Abstract
Background
Ferns, originated about 360 million years ago, are the sister group of seed plants. Despite the remarkable progress in our understanding of fern phylogeny, with conflicting molecular evidence and different morphological interpretations, relationships among major fern lineages remain controversial.
Results
With the aim to obtain a robust fern phylogeny, we carried out a large-scale phylogenomic analysis using high-quality transcriptome sequencing data, which covered 69 fern species from 38 families and 11 orders. Both coalescent-based and concatenation-based methods were applied to both nucleotide and amino acid sequences in species tree estimation. The resulting topologies are largely congruent with each other, except for the placement of Angiopteris fokiensis, Cheiropleuria bicuspis, Diplaziopsis brunoniana, Matteuccia struthiopteris, Elaphoglossum mcclurei, and Tectaria subpedata.
Conclusions
Our result confirmed that Equisetales is sister to the rest of ferns, and Dennstaedtiaceae is sister to eupolypods. Moreover, our result strongly supported some relationships different from the current view of fern phylogeny, including that Marattiaceae may be sister to the monophyletic clade of Psilotaceae and Ophioglossaceae; that Gleicheniaceae and Hymenophyllaceae form a monophyletic clade sister to Dipteridaceae; and that Aspleniaceae is sister to the rest of the groups in eupolypods II. These results were interpreted with morphological traits, especially sporangia characters, and a new evolutionary route of sporangial annulus in ferns was suggested. This backbone phylogeny in ferns sets a foundation for further studies in biology and evolution in ferns, and therefore in plants.
Keywords: phylogenomic, monilophytes, evolution, sporangium, transcriptome
Background
Phylogeny, which reflects natural history, is fundamental to understanding evolution and biodiversity. Ferns (monilophytes), originated about 360 million years (MY) ago, are the sister group of seed plants [1, 2]. With estimated 10 578 extant living species globally [3], they are the second most diverse group of vascular plants. Phylogenetic studies for ferns, especially based on molecular evidence, have been widely carried out in recent decades. These studies have revolutionized our understanding of the evolutionary history of ferns. Milestones included setting ferns as the sister group of seed plants [1, 2], placing Psilotaceae and Equisetaceae within ferns [2, 4, 5], and revealing a major polypods radiation following the rise of angiosperms [6, 7]. Resolutions at shallow phylogenetic depth among families or genera have also been improved remarkably [8–14].
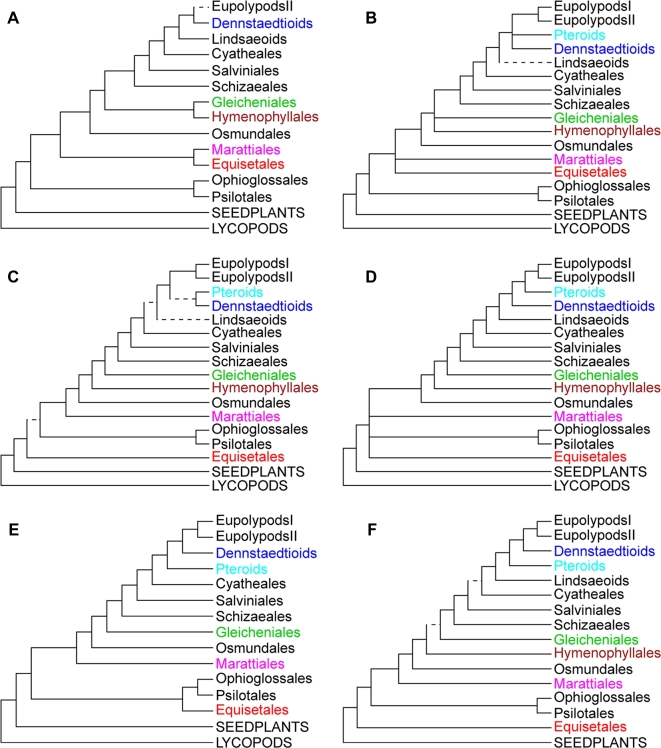
However, previous research on fern phylogeny has mostly relied on plastid genes [10, 12, 13], some combined with a few nuclear genes [4, 5, 14] or morphological traits [5, 11]. Due to incomplete lineage sorting (ILS), genes from different resources often show conflicting evolutionary patterns, especially when based on a limited number of samples, and some deep relationships in fern phylogeny remain controversial (Fig. 1). In the latest PPG I system [3], which has derived from many recent phylogenetic studies, some important nodes remain uncertain, such as (i) what are the relationships among Marattiales, Ophioglossales, and Psilotales?; (ii) are Hymenophyllales and Gleicheniales sister groups?; and (iii) what are the relationships among families in eupolypods II?
Figure 1:
Topologies (a-f) adapted from published results [5, 12–14, 26, 34]. Branches with support <75% were shown using dotted lines, and taxa that differ in their phylogeny locations were shown in different colors.
