+2Csni (targeting CRISPR1-Cas and CRISPR3-Cas simultaneously), then were generated. In general, absolute numbers of BIMs growing in the semisolid agar were lower when STB containing CRISPR-interfering construct pNZ44+Csn2-1i or pNZ44+2Csni was used than when STB WT or STB::pNZ44 was used, although the frequencies of these BIMs were not determined due to the assay-dependent nature of this phenomenon. Following BIM generation, the leader ends (approximately 500 bp) of CRISPR1, CRISPR3, and CRISPR4 repeat/spacer loci in selected BIMs were subjected to Sanger sequencing. CRISPR2 repeat/spacer loci were excluded from this analysis due to their apparent inactivity in this species (26). A summary of BIMs containing CRISPR repeat/spacer loci which showed alterations is shown in Table S3, with detailed analysis in Table S9. It is clear that while the majority (70%) of BIMs generated from the WT strain (and plasmid control) acquired spacers in CRISPR1, those generated from the WT strain containing the Csn2-1-interfering plasmid (pNZ44+Csn2-1i) appeared unable to do so (Fig. 4B). This is consistent with previously published results in which csn2-1 was inactivated (30). Interestingly, the majority (60%) of these STB::pNZ44+Csn2-1i-derived BIMs showed alterations in the CRISPR3 locus, indicating an increase in the ratio of CRISPR3-mediated BIMs to CRISPR1-mediated BIMs in the event of a nonfunctioning CRISPR1 (Fig. 4B). This increase in ratio is reflected in the detection of CRISPR3-mediated BIMs derived from the STB WT::pNZ44+Csn2-1i strain (Table S9).
Thirty-three percent of BIMs generated using STB WT::pNZ44+Csn2-1i (harboring a CRISPR1-interfering plasmid construct) showed no alterations in either of the two analyzed CRISPR loci, demonstrating that the generation of non-CRISPR-mediated BIMs was possible in this strain using this method (Tables S3 and S9). Fifty percent of BIMs generated using the WT strain harboring the pNZ44+2Csni construct (targeting both the CRISPR1-Cas and CRISPR3-Cas systems; Fig. 4C) showed no alterations in any CRISPR locus, indicating that this construct is more effective in reducing the acquisition of CRISPR spacers in response to phages in strain STB. One of the BIMs (here designated STB BIM3) generated using this silencing construct was subjected to further characterization by CRISPR sequencing (Table S2), phage sensitivity assays (Table S4), and adsorption assays (Table S5). This BIM, despite lacking additional spacers in any CRISPR locus, exhibited a reduction in sensitivity to phages 9851 and 9854 (both cos-containing phages) while remaining sensitive to 9853 (a pac-containing phage; Table S4). This BIM can be said to have a wider-ranging phage resistance profile than first-generation CRISPR-mediated BIMs STA BIM1 and STB BIM1, although the reduction in the EOP of phages 9851 and 9854 is slightly less than that exhibited by the CRISPR-based immunity of STB BIM1 to 9854. STB BIM3 was also shown to adsorb phage 9854 at an approximately 20%-reduced level (compared to the wild-type strain; Table S5). Despite this decrease in adsorption level, this result was not statistically significant, rendering us unable to define STB BIM3 as an adsorption-deficient BIM. Nonetheless, in the absence of CRISPR spacer addition as an explanation for the reduction in phage sensitivity (Tables S2 and S4), STB BIM3 may be defined as a non-CRISPR-mediated BIM, establishing the proof of principle of non-CRISPR BIM generation in S. thermophilus using the above-described silencing protocol.
Conclusions.
Several mechanisms of phage resistance have been described in S. thermophilus, with the CRISPR-Cas system being by far the most widely studied. The level of detailed research into these systems has confirmed their integral role in the resistance of S. thermophilus to bacteriophages (26, 29, 30, 39, 43) and now has progressed to the characterization of so-called anti-CRISPR proteins expressed by certain phages (54).
Mills and colleagues (55) generated a selection of BIMs, wherein CRISPR alteration was observed to be an unpredictable process, with different numbers of spacers being acquired (or deleted, indicating an inherent instability in the system) in the same strain, even when similar phages were used in the challenge. This unpredictability, as well as the response of the phages to CRISPR spacer acquisition, namely, the modification of single nucleotides on the phage genomes in order to escape CRISPR targeting (40, 42), suggests that further phage robustness can be achieved by utilizing other phage resistance mechanisms present in S. thermophilus (in combination with CRISPR). Indeed, this has been shown to be extremely effective in S. thermophilus by Dupuis and colleagues using a representative strain (DGCC7710) (43). The purpose of the present study was to develop a useful tool which can be utilized to select non-CRISPR-mediated BIMs, which may not be possible using standard BIM generation methods due to the inherent activity of the system.
