Summary
Antibodies are highly functional glycoproteins capable of providing immune protection through multiple mechanisms, including direct pathogen neutralization and the engagement of their Fc portions with surrounding effector immune cells that induce anti‐pathogenic responses. Small modifications to multiple antibody biophysical features induced by vaccines can significantly alter functional immune outcomes, though it is difficult to predict which combinations confer protective immunity. In order to give insight into the highly complex and dynamic processes that drive an effective humoral immune response, here we discuss recent applications of ‘Systems Serology’, a new approach that uses data‐driven (also called ‘machine learning’) computational analysis and high‐throughput experimental data to infer networks of important antibody features associated with protective humoral immunity and/or Fc functional activity. This approach offers the ability to understand humoral immunity beyond single correlates of protection, assessing the relative importance of multiple biophysical modifications to antibody features with multivariate computational approaches. Systems Serology has the exciting potential to help identify novel correlates of protection from infection and may generate a more comprehensive understanding of the mechanisms behind protection, including key relationships between specific Fc functions and antibody biophysical features (e.g. antigen recognition, isotype, subclass and/or glycosylation events). Reviewed here are some of the experimental and computational technologies available for Systems Serology research and evidence that the application has broad relevance to multiple different infectious diseases including viruses, bacteria, fungi and parasites.
Keywords: antibody, Fab, Fc, Fc receptors, vaccine
Introduction
In 1796, Edward Jenner inoculated a child with matter from a cowpox sore on a milkmaid's hand, and noted that the child was then protected against smallpox infection.1 This event was the beginning of modern‐day vaccines, which have transformed society and saved millions of lives. As the success of vaccines has been wonderfully beneficial, it has influenced our approach to the study and treatment of infectious diseases. Vaccination methods today remain largely based on broad single‐target approaches, similar to those first employed by Jenner more than 200 years ago.2 More specifically, many of the currently licensed vaccines focus on inducing a single immune correlate, with the detection of total binding antigen‐specific antibodies or neutralizing antibodies being the most common assessment for protection against pathogens including polio virus, influenza virus, yellow fever virus, hepatitis viruses, human papillomavirus, Bordetella pertussis and pneumococci.3, 4 However, for many of the world's deadliest pathogens, including Ebola virus, Plasmodium falciparum (malaria) and human immunodeficiency virus (HIV), the development of an effective vaccine has been hindered largely by our inability to elucidate the immune correlates of protection by traditional approaches.
The importance of Fc‐mediated functional antibodies for protection and control of diseases
Antibodies are highly functional glycoproteins that are a vital immune component for protection and control of infectious diseases. For a number of vaccines (e.g. polio, influenza, tetanus) neutralizing antibodies against the pathogen or toxins have been identified as the correlates of protection. Interestingly, for many other vaccines (eg. hepatitis A), total pathogen‐specific binding antibodies have been identified as correlates of protection, yet the specific mechanisms behind these pathogen‐specific binding antibodies remain unclear.4 Beyond neutralization, antibodies are capable of providing immune protection through multiple additional mechanisms, via engagement of their Fc (Fragment crystallizable) portions. To date, only one licensed human vaccine (that for pneumococcus) has identified Fc‐mediated functional antibodies as a correlate of protection.5 However, there is growing evidence that supports the role for Fc functional antibodies in the control of a wide range of pathogens including bacterial, viral, fungal and parasitic infections. These antibodies have the unique capacity to bridge the gap between innate and adaptive immunity, by harnessing both the specificity of the humoral adaptive immune response provided by the antibody's Fab (Fragment antigen‐binding) region, which recognizes the pathogen, as well as by rapidly activating Fc Receptor (FcR) innate immune effector cell responses (e.g. complement) via the antibody's Fc region. Activation can induce a range of anti‐pathogenic immune responses including but not limited to antibody‐dependent cellular cytotoxicity (ADCC), antibody‐dependent cellular phagocytosis (ADCP), antibody‐dependent complement activity and antibody‐dependent cytokine, chemokine and/or enzyme release (Fig. 1). Importantly, FcR innate immune effector cells are abundantly located throughout the body and can be recruited by these non‐neutralizing antibodies without any need for prior antigen sensitization.6, 7
Figure 1.
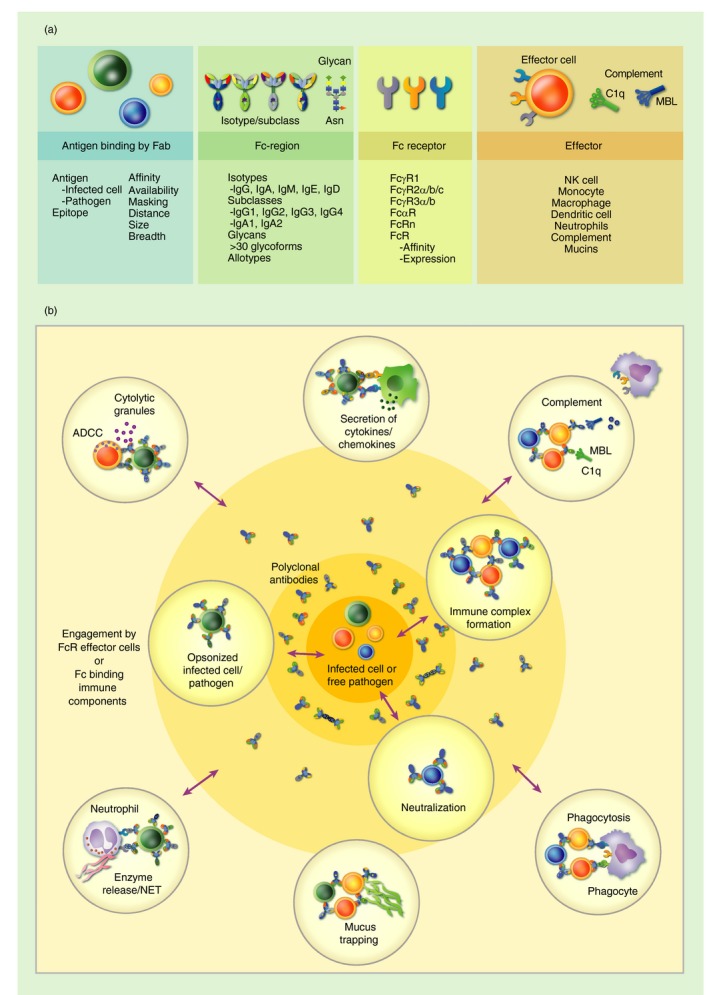
Dynamic complexity of the humoral immune response. (a) The functional capacity of the humoral immune response is determined by complex biophysical antibody features including (i) the pathogen being targeted and the ability of the antibody's Fab to recognize different antigens, (ii) an antibody's Fc region's diversity, which in turn can modulate the antibodies capacity to engage with (iii) Fc receptor/immune molecules and (iv) availability of the Fc receptors on different effector cells/immune molecules in the surrounding environment. (b). The combination of the pathogen targeted (e.g. infected cell versus small infectious particles) and binding by an antibody's Fab determines opsonization, neutralization and immune complex formation. The composition of the Fc‐regions of these antibodies can in turn modulate the functional immune response by surrounding effector cells/immune molecules potentially inducing a range of functions including but not limited to ADCC, antibody‐mediated secretion of cytokines, antibody‐mediated enzyme release/NET (neutrophil extracellular trap) formation, antibody‐dependent phagocytosis, antibody‐mediated complement activity, mucus trapping etc., dependent on the cellular Fc receptor expression or immune components available.
