ABSTRACT
Medium- and long-chain 1-alkanol and α,ω-alkanediols are used in personal care products, in industrial lubricants, and as precursors for polymers synthesized for medical applications. The industrial production of α,ω-alkanediols by alkane hydroxylation primarily occurs at high temperature and pressure using heavy metal catalysts. However, bioproduction has recently emerged as a more economical and environmentally friendly alternative. Among alkane monooxygenases, CYP153A from Marinobacter aquaeolei VT8 (CYP153AM.aq; the strain is also known as Marinobacter hydrocarbonoclasticus VT8) possesses low overoxidation activity and high regioselectivity and thus has great potential for use in terminal hydroxylation. However, the application of CYP153AM.aq is limited because it is encoded by a dysfunctional operon. In this study, we demonstrated that the operon regulator AlkRM.aq is functional, can be induced by alkanes of various lengths, and does not suffer from product inhibition. Additionally, we identified a transposon insertion in the CYP153AM.aq operon. When the transposon was removed, the expression of the operon genes could be induced by alkanes, and the alkanes could then be oxyfunctionalized by the resulting proteins. To increase the accessibility of medium- and long-chain alkanes, we coexpressed a tunable alkane facilitator (AlkL) from Pseudomonas putida GPo1. Using a recombinant Escherichia coli strain, we produced 1.5 g/liter 1-dodecanol in 20 h and 2 g/liter 1-tetradecanol in 50 h by adding dodecane and tetradecane, respectively. Furthermore, in 68 h, we generated 3.76 g/liter of 1,12-dodecanediol by adding a dodecane–1-dodecanol substrate mixture. This study reports a very efficient method of producing C12/C14 alkanols and C12 1,12-alkanediol by whole-cell biotransformation.
IMPORTANCE To produce terminally hydroxylated medium- to long-chain alkane compounds by whole-cell biotransformation, substrate permeability, enzymatic activity, and the control of overoxidability should be considered. Due to difficulties in production, small amounts of 1-dodecanol, 1-tetradecanol, and 1,12-dodecanediol are typically produced. In this study, we identified an alkane-inducible monooxygenase operon that can efficiently catalyze the conversion of alkane to 1-alkanol with no detection of the overoxidation product. By coexpressing an alkane membrane facilitator, high levels of 1-dodecanol, 1-tetradecanol, and 1,12-dodecanediol could be generated. This study is significant for the bioproduction of medium- and long-chain 1-alkanol and α,ω-alkanediols.
KEYWORDS: 1,12-dodecanediol; AlkL; CYP153A operon; fed-batch culture; recombinant E. coli
INTRODUCTION
Medium- and long-chain α,ω-alkanediols have many applications, including use as industrial lubricants, surfactants, detergents, and personal care products (1). Among these molecules, 1,12-dodecanediol also has applications as a component in advanced coatings, plasticizers, suspending agents, and biomedical syntheses. Additionally, 1,12-dodecanediol can replace 1,6-hexanediol in the synthesis of polyester and polyurethane, offering greater chemical resistance and less water absorption (2).
Alkanediols are not present in nature, and the inertness of alkanes makes their oxyfunctionalization difficult. In industry, primary alkanols are mostly synthesized from olefins by using a hydroformylation method (3). Biocatalytic methods have recently gained attention because of their high stereo- and regioselectivities and mild reaction conditions. The oxidation of alkane termini is the first step of aerobic alkane degradation, and the reaction is catalyzed by alkane monooxygenases. There are various alkane monooxygenases distributed across the three domains of life (4). Among them, CYP52, AlkB, and CYP153 are extensively studied. The CYP52 protein family belongs to the membrane-bound eukaryotic class II cytochromes P450, and they are distributed among many fungi such as Candida tropicalis. This protein hydroxylates and further oxidizes terminal carbons on alkanes and ω-carbons on fatty acids into carboxyl groups that can then be utilized for biomass accumulation and energy generation (5, 6). β-Oxidation-deficient C. tropicalis has already been utilized to industrially produce dicarboxylic acids (7). AlkB protein family members are membrane-integrated, nonheme diiron monooxygenases (8). AlkB requires the redox partners rubredoxin and rubredoxin reductase to act as electron donors in the hydroxylation of the AlkB substrates (9). The most studied AlkB, an AlkB from Pseudomonas putida GPo1 (located in the OCT plasmid), has been used to investigate alkanol, alkanoate, and alkanamine production (10–13). However, the overoxidation of alkanols to fatty acids is also an intrinsic activity of AlkB proteins. The CYP153 protein family is composed of prokaryotic soluble cytochromes P450. Most CYP153 systems belong to three-component class I CYPs, comprising ferredoxin, ferredoxin reductase, and P450 (9). In comparison with CYP52 and AlkB, CYP153A exhibits little overoxidation activity and high regioselectivity, and it is flexible in its choice of redox partners (14, 15). In addition, some CYP153A enzymes also have ω-hydroxylation activity that produces α,ω-alkanediol and ω-hydroxylate fatty acids (16). Among the CYP153A enzymes, CYP153AM.aq, which is encoded by M. aquaeolei VT8 (also known as Marinobacter hydrocarbonoclasticus VT8), has a broad substrate range; however, dysfunction of its native operon has limited its applicability (15, 17).
Whole-cell biocatalyst systems provide several advantages over isolated enzyme systems with respect to hydrocarbon oxyfunctionalization. Whole-cell systems offer spatial confinement, a supply of multiple proteins and cofactors, and the maintenance of consistent pH conditions, which are needed for the optimal activity of alkane monooxygenase (18). Moreover, the bulk organic phase, which is composed of alkanes, acts as a reservoir of substrates and can continuously extract toxic alkanol products from the aqueous medium to reduce toxicity and product inhibition (19). However, obstacles to the generation of useful alkane biotransformations include the overoxidation of alcohol products, product inhibition, toxicity from enzyme activity, organic solvent stress, membrane permeability, and the lack of alkane-inducible systems that respond to medium- to long-chain alkanes. A widely used induction system, AlkS (P. putida GPo1), is activated only by C5-C11 alkanes and dicyclopropylketone (20). The AlkR regulator of Acinetobacter baylyi ADP1 is induced by medium- to long-chain alkanes but is subject to product inhibition (21). Recombinant Escherichia coli that carries CYP153A from Acinetobacter produces only low levels of α,ω-alkanediols, the production of which peaked when 722 mg/liter 1,8-octanediol was generated (16).