The cas9-csn2 silencing method outlined here was successful in preventing spacer acquisition in CRISPR loci during standard BIM generation experiments. Introducing a CRISPR1-associated csn2-1 targeting plasmid in STB resulted in 66% of BIMs characterized having acquired spacers in CRISPR3 (and none in CRISPR1), a clear increase in the detection of CRISPR3-mediated BIMs compared to those BIMs produced by the WT strain (of which 90% showed spacer acquisition in the CRISPR1 repeat/spacer locus). To eliminate or at least reduce CRISPR spacer acquisition in either system, it was therefore necessary to target both csn2-1 and csn2-3 using a combinatory, antisense-mediated silencing plasmid construct. While a number of CRISPR1 spacer acquisitions were detected using this construct (possibly due to the increased competition for the resultant antisense mRNA transcript), employing this method also resulted in the production of a number of apparently non-CRISPR-mediated BIMs.
In this study, we present data relating to the generation and characterization of non-CRISPR-mediated BIMs, specifically those mediated by an apparent adsorption inhibition mechanism. The nature of phage receptors in S. thermophilus has been characterized and proposed to be carbohydrate (56, 57). BIMs which are deficient in phage adsorption have previously been described, having been selected by immunolabeling followed by flow cytometry (58), mutant library construction followed by phage challenge (7), or chemical mutagenesis (13). However, these BIMs were not described as having the phenotypic characteristics of the adsorption-deficient BIMs described above, namely, sedimentation and increased cell chain length. The non-CRISPR-mediated BIMs generated in this study displayed some advantages over CRISPR-mediated BIMs, in that they were resistant to a wider range of phages or to a higher level (with no phage escape mutants being observed during standard plaque assays using STA BIM2). Furthermore, it was shown previously that S. thermophilus BIMs mediated by both CRISPR and a restriction/modification (R/M) system are resistant to high levels of phages (43), and it was speculated that CRISPR-Cas systems combined with other antiphage mechanisms also exhibit highly protective effects (also suggested by Viscardi et al. [58]).
The method of BIM generation described above offers a number of advantages over traditional methods of derivation. First, no specialized equipment or chemical mutagenesis is required for its implementation. While the method relies on RNA silencing through plasmid transformation in order to decrease the activity of the CRISPR system(s), the plasmid may be removed prior to industrial implementation. Additionally, the use of antibiotic selection is not required once BIM generation has been carried out. This method may also be used to bias the addition of CRISPR spacers toward either the CRISPR1 or CRISPR3 repeat/spacer locus. This may reduce the risk of spacer deletion or replacement in second-generation CRISPR BIMs, a process which is thought to function in eliminating older (and therefore less useful) spacers, limiting the size of the CRISPR loci (26, 55). In the case of a hypothetical population of BIMs in which the mechanism of resistance is unknown, this method also may be used to rapidly determine if phage insensitivity is conferred by the CRISPR-Cas system alone or by a combination of mechanisms. Lastly, this method may be employed to generate a diverse collection of S. thermophilus BIMs which have become resistant to phage by distinct mechanisms of action for use in industrial strain blends or rotational schemes.
MATERIALS AND METHODS
Isolation, growth, and storage of bacterial strains and bacteriophages.
Bacterial strains and bacteriophages applied in this study are listed in Table 1. S. thermophilus strains were routinely grown from 20% reconstituted skimmed milk (RSM) stocks or from a single colony overnight at 42°C in M17 broth (Oxoid, Hampshire, United Kingdom) supplemented with 0.5% lactose (LM17) or on plates containing 10 g/liter technical agar (Merck, Darmstadt, Germany). Phages were isolated from industrial whey samples and purified by two rounds of single-plaque purification as previously described (59). Phage enumeration assays were performed as previously described (48), using LM17 broth supplemented with 0.25% glycine (Oxoid, United Kingdom), 10 mM CaCl2 (Oxoid, United Kingdom), and either 10 g/liter (solid agar base) or 4 g/liter (semisolid overlay) technical agar. L. lactis NZ9000 (60) was maintained as described above with the following modification: overnight cultures were grown at 30°C with the use of glucose (Sigma-Aldrich, St. Louis, MO) instead of lactose. All transformants were maintained as described above with the addition of chloramphenicol (Cm; Sigma-Aldrich) to a final concentration of 5 μg/ml (LM17+Cm5 or GM17+Cm5).