Emerging evidence from multiple infectious disease models strongly suggest that functional antibodies are important for mediating control and/or protection against viral, bacterial, fungal and parasitic pathogens. Moreover, the fact that several bacterial (e.g. Streptococcus8) and viral (e.g. herpes simplex virus9) pathogens have evolved to encode proteins that specifically protect them from Fc‐mediated antibody functions,10 further supports the notion that these non‐neutralizing anti‐microbial properties of antibodies play a vital role in protection from infectious diseases. Examples of the importance of Fc functional antibodies in the control and/or protection of different pathogens are summarized in Table 1.
Table 1.
Examples of functional antibodies involved in the control of infectious viral, bacterial, fungal and parasitic pathogens
| Antibody function | Virus | Bacteria | Fungus | Parasite |
|---|---|---|---|---|
| Antibody‐dependent cellular cytotoxicity | Human immunodeficiency virus (HIV)12, 15, 18, 48, 49, 50 Influenza virus,51, 52, 53 Ebola virus,54, 55 Herpes simplex virus56 | Salmonella typhi,57 Chlamydia trachomatis,58 Mycobacterium tuberculosis 33 | Cryptococcus neoformans,59 Aspergillus 60 |
Schistosomiasis25
Strongyloides stercoralis,61 Plasmodium 62 |
| Antibody‐mediated phagocytosis | HIV,15, 17, 45 Influenza virus63, 64 | Salmonella paratyphi A, 65 Clostridium difficile toxin A, 66 Mycobacterium tuberculosis 67 | Paracoccidioides brasiliensis,68 Aspergillus fumigatus 69 | Plasmodium,70 Toxoplasma gondii 71 |
| Antibody‐mediated complement | Ebola virus,55 HIV17, 45 | Pseudomonas aeruginosa, Salmonella,72 Borrelia burgdorferi 73 | Aspergillus fumigatus,69 Candida albicans 74 | Strongyloides stercoralis,61 Plasmodium 75 |
| Antibody‐mediated enzyme and/or cytokine release | HIV,15, 18, 45, 48, 76 Influenza virus52, 53 | Mycobacterium tuberculosis 33 | Paracoccidioides brasiliensis 68 |
Schistosoma,25
Leishmania
77, 80
Plasmodium 78, 79 |
| Non‐neutralizing antibody‐mediated pathogen inhibition | HIV81 | Coxiella burnetii,82 Chlamydia 83 | Plasmodium 62, 84 |
Lessons learned from HIV vaccines trials
Despite three decades of intense research, the development of an effective vaccine against HIV continues to produce lacklustre results. To date, only one human Phase III HIV vaccine trial has shown a modest, but significant, level of efficacy (31·2%).11 Surprisingly, this RV144 vaccine trial did not induce CD8+ T‐cell cellular immunity, broadly neutralizing antibody responses or high antigen‐specific antibody‐binding levels.11, 12 Instead immune correlates analysis identified the importance of antibodies targeting the V1V2 region of the HIV envelope and ADCC activity, in the absence of high levels of IgA.12, 13 Follow‐up analyses discovered additional features of the humoral immune response associated with protection, including the preferential induction of IgG3 responses,14, 15 which were able to mediated multiple antibody effector functions including ADCC, antibody‐mediated cytokine and chemokine production from natural killer cells and ADCP in a coordinated manner, otherwise known as polyfunctional antibody immunity.15
Furthermore, multiple non‐human primate (NHP) simian immunodeficiency virus (SIV)/ simian–human immunodeficiency virus (SHIV) vaccine studies have recently been conducted highlighting the complexity of potential correlates of protection. Administration of an adenovirus vector 26 (AD26) prime followed by an envelope protein boost in NHP was able to provide 50% protection against repetitive SIV challenges.16 Interestingly, protective efficacy was not associated with a neutralization, but instead polyfunctional antibody immune responses (incorporating six different antibody Fc functions) were associated with protection.16 Similarly, other NHP studies have correlated both ADCP and antibody‐dependent complement deposition with protective efficacy.17 More recently, partial protection from SHIV infection was observed in NHP when administered with a canary pox prime (ALVAC)/ recombinant pentavalent envelope protein vaccine.18 Multiple humoral immune correlates were associated with decreased risk of infection, including plasma antibody binding to HIV‐infected cells, ADCC antibody titres, natural killer cell‐mediated ADCC and antibody‐mediated activation of macrophage inflammatory protein‐1β.18
These recent human and NHP HIV vaccine studies have highlighted our limited understanding of humoral immune responses and challenges us to shift our analysis of potential humoral immune correlates from being a univariate or ‘one component at a time’ paradigm (e.g. neutralization or total antibody‐binding titres alone) to a multivariate ‘many components at once’, or systems concept for design of new strategies for more difficult to vaccinate diseases, based on systems‐level properties of humoral immunity or as it has been more simply termed ‘Systems Serology’.19, 20
Complexity of functional antibodies
Upon vaccination or infection by a pathogen, the humoral immune response aims to produce diverse, highly polyclonal antibodies to target the foreign pathogens. The functional capacity of the humoral immune response is determined by multiple cumulative factors defined by an antibody's biophysical features that are modulated by genetic, molecular and environmental factors (Fig. 1 and summarized in Table 2). These include the ability of the antibody to effectively recognize the foreign antigen dictated by an antibody's Fab region, along with the capacity of the antibody to engage with surrounding Fc effector cells and immune components (modulated by the antibody Fc portion).
Table 2.