Among the known monooxygenases, CYP153A has high regioselectivity with little overoxidation activity. To increase the usefulness of CYP153A, we performed sequence analyses and discovered that the dysfunction of the CYP153AM.aq operon was due to a transposon insertion. After removal of the transposon, the fully functional operon contains an alkane-inducible regulator that can be induced by a wide range of alkanes and fatty acid methyl esters (FAMEs), and the regulator did not exhibit product inhibition. Additionally, we employed an alkane transporter, AlkL, that was driven by a rhamnose-inducible promoter to facilitate medium- and long-chain alkane uptake and therefore increase biotransformation efficiency (22). By coexpressing this operon and AlkL in E. coli, we successfully produced considerable amounts of 1-dodecanol, 1-tetradecanol, and 1,12-dodecanediol in a 5-liter bioreactor.
RESULTS
Sequence analysis of the putative alkane response regulator and promoter.
CYP153AM.aq has high regioselectivity and produces little overoxidation during terminal hydrocarbon hydroxylation. However, the native CYP153AM.aq operon is dysfunctional. To understand the lack of activity of this operon, we performed DNA sequence analyses. A predicted AraC family alkane response regulator (maqu_0596) is located upstream of the CYP153AM.aq operon. In bacteria, operon regulators are commonly located upstream and in the opposite orientation of the polycistronic genes that they regulate. The CYP153A operon regulator that was identified in Gram-positive bacteria has this feature. Interestingly, the protein sequence identities between the M. aquaeolei CYP153A operon and those from Gram-positive bacteria are only 25% (23) (see Fig. S1 in the supplemental material). Phylogenetic analysis strengthened the possibility of AlkRM.aq belonging to the AraC-type regulator (Fig. S2). A tandem repeat (TTGTCNNNAAATGACC, 5 bp apart) was found 62 bp upstream of the start codon of ferredoxin, within the intergenic region between the regulator and the genes that it regulates. The direct repeat is evolutionarily conserved with the operon operator sequences. We therefore postulated that this AraC-type protein is the alkane response regulator that regulates the CYP153A genes.
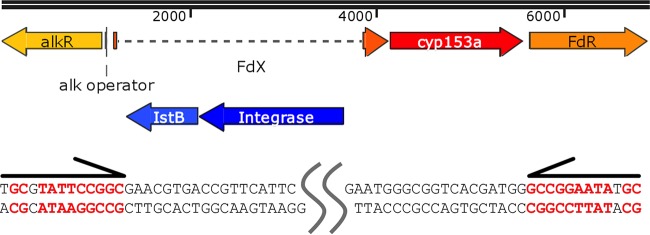
We found a transposon insertion at the N terminus of the ferredoxin coding sequence in the CYP153AM.aq operon by BLAST analysis. The sequence of the CYP153A operon in M. salarius R9SW1 (GenBank accession number NZ_CP007152.1) is almost identical to the M. aquaeolei VT8 CYP153A operon sequence, except for the transposon present in the M. aquaeolei CYP153A operon (Fig. 1). The transposon may cause the loss of function.
FIG 1.
Transposon sequence identification by sequence alignment. A diagram of the native dysfunctional CYP153AM.aq operon is shown in the upper panel. The open reading frames of the transposase and the helper protein are shown in blue. The sequences of these are shown in the lower panel. The transposon belongs to the IS21 family, with 10 other homologs being present in the genome of M. aquaeolei VT8 (90 to 100% identities). All paralogs have the same imperfect terminal inverted repeats (13 bp and 12 bp long, indicated by the arrows above the sequences) and a 6-bp target site duplication. FdX, ferredoxin; FdR, ferredoxin reductase.
An A-rich sequence between the start codon and the ribosome binding site (RBS) of the ferredoxin gene resembles the catabolite activity (CA) motif (AANAANAA) of Pseudomonas and Acinetobacter (24–26). The CA motifs are always located near a start codon and an RBS, and they act as a binding site of the CRC (catabolite repression control) global regulator. In the presence of another preferred carbon source, the CRC regulator posttranscriptionally represses operon expression with the assistance of the Hfq protein (26). A BLAST analysis showed that the A-rich sequence is also present in the CYP153AM.aq operon (Fig. S3). To avoid the possibility of CRC regulation, we expressed the CYP153AM.aq operon in E. coli, which does not have the CRC control mechanism. Moreover, E. coli has less overoxidation activity, and its endogenous aldehyde reductases (ALRs) can even reduce fatty aldehydes to alkanols (27). Therefore, the alkane biotransformation experiments in this study were conducted and optimized in E. coli.
Characterization of the AraC/XylS family alkane response regulator.
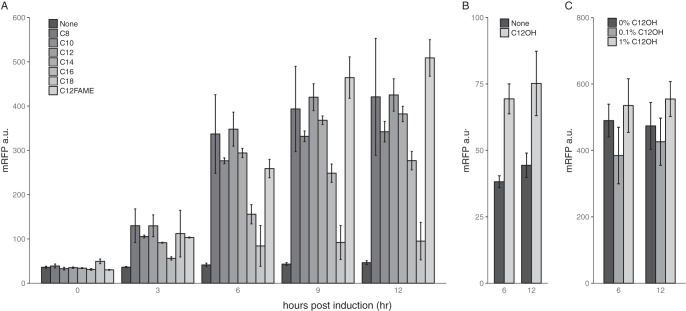
To determine whether the operon regulator AlkR is functional, we fused putative AlkRM.aq with the reporter monomeric red fluorescent protein (mRFP) and assayed reporter expression by induction with different alkanes. As shown in Fig. 2A, n-alkanes between C8 and C16 in size (especially C8 to C14) significantly induced mRFP expression. Methyl laurate (C12-fatty acid methyl ester [FAME]) also has an induction strength similar to that of dodecane. Dodecanol could slightly induce mRFP expression, suggesting that the regulator can be strongly induced by the alkane substrate of the operon but is weakly induced by the alkanol product (Fig. 2B). The presence of 1-dodecanol did not affect induction caused by alkane, indicating that the operon was not subject to product inhibition (Fig. 2C).
FIG 2.
Characterization of the putative CYP153A operon regulator AlkR. (A) AlkR is induced by medium- and long-chain n-alkanes and C12-FAME. (B) AlkR can be weakly induced by 1-dodecanol. (C) The addition of 1-dodecanol does not repress dodecane induction. Abbreviations: None, water for induction control; C8 to C18, alkanes with different carbon numbers; C12-FAME, dodecyl methyl ether; C12OH, 1-dodecanol; a.u., arbitrary units.