BIM generation and validation.
BIMs of S. thermophilus ST47795 (strain STA) and ST64985 (strain STB) were generated by adding 300 μl fresh overnight bacterial culture and 10 μl undiluted phage lysate (at a titer of approximately 1 × 107 PFU/ml) to 4 ml of semisolid agar (as described above) in a modification as outlined in a previously published plaque assay method (48). Colonies visibly growing in the semisolid agar following overnight incubation (where the bacterial lawn failed to grow due to the presence of phages) were twice single-colony purified and subjected to phage challenge (as described above) and selected for further characterization (as described below). In cases where an insufficient number of BIMs were isolated using this method, the amount of culture and/or phage lysate added was increased or the method repeated. Sensitivity of wild-type and derived BIMs to phages was determined using spot and/or plaque assays, as previously described (48). The relative efficiency of plaquing (EOP) of phages on wild-type strains and derived BIMs was determined by dividing the observed titer of the phage on a given BIM host by that on the wild-type strain (each plaque assay having been performed independently three times).
All BIMs generated were subjected to PCR profiling to confirm their relatedness to the relevant parent strain from which they were derived by CRISPR locus sequencing (26). Primers applied in this study are listed in Table 6. CRISPR PCR conditions were the following: 95°C for 10 min, followed by 30 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for either 2 min 45 s (CRISPR1), 1 min (STB CRISPR2, CRISPR3, and CRISPR4), or 1 min 30 s (STA CRISPR2 and CRISPR3), with a final extension step of 72°C for 10 min.
TABLE 6.
PCR primers applied during this study
| Primer nameb | Sequencea (5′–3′) | Target | Size of product (approx; bp) | Source |
|---|---|---|---|---|
| pNZ44F | CTAATGTCACTAACCTGCCCCG | pNZ44 MCS | Template dependent | This study |
| pNZ44R | GCTTTATCAACTGCTGCT | pNZ44 MCS | Template dependent | This study |
| STACas9iF | AGCAGCTCTAGAGTCGTTAGAGGGAGGATTAC | STA Cas9 | 3,366 | This study |
| STACas9iR | AGCAGCCTGCAGTTAAAAATCTAGCTTAGGC | STA Cas9 | This study | |
| STBCas9-1iF | AGCAGCTCTAGAGTTGCGAATTTTCAGATAC | STB CRISPR1-Cas9 | 3,366 | This study |
| STBCas9-1iR | AGCAGCCTGCAGGTAACTGTGTAAGGCGCC | STB CRISPR1-Cas9 | This study | |
| STBCas9-3iF | AGCAGCCTGCAGAAGGAGAAATGTATGACTAAG | STB CRISPR3-Cas9 | 4,167 | This study |
| STBCas9-3iR | AGCAGCCCATGGCTGGCTCTAGTTTAGGGTATT | STB CRISPR3-Cas9 | This study | |
| STBCsn2-1iF | AGCAGCCTGCAGCAGTGATAATAAGTTGGTGGT | STB CRISPR1-Csn2 | 1,053 | This study |
| STBCsn2-1iR | AGCAGCCCATGGCTGTCCTTGTCAATCCTTAC | STB CRISPR1-Csn2 | This study | |
| STBCsn2-3iF | AGCAGCTCTAGAGCCAATTCAGAGGAAAGG | STB CRISPR3-Csn2 | 660 | This study |
| STBCsn2-3iR | AGCAGCCTGCAGCAAGATGTGACTGTCACC | STB CRISPR3-Csn2 | This study | |
| CRISPR1F | TGCTGAGACAACCTAGTCTCTC | CRISPR1 repeat/spacer loci | Template dependent | 26 |
| CRISPR1R | TAAACAGAGCCTCCCTATCC | CRISPR1 repeat/spacer loci | Template dependent | 26 |
| CRISPR2F | TTAGCCCCTACCATAGTGCTG | CRISPR2 repeat/spacer loci | Template dependent | 26 |
| CRISPR2R | TAGTCTAACACTTTCTGGAAGC | CRISPR2 repeat/spacer loci | Template dependent | 26 |
| CRISPR3F | CTGAGATTAATAGTGCGATTACG | CRISPR3 repeat/spacer loci | Template dependent | 26 |
| CRISPR3R | GCTGGATATTCGTATAACATGTC | CRISPR3 repeat/spacer loci | Template dependent | 26 |