Antibody biophysical features that can modulate Fc functionality
| Fab | Examples measurements | Example assays | References |
|---|---|---|---|
| Masking/availability, Antigen density | Abundance of antigen available on pathogen/infected cells | 85, 86 | |
| Size |
Smaller pathogen e.g. virus Larger pathogen e.g. parasite, or infected cell |
Immune complex assays | 87, 88 |
| Antigen target |
Protein Glycoprotein Glycan Glycolipid |
Protein, glycan, glycolipid, glycoprotein screening arrays, | 89, 90, 91, 92 |
| Epitope |
Conformational Linear |
Overlapping peptide arrays Protein scaffold arrays Multiplex ELISAs Intracellular Cytokine Staining (ICS) |
93, 94, 95, 96, 97 |
| Antibody–antigen affinity | Equilibrium constant |
Surface plasmon resonance Chaotrope |
98, 99, 100 |
| Distance | Distance from cell membrane | Assays with variable epitope distances | 101 |
| Breadth | Clades, strains, serotypes |
Protein arrays Multiplex |
102, 103 |
| Fc | Examples | Assays | References |
|---|---|---|---|
| Isotype | IgG, IgA, IgM, IgE, IgD |
Multiplex ELISAs |
95, 104 |
| Subclass | IgG1, IgG2, IgG3, IgG4, IgA1, IgA2 |
Multiplex ELISAs |
95, 104 |
| Glycosylation |
Fucose Galactose Bisecting GlcNAC Sialic acid |
Mass spectrometry HPLC CE Multiplex |
31, 33, 104, 105 |
| Allotype |
IgG1 (six alleles) IgG2 (one allele) IgG3 (13 alleles) IgA (three alleles) |
sequencing ELISAs |
106, 107, 108 |
| FcR/Complement binding | C1q, MBL, FcγRI, FcγRIIa, FcγRIIb, FcγRIIIa, FcRγIIIb, FcaR, FcER (and respective polymorphisms) |
ELISA Multiplex |
102, 104 |
| FcR affinity | FcR binding kinetics | Surface plasmon resonance | 109, 110 |
Despite an antibody's Fc region often being referred to as the ‘constant’ region, the Fc is surprisingly diverse, with subtle modifications having the capacity to significantly alter engagement and affinity to FcRs and/or other Fc‐binding immune components, including complement and mucins. These include differences in immunoglobulin isotypes: IgA, IgD, IgE, IgG and IgM, of which IgG is the most predominant immunoglobulin present in healthy human plasma.21 Although each isotype has its own characteristic properties and functions, IgG is most commonly associated with mediating Fc effector responses, although IgA,22 IgM23 and IgE24 also induce vital roles in protective immunity by activating their respective FcR innate immune cells and/or complement. For example, the importance of IgE and activation of FcεR effector cells for protection against parasitic infections has been well documented.25 As an additional level of complexity, immunoglobulin isotypes also express different subclasses. For example, IgG consists of four subclasses, IgG1, IgG2, IgG3 and IgG4, each binding with varying affinity to different FcγRs.26, 27
Beyond subclass, Fc function is also determined by changes in antibody glycosylation, particularly the glycan structure attached at asparagine 297 (Asn297) of the antibody Fc heavy chain,28, 29 which can have important functional consequences by influencing the affinity of IgGs for their respective FcγRs on effector cells and complement proteins. Complete aglycosylation of an antibody abolishes FcγR and complement binding,30 whereas the presence or absence of particular glycan forms can alternatively inhibit or enhance Fc functionality.31, 32 Table 2 summarizes the many different features of the antigen–Fab antibody and antibody Fc–FcR interactions that can modulate Fc functionality and lists example assays available to allow for the in‐depth assessment of these antibody features. Systems Serology aims to use high‐throughput assays, to collate a holistic assessment of all antibody features that can potentially modulate Fc functionality, providing us with a detailed portrait, or humoral immune ‘signature’ associated with protection or control of infection. Although many of these assays have been developed and optimized for use predominately against viruses (especially HIV18, 19), these assays have the potential to be adapted and optimized for examination of other infectious diseases.33
Generating insights into the complexity of the humoral response: Systems Serology
Given the complexity of antibody biophysical features, a quantitative, systems approach will provide new perspective and insight into key quantitative relationships between the features that characterize a vaccine response, confer protection or underpin a desired functional response. A quantitative understanding of relationships between antibody biophysical features, Fc functional responses and clinical outcomes could enable design of new vaccine regimens specifically targeted to enhance or suppress key parts of this system; altering overall network humoral immunity rather than a single component (Fig. 1b). Though advancements in experimental technologies now enable the measurement of large numbers of biophysical antibody features (detailed in Table 2), a major challenge still remains in determining the relative importance of alterations in these antibody features that occur with vaccination, and key quantitative relationships that drive a desired immune response or confer protection. ‘Data‐driven’ modeling (also called ‘machine learning’) approaches34 hold great promise for better understanding antibody systems, as they enable integration of high‐throughput experimental data to mathematically identify relationships between antibody biophysical features that are associated with important functional outcomes, vaccine regimen, or protection/control of infection (Fig. 2). These approaches can be applied as useful hypothesis‐generating tools for new systems‐level mechanisms involving multiple antibody features, and have the potential to accelerate our understanding of the humoral immune system by helping to define areas of interest for further experimental testing and additional quantitative models. The value of data‐driven approaches in identifying gene and transcriptional signatures correlated with vaccine response has been demonstrated in a wide range of vaccinology applications.35, 36, 37 However, many of these previous studies have specifically focused on identifying genetic and transcriptional correlates of vaccine protection, especially for cellular immunity. In contrast, application of Systems Serology instead aims to focus upon gaining insights to functional humoral immunity.
Figure 2.
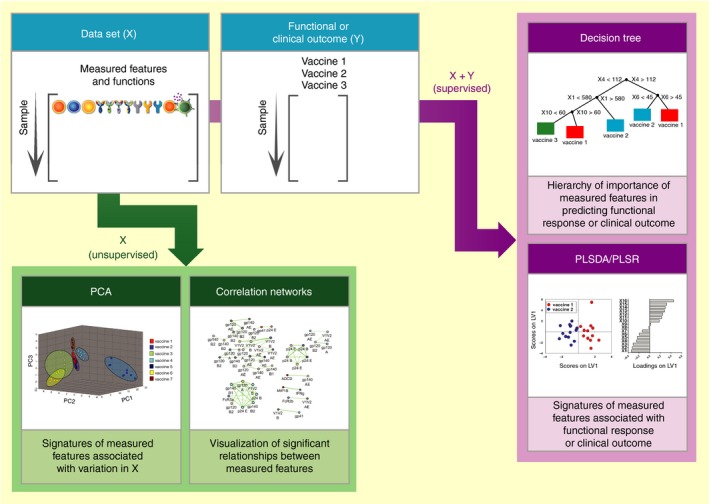
Systems serology data‐driven modeling approaches. Systems Serology involves running high‐throughput experimental assays that measure antibody biophysical and functional data (X) in parallel with functional or clinical outcomes (Y). Upon collation, the data sets can be interrogated by unsupervised and supervised machine learning computational techniques, including principal component analysis (PCA), correlation networks, partial least square discriminant analysis and regression (PLSDA and PLSR), and decision trees. The correlation network figure was kindly contributed by Manu Kumar and Doug Lauffenburger (MIT).
Data‐driven tools: overview and examples
Data‐driven models have the potential to provide both better classification of vaccine responses (e.g. between protective and non‐protective vaccines) as well as give systems‐level insight into networks of antibody biophysical features involved in important functional responses. Altogether, they are able to generate a valuable network ‘picture’ (Fig. 1b) of key events that may contribute to a specific functional immune response or clinical outcome. In general, all data‐driven approaches involve analysis of a large data set (‘X’: Fig. 2). In the case of Systems Serology, this may include measurements of the antibody's biophysical features (e.g. antibody Fab recognition, antibody isotype, glycosylation, Fc receptor; detailed in Table 2) believed to contribute to a particular outcome (e.g. functional response, vaccine regimen, or protection). A subset of data‐driven modelling approaches [including principal component analysis (PCA) and correlation networks] only employ this X data set, searching for significant multivariate relationships between measured features. This subset of approaches is considered ‘unsupervised’ in that they evaluate relationships between features in X without information about an outcome. The strength of unsupervised approaches lies in the ability to search for features involved in the differentiation of outcomes in a completely unbiased way. Systemic, unbiased, examination of broad antibody profiles provides us with a more comprehensive understanding of the mechanisms behind specific functions, potentially revealing novel correlates between antibody features and functions that would not normally be identified by traditional approaches.