To further rule out the possibility that the alkane-responsive promoter may be regulated by endogenous E. coli proteins rather than the AlkR regulator, we partially deleted the regulator and did not observe leaky expression induced by either octane or methyl laurate (Fig. S4). Thus, the regulator is indeed responsible for alkane induction. The characterization of the regulator proved that it is part of the CYP153AM.aq operon.
Reconstitution of the CYP153AM.aq operon.
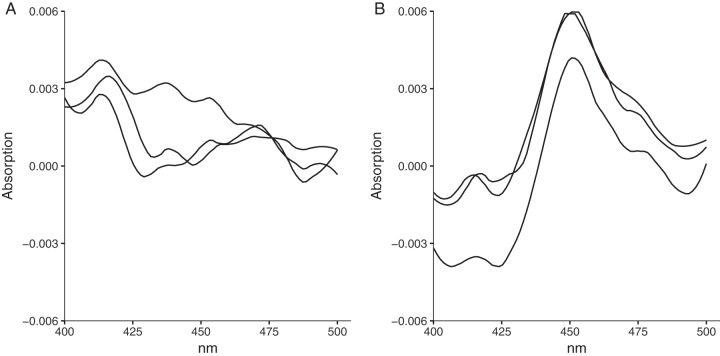
Sequence analysis led us to hypothesize that the dysfunction of the CYP153AM.aq operon might be due to a transposon insertion in the first open reading frame of the polycistronic operon. We first confirmed that the sequence upstream of the transposon contains a functional RBS and a TTG start codon using an enhanced green fluorescent protein (eGFP) translational fusion (Fig. S3 and S5). The CYP153AM.aq operon was then recovered by joining the two fragments that flank the transposon, and the functionality of the reconstituted operon was tested in both P. putida mt-2 and E. coli JM109 cells. When octane was used as the sole carbon source, wild-type P. putida did not grow. In contrast, P. putida carrying the restored CYP153AM.aq operon grew (Fig. S6). This result suggested that P. putida carrying the CYP153AM.aq operon could oxidize octane to 1-octanol, which can be further oxidized to 1-octanoic acid and utilized as a fatty acid. E. coli JM109 could express the CYP153A protein upon methyl laurate induction (Fig. S7). The carbon monoxide (CO) difference spectral assay confirmed that CYP153A was functionally expressed (Fig. 3A and B). Moreover, the E. coli strain carrying the reconstituted CYP153AM.aq operon could convert octane to 113 mg/liter 1-octanol in 12 h. These results also attested that the AlkRM.aq regulator is part of the CYP153AM.aq operon and that the restored operon can be induced by alkane to produce 1-alkanol. The scheme for reconstituted functional CYP153AM.aq is shown in Fig. S8 in the supplemental material.
FIG 3.
Functional expression of the CYP153AM.aq operon in E. coli. Shown are whole-cell CO difference spectra of methyl laurate-induced triplicate cultures. Data for E. coli strains carrying the reporter only (for control experiments) (A) and reconstituted CYP153A (B) are shown.
Alkane transporter AlkL enhances permeability of medium- and long-chain alkanes.
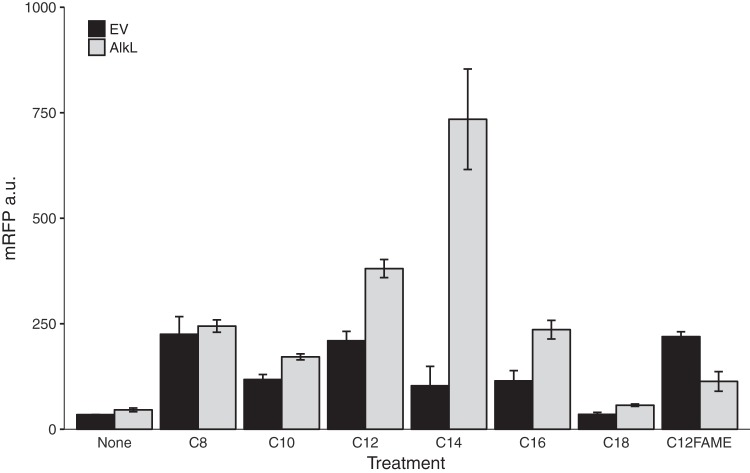
Although both octane and dodecane could activate the CYP153AM.aq operon, only the biotransformation of octane to 1-octanol was observed in flask experiments. The low level of production of dodecanol may be due to the relative impermeability of the E. coli cell envelope to hydrophobic compounds. This property hindered the induction and biotransformation of medium- and long-chain alkanes. To facilitate the biotransformation of these alkanes, we screened several outer membrane facilitators, including the well-characterized alkane transporter AlkL from P. putida (AlkLP.pu) and two putative hydrophobic compound transporters, OmpH and OmpH-0644 from M. aquaeolei. We constructed these transporters under the control of the tunable rhamnose-inducible promoter and a weak RBS to probe their expression levels. We observed that only AlkL could enhance the expression of the mRFP reporter under our experimental conditions (Fig. S9). Increasing the concentration of the inducer might result in stress and reduces mRFP expression. This result is also congruous with the previously reported observation that the AlkL expression level needs to be carefully titrated (11, 13). The expression of AlkL altered the alkane response profile of AlkRM.aq in E. coli, especially increasing induction caused by C12 to C16 alkanes, whereas induction by the C14 alkane was the most improved (Fig. 4). These results were consistent with AlkL facilitating the uptake of C12 to C16 alkanes, and these alkanes are not native substrates of the AlkB system of P. putida GPo1 (11). When the CYP153A operon was coexpressed with the AlkL transporter in a flask culture, 134 mg/liter dodecanol was produced in 12 h. Our results suggested that the combination of the alkane-inducible CYP153AM.aq operon and AlkLP.pu enables E. coli, as a whole-cell biocatalyst, to oxyfunctionalize medium- and long-chain alkanes.
FIG 4.
Expression of AlkL enhances the induction ability of medium- to long-chain alkanes. AlkL facilitated the uptake of different medium- and long-chain alkanes. mRFP signals were measured 6 h after induction. EV, empty vector.
We tested two putative hydrophobic compound transporters from M. aquaeolei in addition to AlkL. These proteins are almost identical to the putative hexadecane transporter of Marinobacter hydrocarbonoclasticus SP17 (28). The putative hydrophobic compound transporter from Marinobacter may have coordinated with the operon from the same strain, resulting in a higher substrate induction efficiency. However, both transporters failed to increase the mRFP signal in E. coli under all tested conditions, and even the downstream lipoprotein (maqu_0477), which may be its operon partner, was coexpressed. We also tried to express these genes behind a stronger RBS, and we found that the expression of the transporter also failed to activate the reporter (data not shown). The nonfunctional expression of these transporters might be due to their improper folding in E. coli or might indicate that more assistance proteins are required.