| CRISPR4F | GATTCAGTTCCTCATAGAGC | CRISPR4 repeat/spacer locus | Template dependent | This study |
| CRISPR4R | GACCTCAACCAATCGATTG | CRISPR4 repeat/spacer locus | Template dependent | This study |
| AC1g1 (internal) | CCTGTCATCTCTGGGAGT | STA CRISPR1 | NA | This study |
| AC1g2 (internal) | CGGTGTTCTATATCGAGGTC | STA CRISPR1 | NA | This study |
| AC1g3 (internal) | GTGAATGGGAAACTGACGGAA | STA CRISPR1 | NA | This study |
| AC3g1 (internal) | CAATCCGTAGCCACACCT | STA CRISPR3 | NA | This study |
| BC1g1 (internal) | CACTTGGCAGGCTTATTACTC | STB CRISPR1 | NA | This study |
| BC1g2 (internal) | CATCCGGTAACTGCTCAAGTG | STB CRISPR1 | NA | This study |
Incorporated restriction sites, where applicable, are underlined.
Internal gap-closing primers are indicated by a “g” and have been applied to all derivatives of the indicated parent S. thermophilus strain.
PCR-generated products were visualized on a 1% agarose (Fisher Scientific, USA) gel and purified using a JetQuick PCR purification spin kit (Genomed, Lohne, Germany). Sanger sequencing (of all PCR products and plasmids) was performed by MWG Biotech (Eurofins, Ebersberg, Germany) to verify the integrity of all plasmid constructs and to compare the sequences of the CRISPR loci of the BIMs to those of the corresponding parent strain. CRISPRs were assembled using the SeqMan program (DNAStar), and the lengths of the arrays (as well as spacer numbers) were determined using the online CRISPR finder program (61). Detailed analyses of CRISPR repeat/spacer loci (including all BIM-acquired spacer sequences generated as part of this study) are provided in Tables S1, S2, and S6 to S9 in the supplemental material.
Sedimentation, microscopic, and adsorption assays.
Relative sedimentation of S. thermophilus parent strains and derived BIMs was observed by growing (in liquid culture) representatives of each strain or BIM under identical conditions (as described above). Taking care not to disturb the liquid broth, visual assessment of the cultures then was performed to observe relevant growth characteristics, such as pelleting, clumping, and adherence to the walls of the tubes. In all cases, this assay was performed using both glass and plastic tubes in order to account for any possible influence of these materials on observed sedimentation patterns.
Morphological assessment and comparison of parent strains and derived BIMs were performed via wet mount. Five microliters of fresh overnight culture was placed on a glass slide (in duplicate), and a coverslip was immediately placed on top of the sample. Each sample then was visualized using ×63 magnification using a confocal laser scanning microscope and an LSM 5 Exciter (excitation, 488 nm; Carl Zeiss, Jena, Germany). The percent increase in chain length or cells per chain (CPC) of derived BIMs relative to the parent strains was calculated first by determining the average number of individual cells per chain in all samples by counting at least 20 chains per strain. The average increase in length then was expressed as a percentage using the following formula: [(CPCmutant − CPCparent)/CPCparent] × 100%. In all cases, the unpaired Student t test was used to determine significant differences between the data sets obtained from the parent and its derived BIMs.
Adsorption assays were performed as described previously (62), in an adaptation of the protocol as described in a prior publication (63).
Construction of antisense plasmid vectors.