Other data‐driven approaches are considered ‘supervised’ [including partial least squares discriminant analysis (PLSDA), partial least squares regression (PLSR) and decision trees, Fig. 2], as they identify key relationships in X that are related to an important functional or clinical outcome (‘Y’; e.g. functional response, vaccine regimen, or clinical outcome; Fig. 2). Supervised approaches are especially useful for gaining mechanistic insight into networks or systems of immune parameters driving an outcome, because they identify direct relationships between the two. Both unsupervised and supervised approaches are useful in Systems Serology research, depending on the question being asked and the nature of the data. One major advantage of all data‐driven approaches is integration, or the ability to merge disparate data sets into a whole. By combining measurements from different sources into the same model, quantitative relationships between biophysical features associated with a clinical or functional outcome can be linked across experimental assays, tissue compartments, and time. Below we give examples of specific data‐driven approaches that have been applied in Systems Serology research. In each case, we leave detailed mathematical descriptions to other published work, but highlight applications, advantages and limitations of each in the context of Systems Serology use.
Unsupervised approaches
Perhaps the simplest way to visualize relationships between many different measured parameters is through the construction of correlation networks (Fig. 2).19, 38 These diagrams allow for the visualization of significant correlative relationships between paired measured features of interest. These networks can be created by first computing either the Pearson (parametric) or Spearman (non‐parametric) correlation coefficient for each pair of measured variables. Relationships across all features can then be visualized through either a web‐like structure or a heat map that indicates the direction and strength of each significant correlation. The main advantage of correlation networks is that they are easy to create and interpret, and so often give useful insight into potential mechanistic relationships between features. One drawback is that they are unsupervised, and do not directly relate identified correlative relationships to a clinical or functional outcome of interest (Y). Therefore they have little use as predictive tools. Additionally, only pairwise relationships between measured features are considered; so, true multivariate signatures involving three or more measured features are unattainable. This approach has been used previously to examine antibody network connectivity between antibody biophysical features and functions associated with the humoral response elicited by four different HIV vaccines (VAX003, RV144, HVTN204 and IPCAVD001).19 Vastly different network topographies or ‘humoral signatures’ were observed between the different vaccines trials and were able to highlight important mechanisms behind the moderately protective RV144 trial. More specifically, IgG1 and IgG3 where highly connected with multiple antibody Fc effector functions including ADCC, ADCP and antibody‐dependent complement deposition, indicating their importance in modulating multiple Fc functions, whereas these interactions were not observed for the other non‐efficacious vaccine trials.
Principal component analysis 39 is an unsupervised approach that can be used to determine signatures of measured features that account for the most variation between samples, in a set of measured features. For example, given data set ‘X’ (Fig. 2) containing measurements of antibody biophysical features, PCA identifies orthogonal, linear combinations (‘signatures’) of these measured features (termed ‘Principal Components’) that account for the most variation in the data, without any information about functional or clinical outcomes (Y). Both advantages and disadvantages of PCA arise from the fact that it is an unsupervised approach – the algorithm receives no information about the outcome. This is advantageous, in that response differences can be visualized in an unbiased way, but disadvantageous in that it is not inherently hypothesis‐driven. Although the identified principal components represent signatures of measured features that account for the most variation in the data, they are not specifically identified to discriminate between outcomes of interest, as a functional or clinical response (Y) is not included in the model. Hence, they can provide insight into important relationships between measured features, but they cannot directly predict how those features are associated with a functional or clinical outcome. Previously Systems Serology application of PCA applied to Mycobacterium tuberculosis serology studies was able to identify the importance of antibody glycosylation in distinguishing latent from active infection.33
Supervised approaches
Partial least squares discriminant analysis and partial least squares regression 40, 41 are supervised methods that identify signatures of measured features (X) quantitatively related to a functional or clinical outcome (Y) (Fig. 2). Thus, both PLSDA and PLSR require input of both a data set of measured antibody features (X), as well as a measured outcome (Y). PLSDA and PLSR are differentiated by the fact that in PLSDA, Y contains a discrete class or label information (e.g. vaccine 1, vaccine 2, etc.) for each outcome, whereas Y for PLSR contains continuous numerical data (e.g. ADCC measurements that can range from 0 to 100% cytotoxicity). Y is often a single column of data (e.g. only one outcome variable), but it can also be a matrix with multiple columns in situations for which there are several outcomes of interest. These algorithms determine orthogonal linear combinations (‘signatures’) of experimentally measured features (X) that best differentiate between outcomes (Y). Each sample can then be scored and plotted on these signatures (termed ‘latent variables’) to determine model accuracy for predicting clinical outcome based on measured features. Each identified latent variable (signature), contains ‘loadings’, or specified amounts of each of the measured features. PLSDA and PLSR are especially useful for hypothesis‐driven Systems Serology research as they specifically search for signatures directly associated with an outcome [in contrast to PCA, which only evaluates overall variation in the data set (X)]. An important consideration in using PLS algorithms is to ensure that models are not ‘overfit’,40 i.e. that the model contains only information about important underlying relationships rather than including random error or noise. This can be avoided by performing cross‐validation (reviewed for PLSDA in ref. 40), whereby a smaller portion of the data is reserved to test a model generated by majority of the data. The ability of the model to accurately predict each sample in the test set can then be used to calculate cross‐validation error, a measure of the model's predictive ability. If cross‐validation error is high, the model can be improved by performing ‘feature selection’ to remove features that contribute to random error. There are a number of different feature selection algorithms that may be used depending on the nature of the data set, some examples of these include use of variable importance projection (VIP) scores42 and the least absolute shrinkage and selection operator (LASSO).43, 44 One key advantage of PLS approaches for Systems Serology research is that loadings on latent variables of a feature‐selected model can give great insight into co‐varying serological features that are most involved in differentiating a functional or clinical outcome. In other words, the ‘minimum signature’ that best defines a vaccine response can give a picture of key antibody features that would be best used to reconstruct the system (Fig. 1b) for theoretical analysis.