Biotransformation of dodecane to 1-dodecanol and tetradecane to 1-tetradecanol using 5-liter bioreactors.
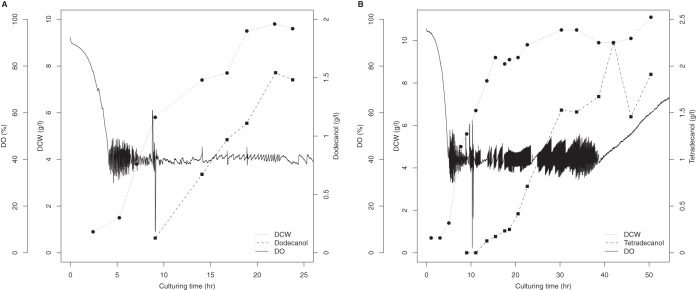
We designed fed-batch biotransformation methods to produce 1-alkanols in a 5-liter bioreactor. When the dissolved oxygen (DO) concentration dropped to 40%, after approximately 9 h, we started feeding the bioreactor continuously and induced biotransformation by the addition of 0.1 g/liter rhamnose. We were able to harvest more than 1.5 g/liter 1-dodecanol after 11 h of induction (Fig. 5A). In contrast, 1-tetradecanol accumulated much more slowly than did 1-dodecanol. After 11 h of induction, only 0.5 g/liter 1-tetradecanol was produced. By extending the culturing time to 50 h, more than 2 g/liter 1-tetradecanol was produced (Fig. 5B). The production rates of 1-dodecanol and 1-tetradodecanl were 85 mg · liter−1 h−1 and 40 mg · liter−1 h−1, respectively.
FIG 5.
1-Alkanol production by fed-batch culture in a 5-liter bioreactor. (A) Bio-oxidation of n-dodecane to 1-dodecanol; (B) bio-oxidation of n-tetradecane to 1-tetradecanol. DCW, dry cell weight.
We were able to oxyfunctionalize dodecane and tetradecane in E. coli using a continuous-growth strategy with the CYP153AM.aq operon and AlkLP.pu. Less overoxidation occurs during oxyfunctionalization with CYP153AM.aq than with AlkB and CYP52 (15). During biotransformation, levels of C12/C14 1-alkanal and 1-alkanoic acid were below the gas chromatography/mass spectrometry (GC/MS) limit of detection. This result indicates that the expression of the CYP153A operon in E. coli JM109 cells results in limited overoxidation activity. In addition, neither 1,12-dodecanediol nor 1,14-tetradecanediol could be detected in the respective cultures by GC/flame ionization detector (FID), indicating that CYP153AM.aq has a higher affinity for alkane than 1-alkanol.
Biotransformation of a dodecane–1-dodecanol mixture into 1,12-dodecanediol in a 5-liter bioreactor.
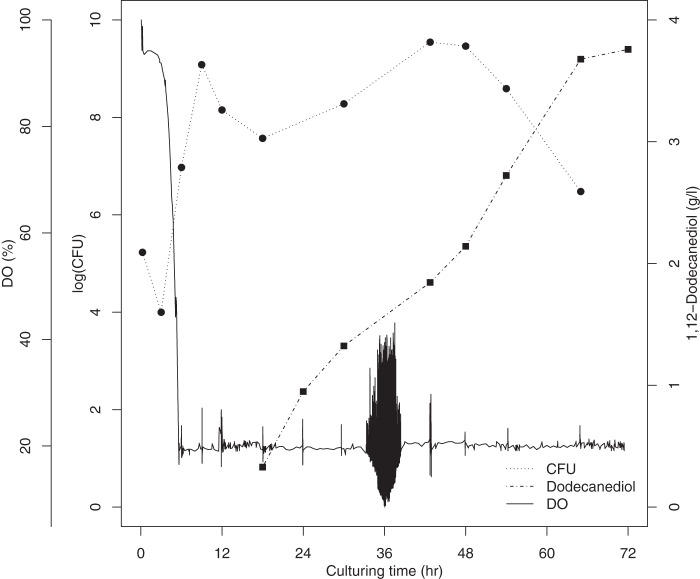
Alkane monooxygenases can utilize not only alkanes but also 1-alkanoic acids as the substrates (5). We proposed that α,ω-alkanediols could also be produced by our constructed strain. However, we could detect only 1-dodecanol when dodecane was added. We then tried to add both dodecane and 1-dodecanol as the inducer and the substrate; the culture produced approximately 800 mg/liter α,ω-dodecanediol after 72 h of fed-batch culturing (data not shown). The number of living cells dramatically decreased upon the addition of 1-dodecanol because of its toxicity. To restore cell growth and boost 1,12-dodecanediol production, we decreased the temperature to 25°C upon induction and added substrates twice; 0.1 g/liter rhamnose, 150 g of dodecane, and 30 g of 1-dodecanol were added at each time. Nine hours after the first induction, we began to feed the bioreactor glycerol and yeast extract continuously at rates of 12.5 and 2.5 g/h, respectively, until the end of culturing. The second induction was performed after 40 h of culturing. This strategy resulted in a level of 1,12-dodecanediol production of 3.76 g/liter at 71.5 h (Fig. 6). The rate of production of 1,12-dodecanediol was 55 mg · liter−1 h−1. The total working volume at the end of the biotransformation was approximately 3 liters. Therefore, approximately 11.28 g of product was produced from 300 g dodecane and 60 g 1-dodecanol.
FIG 6.
1,12-Dodecanediol production by fed-batch culture in a 5-liter bioreactor.
During culturing, the color of the E. coli cell pellet was pinkish red after substrate induction, which may have resulted from the heme prosthetic group in the expressed P450 enzyme (29). During the middle to late stages of the fed-batch 1,12-dodecanediol biotransformation, we observed a distinct milk-colored layer in the E. coli cell pellet, and the CFU counts also dramatically decreased (Fig. 6). In addition, the dry cell weight decreased (data not shown). Thus, we suggest that this layer contained cytotoxic 1-dodecanol, which led to cell lysis. However, the remaining bacteria were capable of reproducing and bioconverting 1,12-dodecanediol as we continued feeding. Given this result, we decided to induce the culture twice to decrease the diminished growth. However, the cytotoxicity of alkanols remains the most difficult challenge in whole-cell biotransformation.
Interestingly, during the late 1,12-dodecandiol biotransformation process, we found a traceable amount of dodecyl acetate during GC/MS analysis; it may be derived from the esterification of dodecanol and acetate. The formation of dodecyl acetate reduced the amount of 1-dodecanol in the medium, which reduced cell toxicity as well as substrate availability. The effect of esterification during the biotransformation process should be further clarified.