In order to distinguish cas genes associated with distinct CRISPR-Cas systems, a specific nomenclature was devised which was generally in accordance with a previously described naming system (34). Where identically named cas genes were found to be associated with distinct CRISPR systems on a single bacterial genome, a suffix was added to denote this (for example, csn2-1 refers to the gene encoding the Csn2 protein associated with the CRISPR1-Cas system in that strain, etc.). The PCR primers used to amplify csn2-1, csn2-3, cas9-1, and cas9-3 from either STA or STB are listed in Table 6. All primers were designed to incorporate the entire relevant gene, including the Shine-Dalgarno (SD) sequence, with the exception of those targeting cas9-3, which were designed to incorporate approximately 3 kb of this gene (of a total gene size of 4,167 bp), due to the insert size constraint of plasmid pNZ44 (this plasmid becomes unstable when large fragments are cloned into it due to the fact that it is a rolling-circle replication plasmid [64]). To ensure that an antisense product was produced, all cas genes were cloned into the pNZ44 plasmid in reverse orientation relative to the p44 promoter (44). In the case of single-gene constructs, the relevant gene was cloned directly behind the p44 promoter in reverse orientation, whereas in the 2Csni construct (simultaneously targeting the csn2 genes of both the CRISPR1-Cas and CRISPR3-Cas systems, i.e., csn2-1 and csn2-3), csn2-1 was cloned directly behind the p44 promoter, followed by csn2-3, both again in reverse orientation relative to the transcriptional direction of the p44 promoter. PCR amplifications were performed as described above using the appropriate extension time for each gene. PCR product purification and sequencing were performed as described above. Restriction of plasmid vectors and inserts was performed using the appropriate restriction enzyme (Roche, Basel, Switzerland) in a total volume of 200 μl overnight at room temperature (RT). Ligations were performed using T4 DNA ligase (New England BioLabs, Ipswich, MA) in a total volume of 10 μl overnight at RT. In preparing the 2Csni construct, 3 μl of each appropriate insert was used in the ligation reaction. Antisense constructs used in this study are listed in Table 1.
Preparation of competent cells, electrotransformation, and transformant screening.
Competent cells were prepared as described previously (65), with the following modifications. A series of tubes containing 10 ml LM17 or GM17 (for L. lactis) broth and various (from 0.2% to 2.4%) concentrations of threonine (Sigma-Aldrich) were prepared and inoculated (1%) with a fresh overnight culture. The tubes were incubated at 42°C overnight and examined for growth. LM17 broth containing 0.5% sucrose (SLM17; Sigma-Aldrich) and supplemented with the highest level of threonine tolerated by the strains was used to prepare competent cells.
Prior to transformation, plasmid constructs, prepared using the GeneJet plasmid miniprep kit (Thermo Scientific, Waltham, MA), were dialyzed using 0.025-μm MF membrane filters (Merck Millipore, Billerica, MA) for 10 min against sterile distilled water (sdH2O). All constructs were generated in L. lactis NZ9000 prior to their subsequent transfer to S. thermophilus STA or STB. Electrotransformation was performed using freshly prepared competent cells as described above, wherein a mixture of cells (50 μl) and plasmid construct (10 μl) was transferred to a prechilled (on ice) 2-mm electroporation cuvette (Cell Projects, Kent, United Kingdom) and subjected to electroporation at 1.75 kV (S. thermophilus) or 2.0 kV (L. lactis)/200 Ω/25 μF. Following this, 950 μl recovery broth (LM17 or GM17 with the addition of 20 mM MgCl2 and 2 mM CaCl2 [Sigma-Aldrich]) was immediately added, and the transformed cells were recovered at 30°C (L. lactis) or 42°C (S. thermophilus) for 2.5 h prior to spread plating (100 μl) on LM17+Cm5 or GM17+Cm5 agar plates as described above. Presumed transformants were purified on LM17+Cm5 or GM17+Cm5 agar plates and subjected to CRISPR sequencing, phage sensitivity assays (as described above), and plasmid sequencing, which was performed using primers designed outside the multiple cloning site of pNZ44 (pNZ44F and pNZ44R) (Table 6).
Plasmid curing.
To cure pNZ44 and pNZ44 derivative vectors from S. thermophilus transformants, such strains were subjected to at least two overnight passages at 42°C in LM17 in the absence of Cm. Overnight cultures then were 10-fold serially diluted in 1/4-strength Ringers solution (Merck) or a culture streak was performed, and individual colonies were assessed for sensitivity to Cm by streaking on LM17+Cm5 agar plates. Colonies incapable of growth on LM17+Cm5 yet capable of growth in the absence of Cm were defined as presumptive cured transformants and subjected to validation by CRISPR PCR and plasmid preparation as described above. In cases where two overnight passages were not sufficient for plasmid curing (as indicated by the ability to grow on LM17+Cm5), passaging was repeated until curing was achieved and cured derivatives validated as outlined above.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the technical assistance of Eoghan Casey and Ian O'Neill in preparing the confocal micrographs. We also acknowledge Emiel Ver-Loren-van-Themaat for helpful discussions on the bioinformatic analysis of the generated mutant derivative strains.