The application of PLSDA/PLSR analysis has been successfully applied in a wide range of Systems Serology settings, including to identify humoral immune correlates of the moderately protective human HIV RV144 vaccine trial, in NHP SIV/SHIV vaccine studies, and to examine the humoral responses induced by topical anti‐retrovirals for pre‐exposure prophylaxis following HIV infection.18, 19, 43, 45 In the study of topical anti‐retrovirals for pre‐exposure prophylaxis following HIV infection,43 a PLSDA model used with least absolute shrinkage and selection operator feature selection identified a signature of seven measured antibody features that differentiated women in the topical anti‐retrovirals and placebo groups with 77% cross‐validation accuracy, indicating that topical anti‐retroviral application was associated with a specific antibody signature including measurements from different time‐points (6 and 12 months) and tissue compartments (plasma and cervicovaginal lavage). Individual antibody measurements did not differentiate between groups. Altogether this illustrates the utility of PLSDA for differentiating functional or clinical outcomes and for integrating antibody measurements to identify new hypotheses for mechanisms that may vary over time or tissue compartments.
Decision trees 38, 46 (Fig. 2) provide unique insight into humoral responses in that they are easy to interpret, and can give useful information about the hierarchy of importance and critical ranges (e.g. concentration, binding affinity) of measured antibody features for a particular functional or clinical outcome. For these reasons, they can be especially useful for giving insight into potential mechanistic relationships between measured serological features. A decision tree algorithm works by performing a series of binary tests on the data set of measured antibody features (X), to split samples into groups based on the functional or clinical outcome (Y). The specific binary test performed is selected by the user, and is termed a ‘split criterion’.46 Each split further purifies samples based on functional or clinical outcomes of interest (e.g. vaccine 1 versus vaccine 2 versus vaccine 3, etc.; Fig. 2). The result is a tree‐like structure that illustrates the hierarchy of importance of measured features based on outcome, with specific measurement ranges required for each node selected by the algorithm. As with other supervised approaches, an important consideration in using decision tree algorithms is cross‐validation to prevent overfitting (described above). If cross‐validation determines a decision tree is overfit, ‘pruning’ may be used to improve the model, whereby peripheral branches of the tree are removed if they contribute little to classification. More detailed information on decision‐tree cross‐validation and pruning is reviewed in.46
Future outlook
Although the data‐driven models used in current Systems Serology applications offer the exciting opportunity to integrate high‐throughput data to identify key antibody features associated with a protective immune response, insight is still limited to multivariate statistical associations, without quantitative understanding of true cause–effect relationships that underpin mechanistic function. Although carefully planned experiments based on data‐driven models give some insight in this direction, they too are limited. Other quantitative approaches will be needed to truly understand the underlying complexity of these systems; moving beyond statistical associations and towards a quantitative systems‐level understanding of mechanism. This will require the use of equation‐based methods, also called ‘theory‐driven’ approaches, where mathematical models are constructed based on previous knowledge of a system. Data‐driven models can provide the underlying framework for these models – used to decide key parameters that should be included for a given question, boundaries and important input/output. Once constructed, these theory‐driven models will provide a valuable hypothesis‐testing tool, lending insight into (i) the importance of key antibody parameters in the formation of immune complexes and (ii) the relative importance and synergistic effects of multiple antibody alterations involved in a functional or clinical outcome. These types of approaches have already been employed to optimize the design of antibodies that trap viruses in mucus of the female reproductive tract, determining optimal quantitative ranges of antibody binding affinities that maximize both virion binding and antibody mobility in mucus.47
Clearly Systems Serology technologies, both experimental assays and the application of analytical technologies, are still in their infancy. Over time, high‐throughput assays to assess biophysical antibody features and functions will continue to be developed and improved, encapsulating a wider range of infectious diseases and allow for the examination of antibody features and functions relevant to different tissue compartments and locations. Furthermore, Systems Serology applications can potentially be expanded to address other diseases associated with humoral immunity, including autoimmune diseases and selective cancers. There is no doubt that Systems Serology will continue to evolve to capture broader applications providing us with an increasingly comprehensive understanding of protective humoral immunity.
Disclosures
KBA and AWC wrote and edited the review and figures. The authors have no conflicts of interest.
Funding
This work was supported by the Australian National Health & Medical Research Centre (NHMRC) (AWC) and the American Foundation for AIDS Research (amfAR) Mathilde Krim Fellowship (AWC).
Acknowledgements
We are grateful to Manu Kumar (MIT) and Doug Lauffenburger (MIT) for contributions to Fig. 2 and to Alison Schroeer for assistance with illustrating Figs 1 and 2. We thank Melissa Lemke, Timon Damelang and Ester Lopez for assistance revising this manuscript.
Contributor Information
Kelly B. Arnold, Email: kbarnold@umich.edu
Amy W. Chung, Email: awchung@unimelb.edu.au.
References
- 1. Baxby D. Jenner's Smallpox Vaccine. London: Heinemann Educational Books Ltd, 1981. [Google Scholar]
- 2. Plotkin SA. Vaccines: the fourth century. Clin Vaccine Immunol 2009; 16:1709–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol 2010; 17:1055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol 2011; 12:509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee LH, Frasch CE, Falk LA, Klein DL, Deal CD. Correlates of immunity for pneumococcal conjugate vaccines. Vaccine 2003; 21:2190–6. [DOI] [PubMed] [Google Scholar]
- 6. Cheeseman HM, Carias AM, Evans AB, Olejniczak NJ, Ziprin P, King DF, et al Expression profile of human Fc receptors in mucosal tissue: implications for antibody‐dependent cellular effector functions targeting HIV‐1 transmission. PLoS One 2016; 11:e0154656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sips M, Krykbaeva M, Diefenbach TJ, Ghebremichael M, Bowman BA, Dugast AS, et al Fc receptor‐mediated phagocytosis in tissues as a potent mechanism for preventive and therapeutic HIV vaccine strategies. Mucosal Immunol 2016; 9:1584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vincents B, von Pawel‐Rammingen U, Bjorck L, Abrahamson M. Enzymatic characterization of the streptococcal endopeptidase, IdeS, reveals that it is a cysteine protease with strict specificity for IgG cleavage due to exosite binding. Biochemistry 2004; 43:15540–9. [DOI] [PubMed] [Google Scholar]