DISCUSSION
CYP153AM.aq has gained attention for terminal hydroxylation of medium- and long-chain alkanes due to its low overoxidation activity and high regioselectivity. However, the native CYP153AM.aq operon is dysfunctional; therefore, CYP153AM.aq is regulated by an inducible promoter, and its monooxygenase activity functions with redox partners from other species (12, 15). This work demonstrated that a transposon insertion caused the native CYP153AM.aq operon to be dysfunctional (Fig. 1). By the removal of the transposon, the operon became fully functional in E. coli (Fig. 2 and 3). Subsequently, by combining AlkLP.pu coexpression, the operon could oxyfunctionalize dodecane and tetradecane. Fed-batch culturing and the presence of the respective substrates in a 5-liter bioreactor resulted in recombinant E. coli producing 1.5 g/liter 1-dodecanol in 20 h and 2 g/liter tetradecanol in 50 h (Fig. 5). Moreover, the E. coli strain generated 3.76 g/liter 1,12-dodecanediol in 60 h by the addition of a dodecane–1-dodecanol substrate mixture (Fig. 6). This study reports an efficient way to produce medium- and long-chain 1-alkanol and α,ω-alkanediols.
Sequence analysis showed that AlkRM.aq was quite different from AlkR in Gram-positive bacteria, but the CYP153A operon was highly conserved among all species (see Fig. S1 in the supplemental material). These findings indicate that these regulators independently evolved or had diverged for a long time after their common ancestor first emerged, and the CYP153A enzymatic mechanisms are more conserved than those for expression regulation. Considering the phylogenetic relationship among Pseudomonas, Acinetobacter, and Marinobacter, we cannot eliminate the possibility that the operon contains nonconserved CA motifs and is CRC regulated (30). Further studies should test whether the operon is regulated by CRC. If it is free from CRC regulation, the operon can be used for alkane oxyfunctionalization for an even broader range of hosts.
C12/C14 alkane hydroxylation is important for the production of alcohols, carboxylic acid, and amines. However, the difficulties of producing these compounds are different. Products with higher melting points than the culturing temperature, which may have little toxicity toward cells, can accumulate to massive amounts during culturing. For example, the levels of production of 1,12-dodecanedionic acid and 14-hydroxytetradecanoic acid were reported to be 140 and 174 g/liter, respectively (6, 31). In contrast, for products containing hydroxyl groups or amine groups, which remain as a liquid during culturing, production is drastically reduced. For example, the conversion of dodecane to 800 mg/liter 1-dodecanol and 79 mg/liter 1,12-dodecanediol using AlkB monooxygenase or CYP153A with heterologous redox partners was reported (14–16, 32). In addition, only the small-scale biotransformation of alkanamine was tested (12, 33). In this study, we improved the levels of production to 1.5 g/liter 1-dodecanol and 3.76 g/liter 1,12 dodecanediol. The improvements came from the combination of the coexpression of the highly regioselectivity monooxygenase with its redox partners and a long-chain alkane membrane facilitator. To increase production, increased cell viability may be necessary (13). Future investigations will be conducted to screen expression strains and apply a continuous extraction method to immediately remove the product.
In addition to whole-cell biotransformation, cell-free systems are usually used to produce toxic compounds. However, cell-free enzyme conversion is not used for alkanol production currently because the generation of the cofactors for alkane monooxygenase is problematic. Moreover, when agitated, the excess alkane substrate can form small micelles and become evenly distributed into the water phase. These micelles may have larger surface areas and may be more accessible to E. coli cells. Once the toxic alkanol is produced, the alkane micelles can extract it more effectively. These are the advantages that whole-cell biotransformation has over the cell-free system. In the future, we will attempt to compare different production processes, such as a resting-cell strategy and a continuous-extraction strategy.
With respect to 1,12-dodecanediol production, only 11.28 g of product was generated from 300 g of dodecane and 60 g of 1-dodecanol. Although the highest level of production of 1,12-dodecanediol reported to date was observed in this study, the conversion rate can still be improved. However, with our proposed biotransformation method, the remaining substrates can be easily isolated from the aqueous phase and be recycled, lowering the cost of substrates. Thus, the biotransformation method used in this study has market competitiveness.
Bioproduction of medium- and long-chain 1-alkanols and α,ω-alkanediols has many advantages over chemical synthesis. Our study provides a comprehensive design for terminal hydroxylation by E. coli biotransformation, which can be used extensively to further improve the bioproduction process.
MATERIALS AND METHODS
Bacterial strains, genomic DNA extraction, primers, and vectors.
M. aquaeolei VT8 ATCC 700491, purchased from the ATCC, was maintained in Halomonas medium. Pseudomonas putida mt-2 ATCC 33015, obtained from the BCRC (Bioresource Collection and Research Center, Taiwan), was cultured in LB medium. Bacterial genomic DNA was extracted by using a Wizard Genomic DNA purification kit according to the manufacturer's instructions (Promega). BioBrick plasmids were kindly provided by Yen-Rong Chen. All primers and vectors used in this study are listed in Tables 1 and 2, respectively.
TABLE 1.