We gratefully acknowledge the financial support of DSM Food Specialties. T.R.H.M.K. and L.H. are employees of DSM Food Specialties. J.M. is in receipt of a Starting Investigator Research Grant (SIRG) (Ref. No. 15/SIRG/3430) funded by Science Foundation Ireland (SFI). D.V.S. is supported by a Principal Investigator award (Ref. No. 13/IA/1953) through SFI. B.M. is funded by DSM Food Specialties.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01733-17.
REFERENCES
- 1.Mora D, Fortina MG, Parini C, Ricci G, Gatti M, Giraffa G, Manachini PL. 2002. Genetic diversity and technological properties of Streptococcus thermophilus strains isolated from dairy products. J Appl Microbiol 93:278–287. doi: 10.1046/j.1365-2672.2002.01696.x. [DOI] [PubMed] [Google Scholar]
- 2.Quiberoni A, Tremblay D, Ackermann HW, Moineau S, Reinheimer JA. 2006. Diversity of Streptococcus thermophilus phages in a large-production cheese factory in Argentina. J Dairy Sci 89:3791–3799. doi: 10.3168/jds.S0022-0302(06)72420-1. [DOI] [PubMed] [Google Scholar]
- 3.Garneau JE, Moineau S. 2011. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb Cell Fact 10(Suppl 1):S20. doi: 10.1186/1475-2859-10-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guglielmotti DM, Mercanti DJ, Reinheimer JA, Quiberoni ADL. 2011. Review: efficiency of physical and chemical treatments on the inactivation of dairy bacteriophages. Front Microbiol 2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucchini S, Sidoti J, Brussow H. 2000. Broad-range bacteriophage resistance in Streptococcus thermophilus by insertional mutagenesis. Virology 275:267–277. doi: 10.1006/viro.2000.0499. [DOI] [PubMed] [Google Scholar]
- 6.Viscardi M, Capparelli R, Iannelli D. 2003. Rapid selection of phage-resistant mutants in Streptococcus thermophilus by immunoselection and cell sorting. Int J Food Microbiol 89:223–231. doi: 10.1016/S0168-1605(03)00151-X. [DOI] [PubMed] [Google Scholar]
- 7.Duplessis M, Levesque CM, Moineau S. 2006. Characterization of Streptococcus thermophilus host range phage mutants. Appl Environ Microbiol 72:3036–3041. doi: 10.1128/AEM.72.4.3036-3041.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binetti A, Bailo N, Reinheimer J. 2007. Spontaneous phage-resistant mutants of Streptococcus thermophilus: isolation and technological characteristics. Int Dairy J 17:343–349. doi: 10.1016/j.idairyj.2006.05.002. [DOI] [Google Scholar]
- 9.Mills S, Coffey A, McAuliffe OE, Meijer WC, Hafkamp B, Ross RP. 2007. Efficient method for generation of bacteriophage insensitive mutants of Streptococcus thermophilus yoghurt and mozzarella strains. J Microbiol Methods 70:159–164. doi: 10.1016/j.mimet.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Chirico D, Gorla A, Verga V, Pedersen PD, Polgatti E, Cava A, Dal Bello F. 2014. Bacteriophage-insensitive mutants for high quality Crescenza manufacture. Front Microbiol 5:201. doi: 10.3389/fmicb.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee AN. 1969. Use of bacteriophage-resistant mutants to study the nature of the bacteriophage receptor site of Staphylococcus aureus. J Bacteriol 98:519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassford PJ Jr, Diedrich DL, Schnaitman CL, Reeves P. 1977. Outer membrane proteins of Escherichia coli. VI. Protein alteration in bacteriophage-resistant mutants. J Bacteriol 131:608–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez C, Van der Meulen R, Vaningelgem F, Font de Valdez G, Raya R, De Vuyst L, Mozzi F. 2008. Sensitivity of capsular-producing Streptococcus thermophilus strains to bacteriophage adsorption. Lett Appl Microbiol 46:462–468. doi: 10.1111/j.1472-765X.2008.02341.x. [DOI] [PubMed] [Google Scholar]
- 14.Heap H, Lawrence R. 1976. Selection of starter strains for cheesemaking. New Zeal J Dairy Sci 11:16–20. [Google Scholar]
- 15.Burrus V, Bontemps C, Decaris B, Guedon G. 2001. Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl Environ Microbiol 67:1522–1528. doi: 10.1128/AEM.67.4.1522-1528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guimont C, Henry P, Linden G. 1993. Restriction/modification in Streptococcus thermophilus: isolation and characterization of a type II restriction endonuclease Sth455I. Appl Microbiol Biotechnol 39:216–220. doi: 10.1007/BF00228609. [DOI] [PubMed] [Google Scholar]