- 9. Nagashunmugam T, Lubinski J, Wang L, Goldstein LT, Weeks BS, Sundaresan P, et al In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J Virol 1998; 72:5351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brezski RJ, Vafa O, Petrone D, Tam SH, Powers G, Ryan MH, et al Tumor‐associated and microbial proteases compromise host IgG effector functions by a single cleavage proximal to the hinge. Proc Natl Acad Sci USA 2009; 106:17864–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rerks‐Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al Vaccination with ALVAC and AIDSVAX to prevent HIV‐1 infection in Thailand. N Engl J Med 2009; 361:2209–20. [DOI] [PubMed] [Google Scholar]
- 12. Haynes BF, Gilbert PB, McElrath MJ, Zolla‐Pazner S, Tomaras GD, Alam SM, et al Immune‐correlates analysis of an HIV‐1 vaccine efficacy trial. N Engl J Med 2012; 366:1275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zolla‐Pazner S, deCamp A, Gilbert PB, Williams C, Yates NL, Williams WT, et al Vaccine‐induced IgG antibodies to V1V2 regions of multiple HIV‐1 subtypes correlate with decreased risk of HIV‐1 infection. PLoS One 2014; 9:e87572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, et al Vaccine‐induced Env V1‐V2 IgG3 correlates with lower HIV‐1 infection risk and declines soon after vaccination. Sci Transl Med 2014; 6:228ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, et al Polyfunctional Fc‐effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci Transl Med 2014a; 6:228ra38. [DOI] [PubMed] [Google Scholar]
- 16. Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, et al Vaccine protection against acquisition of neutralization‐resistant SIV challenges in rhesus monkeys. Nature 2012; 482:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, et al Protective efficacy of a global HIV‐1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 2013; 155:531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bradley T, Pollara J, Santra S, Vandergrift N, Pittala S, Bailey‐Kellogg C, et al Pentavalent HIV‐1 vaccine protects against simian‐human immunodeficiency virus challenge. Nat Commun 2017; 8:15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, et al Dissecting polyclonal vaccine‐induced humoral immunity against HIV using systems serology. Cell 2015; 163:988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ackerman ME, Barouch DH, Alter G. Systems serology for evaluation of HIV vaccine trials. Immunol Rev 2017; 275:262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schroeder HW Jr, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol 2010; 125:S41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Woof JM, Russell MW. Structure and function relationships in IgA. Mucosal Immunol 2011; 4:590–7. [DOI] [PubMed] [Google Scholar]
- 23. Choi SC, Wang H, Tian L, Murakami Y, Shin DM, Borrego F, et al Mouse IgM Fc receptor, FCMR, promotes B cell development and modulates antigen‐driven immune responses. J Immunol 2013; 190:987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang M, Murphy RF, Agrawal DK. Decoding IgE Fc receptors. Immunol Res 2007; 37:1–16. [DOI] [PubMed] [Google Scholar]
- 25. Capron M, Capron A. Immunoglobulin E and effector cells in schistosomiasis. Science 1994; 264:1876–7. [DOI] [PubMed] [Google Scholar]
- 26. Chung AW, Alter G. Dissecting the antibody constant region protective immune parameters in HIV infection. Future Virol 2014; 9:397–414. [Google Scholar]
- 27. Hogarth PM, Pietersz GA. Fc receptor‐targeted therapies for the treatment of inflammation, cancer and beyond. Nat Rev Drug Discov 2012; 11:311–31. [DOI] [PubMed] [Google Scholar]
- 28. Jefferis R. Glycosylation as a strategy to improve antibody‐based therapeutics. Nat Rev Drug Discovery 2009; 8:226–34. [DOI] [PubMed] [Google Scholar]
- 29. Jefferis R. Isotype and glycoform selection for antibody therapeutics. Arch Biochem Biophys 2012; 526:159–66. [DOI] [PubMed] [Google Scholar]
- 30. Hristodorov D, Fischer R, Linden L. With or without sugar? (A)glycosylation of therapeutic antibodies. Mol Biotechnol 2013; 54:1056–68. [DOI] [PubMed] [Google Scholar]
- 31. Chung AW, Crispin M, Pritchard L, Robinson H, Gorny MK, Yu X, et al Identification of antibody glycosylation structures that predict monoclonal antibody Fc‐effector function. AIDS 2014b; 28:2523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Satoh M, Iida S, Shitara K. Non‐fucosylated therapeutic antibodies as next‐generation therapeutic antibodies. Expert Opin Biol Ther 2006; 6:1161–73. [DOI] [PubMed] [Google Scholar]
- 33. Lu LL, Chung AW, Rosebrock TR, Ghebremichael M, Yu WH, Grace PS, et al A functional role for antibodies in tuberculosis. Cell 2016; 167:433–43.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benedict KF, Lauffenburger DA. Insights into proteomic immune cell signaling and communication via data‐driven modeling. Curr Top Microbiol Immunol 2013; 363:201–33. [DOI] [PubMed] [Google Scholar]
- 35. Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, et al Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 2009; 10:116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kazmin D, Nakaya HI, Lee EK, Johnson MJ, van der Most R, van den Berg RA, et al Systems analysis of protective immune responses to RTS, S malaria vaccination in humans. Proc Natl Acad Sci USA 2017; 114:2425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li S, Rouphael N, Duraisingham S, Romero‐Steiner S, Presnell S, Davis C, et al Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 2014; 15:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi I, Chung AW, Suscovich TJ, Rerks‐Ngarm S, Pitisuttithum P, Nitayaphan S, et al Machine learning methods enable predictive modeling of antibody feature:function relationships in RV144 vaccinees. PLoS Comput Biol 2015; 11:e1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci 2016; 374:20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brereton RG, Lloyd GR. Partial least squares discriminant analysis: taking the magic away. J Chemom 2014; 28:213–25. [Google Scholar]
- 41. Barker M, Rayens W. Partial least squares for discrimination. J Chemom 2003; 17:166–73. [Google Scholar]
- 42. Arnold KB, Szeto GL, Alter G, Irvine DJ, Lauffenburger DA. CD4+ T cell‐dependent and CD4+ T cell‐independent cytokine‐chemokine network changes in the immune responses of HIV‐infected individuals. Sci Signal 2015; 8:ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Archary D, Seaton KE, Passmore JS, Werner L, Deal A, Dunphy LJ, et al Distinct genital tract HIV‐specific antibody profiles associated with tenofovir gel. Mucosal Immunol 2016; 9:821–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tibshirani R. The LASSO method for variable selection in the Cox model. Stat Med 1997; 16:385–95. [DOI] [PubMed] [Google Scholar]
- 45. Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, et al Protective efficacy of adenovirus‐protein vaccines against SIV challenges in rhesus monkeys. Science 2015; 349:320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Geurts P, Irrthum A, Wehenkel L. Supervised learning with decision tree‐based methods in computational and systems biology. Mol BioSyst 2009; 5:1593–605. [DOI] [PubMed] [Google Scholar]