Primers used in this studya
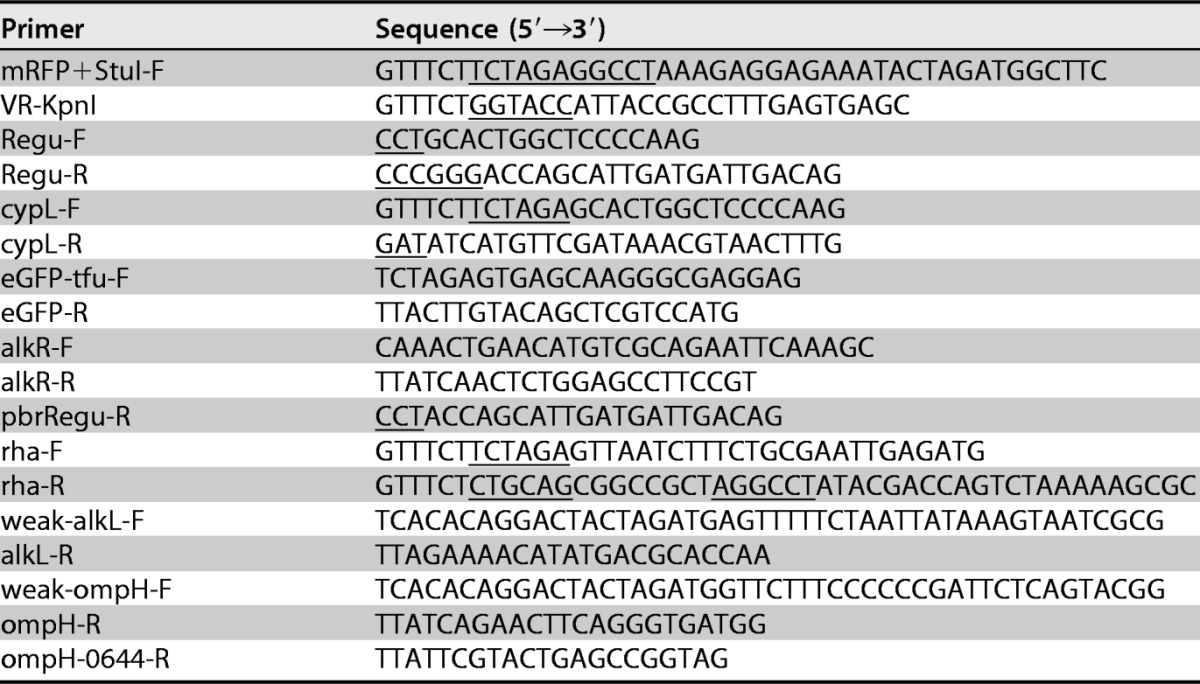
Underlined nucleotide sequences indicate sequences that are required to meet cloning needs, such as restriction enzyme sites or codon completion.
TABLE 2.
Plasmids constructed and used in this worka
| Bacterial strain | Plasmid | Purpose |
|---|---|---|
| E. coli S17-1 | ||
| E. coli JM109 | psb4K5-mRFP | Regulator induction test control |
| E. coli JM109 | psb4K5-AlkR-mRFP | Regulator induction test |
| E. coli JM109 | pBBR1-CYPL-tfu-eGFP | Evaluation of transposon edge |
| E. coli JM109 | pBBR1-CYPLR | Operon functionality |
| E. coli JM109 | pBBR1-Tn::CYPLR | Operon functionality control |
| E. coli JM109 | pBBR1-AlkR | Operon functionality control |
| E. coli JM109 | pBBR1-rha-operon | Operon functionality |
| E. coli JM109 | psb3C5-rha-weak-alkL | Alkane transporter selection and titration |
| E. coli JM109 | psb3C5-rha-weak-ompH | Alkane transporter selection and titration |
| E. coli JM109 | psb3C5-rha-weak-ompH-0644 | Alkane transporter selection and titration |
| P. putida mt-2 | pBBR1-CYPLR | Operon functionality |
| P. putida mt-2 | pBBR1-Tn::CYPLR | Operon functionality control |
| P. putida mt-2 | pBBR1-AlkR | Operon functionality control |
| P. putida mt-2 | pBBR1-alkR-operon | Operon functionality control |
All the listed plasmids were generated for this study.
Gene construction.
Red fluorescent protein (mRFP) was PCR amplified from the BioBrick BBa_J04450 plasmid by using primers mRFP+StuI-F and VR-KpnI and cloned into the low-copy-number BioBrick vector psb4K5 (pSC101 replicon), resulting in plasmid psb4K5-mRFP. The putative alkane response regulator (Maqu_0596) was cloned with primers Regu-F and Regu-R and ligated into plasmid psb4K5-mRFP to create plasmid psb4K5-CypR-mRFP.
The complete, transposonless CYP153AM.aq operon was constructed by ligating together the sequences that were upstream and downstream of the transposon. The upstream region was amplified by using primers cypL-F and cypL-R, and the product was then ligated into the pBBR1-rha vector, resulting in the pBBR1-CYPL plasmid. To evaluate the tail end of this fragment, we cloned eGFP using primers eGFP-tfu-F and eGFP-R and ligated the product with the pBBR1-CYPL vector, resulting in plasmid pBBR1-CYPL-tfueGFP. To assay the functionality of the operon, the fragment that was downstream of the transposon was PCR amplified by using primers cypR-F and cypR-R, and the product was ligated into pBBR1-CYPL to create pBBR1-CYPLR. This plasmid was used for protein expression and whole-cell biotransformation. For control experiments, a transposon-containing operon was cloned by using primers cypL-F and cypR-R, and the product was ligated into the pBBR1-rha vector, resulting in pBBR1-Tn::CYPLR. A regulator-only plasmid, pBBR1-CypR, was created by amplifying the template DNA with primers cypL-F and pbrRegu-R and ligating the produced fragment into the pBBR1-rha vector.
The nucleotide fragment containing rhaR, rhaS, and the rhaPBAD promoter was amplified from E. coli JM109 genomic DNA by using primers rha-F and rha-R. The PCR product was ligated into pBBR1 MCS-2 to create broad-host-range rhamnose-inducible plasmid pBBR1-rha.
Alkane induction assay.
E. coli JM109 cells containing plasmids were grown in 100 ml of LB broth supplemented with 50 mg/liter kanamycin. After the optical density at 600 nm (OD600) of the culture had reached 0.6, 1% (vol/vol) hydrocarbons of different lengths and rhamnose (when AlkL was present) were added to the cells. One milliliter of each sample was collected 0, 3, 6, 9, and 12 h after hydrocarbon induction. Cells were harvested, washed, and resuspended in phosphate-buffered saline (PBS). The mRFP fluorescent signals were detected and quantified by using flow cytometry (BD FACSCanto II).
CYP153AM.aq operon expression and functional assays.
Cultures of E. coli JM109 cells harboring pBBR1-CYPLR, pBBR1-CypR, pBBR1-rha-operon, and pBBR1-rha grown overnight were subcultured in LB with 50 mg/liter kanamycin. When the OD600 of the cultures reached 0.6, the cultures were induced with either 1% (vol/vol) methyl laurate or 2 g/liter rhamnose and then incubated at 30°C for 12 h. The samples were then centrifuged, washed, and resuspended in PBS. After the samples had been boiled in sample buffer (dithiothreitol [DTT]) for 10 min, protein expression was analyzed by SDS-PAGE.
P450 expression was confirmed by whole-cell CO difference spectral assays (34, 35). The bacterial cultures were chilled on ice for 10 min, pelleted by centrifugation at 3,000 × g for 5 min, washed twice with PBS, and then resuspended in P450 spectrum buffer (100 mM Tris-HCl, 20% [vol/vol] glycerol, and 1 mM EDTA [pH 7.4]). A few grains of Na2S2O4 (sodium dithionite) were added to the samples, which were then mixed by gentle inversion. After 10 min of reaction, the baseline was set by scanning the samples from 400 to 500 nm with a photometer. CO gas was then bubbled through samples at a speed of 1 bubble per s for a minute, and the samples were scanned again from 500 to 400 nm. P450 protein expression can be corroborated by the presence of an absorption peak near 450 nm.