- 17.Benbadis L, Garel JR, Hartley DL. 1991. Purification, properties, and sequence specificity of SslI, a new type II restriction endonuclease from Streptococcus salivarius subsp. thermophilus. Appl Environ Microbiol 57:3677–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young JC, Dill BD, Pan C, Hettich RL, Banfield JF, Shah M, Fremaux C, Horvath P, Barrangou R, Verberkmoes NC. 2012. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS One 7:e38077. doi: 10.1371/journal.pone.0038077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labrie SJ, Tremblay DM, Plante PL, Wasserscheid J, Dewar K, Corbeil J, Moineau S. 2015. Complete genome sequence of Streptococcus thermophilus SMQ-301, a model strain for phage-host interactions. Genome Announc 3:e00480-15. doi: 10.1128/genomeA.00480-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solow BT, Somkuti GA. 2001. Molecular properties of Streptococcus thermophilus plasmid pER35 encoding a restriction modification system. Curr Microbiol 42:122–128. [DOI] [PubMed] [Google Scholar]
- 21.Moineau S, Walker SA, Holler BJ, Vedamuthu ER, Vandenbergh PA. 1995. Expression of a Lactococcus lactis phage resistance mechanism by Streptococcus thermophilus. Appl Environ Microbiol 61:2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tangney M, Fitzgerald GF. 2002. AbiA, a lactococcal abortive infection mechanism functioning in Streptococcus thermophilus. Appl Environ Microbiol 68:6388–6391. doi: 10.1128/AEM.68.12.6388-6391.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larbi D, Decaris B, Simonet JM. 1992. Different bacteriophage resistance mechanisms in Streptococcus salivarius subsp. thermophilus. J Dairy Res 59:349–357. doi: 10.1017/S0022029900030624. [DOI] [PubMed] [Google Scholar]
- 24.Ali Y, Koberg S, Hessner S, Sun X, Rabe B, Back A, Neve H, Heller KJ. 2014. Temperate Streptococcus thermophilus phages expressing superinfection exclusion proteins of the Ltp type. Front Microbiol 5:98. doi: 10.3389/fmicb.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X, Gohler A, Heller KJ, Neve H. 2006. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 confers superinfection exclusion to Streptococcus thermophilus and Lactococcus lactis. Virology 350:146–157. doi: 10.1016/j.virol.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. 2013. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J 32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, Barrangou R, Banfield JF. 2015. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. mBio 6:e00262-. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. 2013. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun 4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- 30.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 31.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 32.Chylinski K, Makarova KS, Charpentier E, Koonin EV. 2014. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 36.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arslan Z, Wurm R, Brener O, Ellinger P, Nagel-Steger L, Oesterhelt F, Schmitt L, Willbold D, Wagner R, Gohlke H, Smits SH, Pul U. 2013. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res 41:6347–6359. doi: 10.1093/nar/gkt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. 2015. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carte J, Christopher RT, Smith JT, Olson S, Barrangou R, Moineau S, Glover CV III, Graveley BR, Terns RM, Terns MP. 2014. The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol Microbiol 93:98–112. doi: 10.1111/mmi.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 42.Sun CL, Barrangou R, Thomas BC, Horvath P, Fremaux C, Banfield JF. 2013. Phage mutations in response to CRISPR diversification in a bacterial population. Environ Microbiol 15:463–470. doi: 10.1111/j.1462-2920.2012.02879.x. [DOI] [PubMed] [Google Scholar]
- 43.Dupuis ME, Villion M, Magadan AH, Moineau S. 2013. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat Commun 4:2087. doi: 10.1038/ncomms3087. [DOI] [PubMed] [Google Scholar]