- 47. Wessler T, Chen A, McKinley SA, Cone R, Forest G, Lai SK. Using computational modeling to optimize the design of antibodies that trap viruses in mucus. ACS Infect Dis 2016; 2:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chung AW, Navis M, Isitman G, Wren L, Silvers J, Amin J, et al Activation of NK cells by ADCC antibodies and HIV disease progression. J Acquir Immune Defic Syndr 2011a; 58:127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baum LL, Cassutt KJ, Knigge K, Khattri R, Margolick J, Rinaldo C, et al HIV‐1 gp120‐specific antibody‐dependent cell‐mediated cytotoxicity correlates with rate of disease progression. J Immunol 1996; 157:2168–73. [PubMed] [Google Scholar]
- 50. Chung A, Rollman E, Johansson S, Kent SJ, Stratov I. The utility of ADCC responses in HIV infection. Curr HIV Res 2008; 6:515–9. [DOI] [PubMed] [Google Scholar]
- 51. Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody‐dependent NK cell activity. J Immunol 2004; 172:5598–605. [DOI] [PubMed] [Google Scholar]
- 52. Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, et al Cross‐reactive influenza‐specific antibody‐dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol 2013; 190:1837–48. [DOI] [PubMed] [Google Scholar]
- 53. Vanderven HA, Liu L, Ana‐Sosa‐Batiz F, Nguyen TH, Wan Y, Wines B, et al Fc functional antibodies in humans with severe H7N9 and seasonal influenza. JCI Insight 2017; 2:pii: 92750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Q, Fan C, Li Q, Zhou S, Huang W, Wang L, et al Antibody‐dependent‐cellular‐cytotoxicity‐inducing antibodies significantly affect the post‐exposure treatment of Ebola virus infection. Sci Rep 2017; 7:45552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. Ebola virus‐like particle‐based vaccine protects nonhuman primates against lethal Ebola virus challenge. J Infect Dis 2007; 196(Suppl 2):S430–7. [DOI] [PubMed] [Google Scholar]
- 56. Kohl S. Role of antibody‐dependent cellular cytotoxicity in neonatal infection with herpes simplex virus. Rev Infect Dis 1991; 13(Suppl 11):S950–2. [DOI] [PubMed] [Google Scholar]
- 57. Das S, Chowdhury R, Ghosh S, Das S. A recombinant protein of Salmonella Typhi induces humoral and cell‐mediated immune responses including memory responses. Vaccine 2017; 35:4523–31. [DOI] [PubMed] [Google Scholar]
- 58. Moore T, Ananaba GA, Bolier J, Bowers S, Belay T, Eko FO, et al Fc receptor regulation of protective immunity against Chlamydia trachomatis . Immunology 2002; 105:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miller GP, Kohl S. Antibody‐dependent leukocyte killing of Cryptococcus neoformans . J Immunol 1983; 131:1455–9. [PubMed] [Google Scholar]
- 60. Kurup VP, Nair MP, Schwartz SA, Fink JN. Serum antibodies and their role in antibody‐dependent cell‐mediated cytotoxicity in aspergillosis. Immunobiology 1985; 169:362–71. [DOI] [PubMed] [Google Scholar]
- 61. Ligas JA, Kerepesi LA, Galioto AM, Lustigman S, Nolan TJ, Schad GA, et al Specificity and mechanism of immunoglobulin M (IgM)‐ and IgG‐dependent protective immunity to larval Strongyloides stercoralis in mice. Infect Immun 2003; 71:6835–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jafarshad A, Dziegiel MH, Lundquist R, Nielsen LK, Singh S, Druilhe PL. A novel antibody‐dependent cellular cytotoxicity mechanism involved in defense against malaria requires costimulation of monocytes FcγRII and FcγRIII. J Immunol 2007; 178:3099–106. [DOI] [PubMed] [Google Scholar]
- 63. Tamura M, Webster RG, Ennis FA. Antibodies to HA and NA augment uptake of influenza A viruses into cells via Fc receptor entry. Virology 1991; 182:211–9. [DOI] [PubMed] [Google Scholar]
- 64. Ana‐Sosa‐Batiz F, Vanderven H, Jegaskanda S, Johnston A, Rockman S, Laurie K, et al Influenza‐specific antibody‐dependent phagocytosis. PLoS One 2016; 11:e0154461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gat O, Galen JE, Tennant S, Simon R, Blackwelder WC, Silverman DJ, et al Cell‐associated flagella enhance the protection conferred by mucosally‐administered attenuated Salmonella Paratyphi A vaccines. PLoS Negl Trop Dis 2011; 5:e1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. He X, Sun X, Wang J, Wang X, Zhang Q, Tzipori S, et al Antibody‐enhanced, Fcγ receptor‐mediated endocytosis of Clostridium difficile toxin A. Infect Immun 2009; 77:2294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Armstrong JA, Hart PD. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med 1971; 134:713–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bueno RA, Thomaz L, Munoz JE, da Silva CJ, Nosanchuk JD, Pinto MR, et al Antibodies against glycolipids enhance antifungal activity of macrophages and reduce fungal burden after infection with Paracoccidioides brasiliensis . Front Microbiol 2016; 7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Braem SG, Rooijakkers SH, van Kessel KP, de Cock H, Wosten HA, van Strijp JA, et al Effective neutrophil phagocytosis of Aspergillus fumigatus is mediated by classical pathway complement activation. J Innate Immun 2015; 7:364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Teo A, Feng G, Brown GV, Beeson JG, Rogerson SJ. Functional antibodies and protection against blood‐stage malaria. Trends Parasitol 2016; 32:887–98. [DOI] [PubMed] [Google Scholar]
- 71. Joiner KA, Fuhrman SA, Miettinen HM, Kasper LH, Mellman I. Toxoplasma gondii: fusion competence of parasitophorous vacuoles in Fc receptor‐transfected fibroblasts. Science 1990; 249:641–6. [DOI] [PubMed] [Google Scholar]
- 72. Ramachandran G, Tennant SM, Boyd MA, Wang JY, Tulapurkar ME, Pasetti MF, et al Functional activity of antibodies directed towards flagellin proteins of non‐typhoidal salmonella. PLoS One 2016; 11:e0151875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nowling JM, Philipp MT. Killing of Borrelia burgdorferi by antibody elicited by OspA vaccine is inefficient in the absence of complement. Infect Immun 1999; 67:443–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Han Y, Kozel TR, Zhang MX, MacGill RS, Carroll MC, Cutler JE. Complement is essential for protection by an IgM and an IgG3 monoclonal antibody against experimental, hematogenously disseminated candidiasis. J Immunol 2001; 167:1550–7. [DOI] [PubMed] [Google Scholar]
- 75. Boyle MJ, Reiling L, Feng G, Langer C, Osier FH, Aspeling‐Jones H, et al Human antibodies fix complement to inhibit Plasmodium falciparum invasion of erythrocytes and are associated with protection against malaria. Immunity 2015; 42:580–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chung AW, Rollman E, Center RJ, Kent SJ, Stratov I. Rapid degranulation of NK cells following activation by HIV‐specific antibodies. J Immunol 2009; 182:1202–10. [DOI] [PubMed] [Google Scholar]