To assay biotransformation in flasks, E. coli JM109 cells harboring the vectors being tested were cultured in 100 ml LB with appropriate antibiotics until the OD600 reached 0.6, at which point 1% (vol/vol) alkane and 0.1 g/liter of rhamnose (when AlkL was present) were added. Biotransformation was carried out at 25°C for 12 h. After conversion, the medium was extracted for 1-alkanol and α,ω-alkanediol detection.
Alkane transporter titration.
An RBS with weak transcriptional activity was adapted from BioBrick BBa_B0033 plasmids and incorporated into the forward primers. The transporter and putative transporter AlkL genes, which were cloned from the P. putida GPo1 AlkB operon, were PCR amplified by using primers weak-alkL-F and alkL-R. The M. aquaeolei VT8 homologs of the putative hexadecane transporters OmpH and OmpH-0644 were also cloned by using primers weak-ompH-F, ompH-R, and ompH-0644-R. The PCR products were fused with the RBS and cloned into the psb3C5-rha plasmid. Cell growth and induction using different concentrations of rhamnose were performed as described above. The mRFP reporter plasmid and signal detection are described above.
Quantification of 1-alkanols and α,ω-alkanediols.
An internal standard (1-octanol for C12/C14 biotransformation or 1-dodecanol for C8 biotransformation) was added to the sample, and the sample was extracted with ethyl acetate. To quantify alkanols and alkanediols, 60 μl of the ethyl acetate phase was added to 60 μl of 1% trimethylchlorosilane in N,O-bis(trimethylsilyl) trifluoroacetamide, and the sample was incubated at 75°C for 30 min for derivatization to be completed. Samples were analyzed on a GC/FID or GC/MS instrument equipped with an HP-5ms column (Agilent Technologies). The column length and internal diameter were 30 m and 0.25 μm, respectively. Nitrogen was used as the carrier gas at a flow rate of 1 ml/min. The injector temperature was set at 300°C. The column oven was set at 50°C for 2 min and then raised to 300°C at 20°C min−1.
Production of 1-dodecanol, 1-tetradecanol, and 1,12-dodecanediol by fed-batch culture in 5-liter bioreactors.
In total, 100 ml of a culture grown overnight was inoculated into a 5-liter bioreactor (catalog number FS-01-110; Major Science) that contained 2 liters of TB (terrific broth) medium with antibiotics to produce 1-dodecanol and 1-tetradecanol. The temperature was set at 30°C, the pH value was maintained at 7.2 by the addition of sodium hydroxide and hydrogen chloride, and the DO level was maintained at 40% (±2%) by varying the stirring speed between 300 and 1,000 rpm. When the DO level dropped to 40%, 0.1 g/liter rhamnose was added to induce AlkL expression. After 1.5 h of induction, 200 ml of dodecane or tetradecane was added to initiate biotransformation. When the DO level started to rise, glycerol was continuously fed at a rate of 12.5 g/h. In contrast, 1.5 liters of TB medium was used as the starting medium to produce 1,12-dodecanediol. The temperature, pH, and DO level were controlled as described above. When the DO level rose, glycerol and yeast extract were continuously fed to the reactor at rates of 12.5 and 2.5 g/h, respectively. On the 12th hour of culturing, the temperature was reset to 25°C, and 200 ml of dodecane, 30 ml of 1-dodecanediol, and 0.1 g/liter rhamnose were added to initiate biotransformation. On the 40th hour of culturing, induction was repeated.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the excellent technical assistance provided by the Technology Commons in the College of Life Science and Center for Systems Biology, National Taiwan University, for GC/MS analysis.
This work was funded by grants from the Ministry of Science and Technology (grant no. 102-2622-B-002-005 and 103-2622-B-002-003) and the Ho Tung Chemical Corporation, Taiwan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01806-17.
REFERENCES
- 1.Youngquist JT, Schumacher MH, Rose JP, Raines TC, Politz MC, Copeland MF, Pfleger BF. 2013. Production of medium chain length fatty alcohols in Escherichia coli. Metab Eng 20:177–186. doi: 10.1016/j.ymben.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griehl W, Ruestem D. 1970. Nylon-12—preparation, properties, and applications. Ind Eng Chem 62:16–22. doi: 10.1021/ie50723a005. [DOI] [Google Scholar]
- 3.Huybrechts DRC, Debruycker L, Jacobs PA. 1990. Oxyfunctionalization of alkanes with hydrogen peroxide on titanium silicalite. Nature 345:240–242. doi: 10.1038/345240a0. [DOI] [Google Scholar]
- 4.Fathepure BZ. 2014. Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front Microbiol 5:173. doi: 10.3389/fmicb.2014.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eschenfeldt WH, Zhang YY, Samaha H, Stols L, Eirich LD, Wilson CR, Donnelly MI. 2003. Transformation of fatty acids catalyzed by cytochrome P450 monooxygenase enzymes of Candida tropicalis. Appl Environ Microbiol 69:5992–5999. doi: 10.1128/AEM.69.10.5992-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu WH, Ness JE, Xie WC, Zhang XY, Minshull J, Gross RA. 2010. Biosynthesis of monomers for plastics from renewable oils. J Am Chem Soc 132:15451–15455. doi: 10.1021/ja107707v. [DOI] [PubMed] [Google Scholar]
- 7.Craft DL, Madduri KM, Eshoo M, Wilson CR. 2003. Identification and characterization of the CYP52 family of Candida tropicalis ATCC 20336, important for the conversion of fatty acids and alkanes to alpha, omega-dicarboxylic acids. Appl Environ Microbiol 69:5983–5991. doi: 10.1128/AEM.69.10.5983-5991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Beilen JB, Funhoff EG. 2007. Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21. doi: 10.1007/s00253-006-0748-0. [DOI] [PubMed] [Google Scholar]