- 44.McGrath S, Fitzgerald GF, van Sinderen D. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl Environ Microbiol 67:608–616. doi: 10.1128/AEM.67.2.608-616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SG, Batt CA. 1991. Antisense mRNA-mediated bacteriophage resistance in Lactococcus lactis subsp. lactis. Appl Environ Microbiol 57:1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim SG, Bor Y-C, Batt CA. 1992. Bacteriophage resistance in Lactococcus lactis ssp. lactis using antisense ribonucleic acid. J Dairy Sci 75:1761–1767. doi: 10.3168/jds.S0022-0302(92)77935-1. [DOI] [PubMed] [Google Scholar]
- 47.Sturino JM, Klaenhammer TR. 2002. Expression of antisense RNA targeted against Streptococcus thermophilus bacteriophages. Appl Environ Microbiol 68:588–596. doi: 10.1128/AEM.68.2.588-596.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol 83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 49.McDonnell B, Mahony J, Hanemaaijer L, Neve H, Noben JP, Lugli GA, Ventura M, Kouwen TR, van Sinderen D. 2017. Global survey and genome exploration of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus. Front Microbiol 8:1754. doi: 10.3389/fmicb.2017.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dupont K, Janzen T, Vogensen FK, Josephsen J, Stuer-Lauridsen B. 2004. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl Environ Microbiol 70:5825–5832. doi: 10.1128/AEM.70.10.5825-5832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapot-Chartier M-P, Vinogradov E, Sadovskaya I, Andre G, Mistou M-Y, Trieu-Cuot P, Furlan S, Bidnenko E, Courtin P, Péchoux C. 2010. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J Biol Chem 285:10464–10471. doi: 10.1074/jbc.M109.082958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Layec S, Gerard J, Legue V, Chapot-Chartier MP, Courtin P, Borges F, Decaris B, Leblond-Bourget N. 2009. The CHAP domain of Cse functions as an endopeptidase that acts at mature septa to promote Streptococcus thermophilus cell separation. Mol Microbiol 71:1205–1217. doi: 10.1111/j.1365-2958.2009.06595.x. [DOI] [PubMed] [Google Scholar]
- 53.Magadan AH, Dupuis ME, Villion M, Moineau S. 2012. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One 7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hynes AP, Rousseau GM, Lemay M-L, Horvath P, Romero DA, Fremaux C, Moineau S. 2017. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat Microbiol 2:1374–1380. doi: 10.1038/s41564-017-0004-7. [DOI] [PubMed] [Google Scholar]
- 55.Mills S, Griffin C, Coffey A, Meijer WC, Hafkamp B, Ross RP. 2010. CRISPR analysis of bacteriophage-insensitive mutants (BIMs) of industrial Streptococcus thermophilus–implications for starter design. J Appl Microbiol 108:945–955. doi: 10.1111/j.1365-2672.2009.04486.x. [DOI] [PubMed] [Google Scholar]
- 56.Quiberoni A, Stiefel JI, Reinheimer JA. 2000. Characterization of phage receptors in Streptococcus thermophilus using purified cell walls obtained by a simple protocol. J Appl Microbiol 89:1059–1065. doi: 10.1046/j.1365-2672.2000.01214.x. [DOI] [PubMed] [Google Scholar]
- 57.Binetti A, Quiberoni A, Reinheimer J. 2002. Phage adsorption to Streptococcus thermophilus. Influence of environmental factors and characterization of cell-receptors. Food Res Int 35:73–83. [Google Scholar]
- 58.Viscardi M, Capparelli R, Di Matteo R, Carminati D, Giraffa G, Iannelli D. 2003. Selection of bacteriophage-resistant mutants of Streptococcus thermophilus. J Microbiol Methods 55:109–119. doi: 10.1016/S0167-7012(03)00146-5. [DOI] [PubMed] [Google Scholar]
- 59.Moineau S, Pandian S, Klaenhammer TR. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl Environ Microbiol 60:1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuipers OP, de Ruyter PG, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
- 61.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDonnell B, Mahony J, Neve H, Hanemaaijer L, Noben JP, Kouwen T, van Sinderen D. 2016. Identification and analysis of a novel group of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus. Appl Environ Microbiol 82:5153–5165. doi: 10.1128/AEM.00835-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garvey P, Hill C, Fitzgerald GF. 1996. The lactococcal plasmid pNP40 encodes a third bacteriophage resistance mechanism, one which affects phage DNA penetration. Appl Environ Microbiol 62:676–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiewiet R, Kok J, Seegers JF, Venema G, Bron S. 1993. The mode of replication is a major factor in segregational plasmid instability in Lactococcus lactis. Appl Environ Microbiol 59:358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.