- 77. Vouldoukis I, Riveros‐Moreno V, Dugas B, Ouaaz F, Becherel P, Debre P, et al The killing of Leishmania major by human macrophages is mediated by nitric oxide induced after ligation of the FcεRII/CD23 surface antigen. Proc Natl Acad Sci USA 1995; 92:7804–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Joos C, Marrama L, Polson HE, Corre S, Diatta AM, Diouf B, et al Clinical protection from falciparum malaria correlates with neutrophil respiratory bursts induced by merozoites opsonized with human serum antibodies. PLoS One 2010; 5:e9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Llewellyn D, Miura K, Fay MP, Williams AR, Murungi LM, Shi J, et al Standardization of the antibody‐dependent respiratory burst assay with human neutrophils and Plasmodium falciparum malaria. Sci Rep 2015; 5:14081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gibson‐Corley KN, Bockenstedt MM, Li H, Boggiatto PM, Phanse Y, Petersen CA, et al An in vitro model of antibody‐enhanced killing of the intracellular parasite Leishmania amazonensis . PLoS One 2014; 9:e106426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Forthal DN, Gilbert PB, Landucci G, Phan T. Recombinant gp120 vaccine‐induced antibodies inhibit clinical strains of HIV‐1 in the presence of Fc receptor‐bearing effector cells and correlate inversely with HIV infection rate. J Immunol 2007; 178:6596–603. [DOI] [PubMed] [Google Scholar]
- 82. Shannon JG, Cockrell DC, Takahashi K, Stahl GL, Heinzen RA. Antibody‐mediated immunity to the obligate intracellular bacterial pathogen Coxiella burnetii is Fc receptor‐ and complement‐independent. BMC Immunol 2009; 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moore T, Ekworomadu CO, Eko FO, MacMillan L, Ramey K, Ananaba GA, et al Fc receptor‐mediated antibody regulation of T cell immunity against intracellular pathogens. J Infect Dis 2003; 188:617–24. [DOI] [PubMed] [Google Scholar]
- 84. Bouharoun‐Tayoun H, Attanath P, Sabchareon A, Chongsuphajaisiddhi T, Druilhe P. Antibodies that protect humans against Plasmodium falciparum blood stages do not on their own inhibit parasite growth and invasion in vitro, but act in cooperation with monocytes. J Exp Med 1990; 172:1633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lewis GK, Finzi A, DeVico AL, Pazgier M. Conformational masking and receptor‐dependent unmasking of highly conserved Env epitopes recognized by non‐neutralizing antibodies that mediate potent ADCC against HIV‐1. Viruses 2015; 7:5115–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, et al CD4 mimetics sensitize HIV‐1‐infected cells to ADCC. Proc Natl Acad Sci USA 2015; 112:E2687–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vanham G, Bloemmen FJ, Ceuppens JL, Stevens EA. Influence of immune‐complex size and antigen‐antibody ratio on immune complex detection with monoclonal rheumatoid factor and C1q. J Clin Lab Immunol 1984; 15:63–8. [PubMed] [Google Scholar]
- 88. Lux A, Yu X, Scanlan CN, Nimmerjahn F. Impact of immune complex size and glycosylation on IgG binding to human FcγRs. J Immunol 2013; 190:4315–23. [DOI] [PubMed] [Google Scholar]
- 89. Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discov 2006; 5:310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schwarz M, Spector L, Gargir A, Shtevi A, Gortler M, Altstock RT, et al A new kind of carbohydrate array, its use for profiling antiglycan antibodies, and the discovery of a novel human cellulose‐binding antibody. Glycobiology 2003; 13:749–54. [DOI] [PubMed] [Google Scholar]
- 91. Oyelaran O, Gildersleeve JC. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol 2009; 13:406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Angenendt P. Progress in protein and antibody microarray technology. Drug Discov Today 2005; 10:503–11. [DOI] [PubMed] [Google Scholar]
- 93. Reineke U, Ivascu C, Schlief M, Landgraf C, Gericke S, Zahn G, et al Identification of distinct antibody epitopes and mimotopes from a peptide array of 5520 randomly generated sequences. J Immunol Methods 2002; 267:37–51. [DOI] [PubMed] [Google Scholar]
- 94. Stratov I, Chung A, Kent SJ. Robust NK cell‐mediated human immunodeficiency virus (HIV)‐specific antibody‐dependent responses in HIV‐infected subjects. J Virol 2008; 82:5450–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brown EP, Licht AF, Dugast AS, Choi I, Bailey‐Kellogg C, Alter G, et al High‐throughput, multiplexed IgG subclassing of antigen‐specific antibodies from clinical samples. J Immunol Methods 2012; 386:117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, et al Plasma IgG to linear epitopes in the V2 and V3 regions of HIV‐1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One 2013; 8:e75665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chung AW, Isitman G, Navis M, Kramski M, Center RJ, Kent SJ, et al Immune escape from HIV‐specific antibody‐dependent cellular cytotoxicity (ADCC) pressure. Proc Natl Acad Sci USA 2011b; 108:7505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Malmqvist M. Surface plasmon resonance for detection and measurement of antibody‐antigen affinity and kinetics. Curr Opin Immunol 1993; 5:282–6. [DOI] [PubMed] [Google Scholar]
- 99. Klasse PJ. How to assess the binding strength of antibodies elicited by vaccination against HIV and other viruses. Expert Rev Vaccines 2016; 15:295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Madhavi V, Wines BD, Amin J, Emery S, Lopez E, Kelleher A., et al HIV‐1 Env‐ and Vpu‐specific antibody‐dependent cellular cytotoxicity responses associated with elite control of HIV. J Virol 2017; 91(18):pii: e00700–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cleary KLS, Chan HTC, James S, Glennie MJ, Cragg MS. Antibody distance from the cell membrane regulates antibody effector mechanisms. J Immunol 2017; 198:3999–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McLean MR, Madhavi V, Wines BD, Hogarth PM, Chung AW, Kent SJ. Dimeric Fcγ receptor enzyme‐linked immunosorbent assay to study hiv‐specific antibodies: a new look into breadth of Fcγ receptor antibodies induced by the RV144 vaccine trial. J Immunol 2017; 199:816–29. [DOI] [PubMed] [Google Scholar]
- 103. Madhavi V, Wren LH, Center RJ, Gonelli C, Winnall WR, Parsons MS, et al Breadth of HIV‐1 Env‐specific antibody‐dependent cellular cytotoxicity: relevance to global HIV vaccine design. AIDS 2014; 28:1859–70. [DOI] [PubMed] [Google Scholar]
- 104. Brown EP, Dowell KG, Boesch AW, Normandin E, Mahan AE, Chu T, et al Multiplexed Fc array for evaluation of antigen‐specific antibody effector profiles. J Immunol Methods 2017; 443:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mahan AE, Jennewein MF, Suscovich T, Dionne K, Tedesco J, Chung AW, et al Antigen‐specific antibody glycosylation is regulated via vaccination. PLoS Pathog 2016; 12:e1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pandey JP, Namboodiri AM. Genetic variants of IgG1 antibodies and FcγRIIIa receptors influence the magnitude of antibody‐dependent cell‐mediated cytotoxicity against prostate cancer cells. Oncoimmunology 2014; 3:e27317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol 2014; 5:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lefranc MP, Lefranc G. Human Gm, Km, and Am allotypes and their molecular characterization: a remarkable demonstration of polymorphism. Methods Mol Biol 2012; 882:635–80. [DOI] [PubMed] [Google Scholar]
- 109. Suzuki T, Ishii‐Watabe A, Tada M, Kobayashi T, Kanayasu‐Toyoda T, Kawanishi T, et al Importance of neonatal FcR in regulating the serum half‐life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc‐fusion proteins to human neonatal FcR. J Immunol 2010; 184:1968–76. [DOI] [PubMed] [Google Scholar]
- 110. Nimmerjahn F, Ravetch JV. Analyzing antibody‐Fc‐receptor interactions. Methods Mol Biol 2008; 415:151–62. [DOI] [PubMed] [Google Scholar]