- 9.Nie Y, Chi CQ, Fang H, Liang JL, Lu SL, Lai GL, Tang YQ, Wu XL. 2014. Diverse alkane hydroxylase genes in microorganisms and environments. Sci Rep 4:4968. doi: 10.1038/srep04968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant C, Woodley JM, Baganz F. 2011. Whole-cell bio-oxidation of n-dodecane using the alkane hydroxylase system of P. putida GPo1 expressed in E. coli. Enzyme Microb Technol 48:480–486. doi: 10.1016/j.enzmictec.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Grant C, Deszcz D, Wei YC, Martinez-Torres RJ, Morris P, Folliard T, Sreenivasan R, Ward J, Dalby P, Woodley JM, Baganz F. 2014. Identification and use of an alkane transporter plug-in for applications in biocatalysis and whole-cell biosensing of alkanes. Sci Rep 4:5844. doi: 10.1038/srep05844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladkau N, Assmann M, Schrewe M, Julsing MK, Schmid A, Buhler B. 2016. Efficient production of the nylon 12 monomer omega-aminododecanoic acid methyl ester from renewable dodecanoic acid methyl ester with engineered Escherichia coli. Metab Eng 36:1–9. doi: 10.1016/j.ymben.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Kadisch M, Julsing MK, Schrewe M, Jehmlich N, Scheer B, von Bergen M, Schmid A, Buhler B. 2017. Maximization of cell viability rather than biocatalyst activity improves whole-cell oxyfunctionalization performance. Biotechnol Bioeng 114:874–884. doi: 10.1002/bit.26213. [DOI] [PubMed] [Google Scholar]
- 14.van Beilen JB, Funhoff EG, van Loon A, Just A, Kaysser L, Bouza M, Holtackers R, Rothlisberger M, Li Z, Witholt B. 2006. Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl Environ Microbiol 72:59–65. doi: 10.1128/AEM.72.1.59-65.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheps D, Honda Malca S, Richter SM, Marisch K, Nestl BM, Hauer B. 2013. Synthesis of omega-hydroxy dodecanoic acid based on an engineered CYP153A fusion construct. Microb Biotechnol 6:694–707. doi: 10.1111/1751-7915.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii T, Narikawa T, Sumisa F, Arisawa A, Takeda K, Kato J. 2006. Production of alpha, omega-alkanediols using Escherichia coli expressing a cytochrome p450 from Acinetobacter sp OC4. Biosci Biotechnol Biochem 70:1379–1385. doi: 10.1271/bbb.50656. [DOI] [PubMed] [Google Scholar]
- 17.Honda Malca S, Scheps D, Kuhnel L, Venegas-Venegas E, Seifert A, Nestl BM, Hauer B. 2012. Bacterial CYP153A monooxygenases for the synthesis of omega-hydroxylated fatty acids. Chem Commun (Camb) 48:5115–5117. doi: 10.1039/c2cc18103g. [DOI] [PubMed] [Google Scholar]
- 18.Schrewe M, Julsing MK, Buhler B, Schmid A. 2013. Whole-cell biocatalysis for selective and productive C-O functional group introduction and modification. Chem Soc Rev 42:6346–6377. doi: 10.1039/c3cs60011d. [DOI] [PubMed] [Google Scholar]
- 19.Leon R, Fernandes P, Pinheiro HM, Cabral JMS. 1998. Whole-cell biocatalysis in organic media. Enzyme Microb Technol 23:483–500. doi: 10.1016/S0141-0229(98)00078-7. [DOI] [Google Scholar]
- 20.Grund A, Shapiro J, Fennewald M, Bacha P, Leahy J, Markbreiter K, Nieder M, Toepfer M. 1975. Regulation of alkane oxidation in Pseudomonas putida. J Bacteriol 123:546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratajczak A, Geissdorfer W, Hillen W. 1998. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J Bacteriol 180:5822–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julsing MK, Schrewe M, Cornelissen S, Hermann I, Schmid A, Buhler B. 2012. Outer membrane protein AlkL boosts biocatalytic oxyfunctionalization of hydrophobic substrates in Escherichia coli. Appl Environ Microbiol 78:5724–5733. doi: 10.1128/AEM.00949-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang JL, JiangYang JH, Nie Y, Wu XL. 2015. Regulation of the alkane hydroxylase CYP153 gene in a Gram-positive alkane-degrading bacterium, Dietzia sp. strain DQ12-45-1b. Appl Environ Microbiol 82:608–619. doi: 10.1128/AEM.02811-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno R, Ruiz-Manzano A, Yuste L, Rojo F. 2007. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol Microbiol 64:665–675. doi: 10.1111/j.1365-2958.2007.05685.x. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann T, Sorg T, Siehler SY, Gerischer U. 2009. Role of Acinetobacter baylyi Crc in catabolite repression of enzymes for aromatic compound catabolism. J Bacteriol 191:2834–2842. doi: 10.1128/JB.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno R, Hernandez-Arranz S, La Rosa R, Yuste L, Madhushani A, Shingler V, Rojo F. 2015. The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ Microbiol 17:105–118. doi: 10.1111/1462-2920.12499. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez GM, Atsumi S. 2014. Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab Eng 25:227–237. doi: 10.1016/j.ymben.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaysse PJ, Prat L, Mangenot S, Cruveiller S, Goulas P, Grimaud R. 2009. Proteomic analysis of Marinobacter hydrocarbonoclasticus SP17 biofilm formation at the alkane-water interface reveals novel proteins and cellular processes involved in hexadecane assimilation. Res Microbiol 160:829–837. doi: 10.1016/j.resmic.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Mak PJ, Denisov IG. 2018. Spectroscopic studies of the cytochrome P450 reaction mechanisms. Biochim Biophys Acta 1866:178–204. doi: 10.1016/j.bbapap.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW. 2010. Phylogeny of Gammaproteobacteria. J Bacteriol 192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picataggio S, Rohrer T, Deanda K, Lanning D, Reynolds R, Mielenz J, Eirich LD. 1992. Metabolic engineering of Candida tropicalis for the production of long-chain dicarboxylic acids. Biotechnology 10:894–898. [DOI] [PubMed] [Google Scholar]
- 32.Fujii T, Narikawa T, Takeda K, Kato J. 2004. Biotransformation of various alkanes using the Escherichia coli expressing an alkane hydroxylase system from Gordonia sp TF6. Biosci Biotechnol Biochem 68:2171–2177. doi: 10.1271/bbb.68.2171. [DOI] [PubMed] [Google Scholar]
- 33.Wood AJL, Weise NJ, Frampton JD, Dunstan MS, Hollas MA, Derrington SR, Lloyd RC, Quaglia D, Parmeggiani F, Leys D, Turner NJ, Flitsch SL. 11 October 2017. Adenylation activity of carboxylic acid reductases enables the synthesis of amides. Angew Chem Int Ed Engl doi: 10.1002/anie.201707918. [DOI] [PubMed] [Google Scholar]
- 34.Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem 239:2379–2385. [PubMed] [Google Scholar]
- 35.Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.