Transcriptome sequencing (RNA-Seq) provides massive transcript information from the genome. Phylogenetic reconstructions based on RNA-Seq are more efficient and cost-effective than traditional polymerase chain reaction–based or expressed sequence tags (EST)-based methods when lacking whole-genome data [15]. Successful cases in recent years include mollusks [16], insects [17], the grape family [18], angiosperms [19], and land plants, including 6 ferns [20]. Here, with the aim to reconstruct the framework of fern phylogeny, we sampled abundant fern species representing all important linages and applied the latest phylogenomic analyses based on RNA-Seq.
To reconstruct a robust and well-resolved phylogeny in ferns, applying multiple methods of phylogenomic analysis is extremely important. Since concatenation-based estimations of species trees usually have good accuracy under a low level of ILS, while coalescent-based methods are developed to overcome the effect of ILS but are sensitive to gene tree estimation error [21], both concatenation-based and coalescent-based estimations are applied. Nucleotide sequence, with higher variability than amino acid sequence, usually brings more useful information in phylogeny reconstruction, especially for closely related taxa. However, the substitutional saturation and compositional bias in nucleotide sequence, especially in the third codon position, may lead to a deviation from the true phylogeny. Here, both nucleotide and amino acid sequences are used in phylogeny reconstruction.
Morphologically, the fern sporangium is an organ for enclosing and dispersing spores, most of which function like a unique catapult with the annulus [22]. During the last centuries, Bower's hypothesis on the evolution of sporangia with a focus on annulus [23] has been one of the most important cornerstones to fern phylogeny based on morphology [24, 25]. However, this hypothesis has been challenged by somewhat conflicting frameworks of fern phylogeny [4, 10, 12, 14, 26]. A robust framework in fern phylogeny that reflects the evolutionary history will improve our understanding of the evolution of fern sporangia as well as other characters.
Data Description
Taxa sampling and RNA-Seq
We chose 69 fern species from 38 families according to the PPG I system (48 fern families in total), covering all the 11 orders (Equisetales, Psilotales, Ophioglossales, Marattiales, Osmundales, Hymenophyllales, Gleicheniales, Schizaeales, Salviniales, Cyatheales, and Polypodiales). Information about the location and time for sampling is given in Table S1. All the sampled species were collected under the permissions of the natural reserves and Shanghai Chenshan Botanical Garden in China.
Sporophyll or/and trophophyll were collected and frozen in liquid nitrogen immediately, and preserved in an ultra-low-temperature refrigerator at –80°C before RNA extraction. Total RNA was extracted using TRIzol (Life Technologies Corp., Carlsbad, California, USA) according to the manufacturer's protocols. The RNA concentration was determined using a NanoDrop spectrophotometer, and RNA quality was assessed with an Agilent Bioanalyzer. Paired-end reads were generated by Majorbio Company (Shanghai, China) using the HiSeq 2500 system. Raw reads were deposited in NCBI [27].
Transcriptomes assembly and orthology assignment
Transcriptomes data were generated from 69 fern species (Table 1). After filtering, about 2726.9 million paired-end DNA sequence reads (about 313 Gbp) were retained. We assembled these reads de novo and obtained a total of 5 449 842 contigs [28].
Table 1:
Sequencing and assembly information of the transcriptome data
| ID | Species | Clean data, G | Total reads (clean) | Q30 % | Number of contigs | N50, bp | Mean, bp | Genes in Matrix 1 | Genes in Matrix 2 |
|---|---|---|---|---|---|---|---|---|---|
| RS1 | Pronephrium simplex | 4.7 | 38 045 864 | 91.24 | 151 319 | 887 | 581.07 | 2168 | 1254 |
| RS10 | Antrophyum callifolium | 4.0 | 32 745 384 | 91.76 | 64 107 | 1819 | 998.73 | 2226 | 1305 |
| RS101 | Oleandra musifolia | 4.5 | 36 487 068 | 91.45 | 37 075 | 1493 | 919.3 | 2093 | 1248 |
| RS103 | Woodsia polystichoides | 3.9 | 31 465 870 | 90.91 | 47 812 | 1348 | 811.3 | 2287 | 1310 |
| RS107 | Equisetum diffusum | 4.4 | 35 693 238 | 90.21 | 88 932 | 1154 | 655.64 | 1811 | 1254 |
| RS108 | Oreogrammitis dorsipila | 4.6 | 37 037 324 | 90.57 | 266 540 | 591 | 485.1 | 2141 | 1273 |
| RS11 | Vandenboschia striata | 4.8 | 38 639 790 | 90.3 | 261 724 | 460 | 422.76 | 1959 | 1276 |
| RS111 | Pleurosoriopsis makinoi | 4.8 | 38 983 796 | 90.13 | 98 187 | 1145 | 632.29 | 2182 | 1277 |
| RS112 | Azolla pinnata subsp. asiatica | 4.4 | 35 735 206 | 90.57 | 78 295 | 1348 | 777.92 | 1418 | 839 |
| RS114 | Taenitis blechnoides | 4.1 | 32 898 682 | 90.98 | 70 495 | 1262 | 711.3 | 2186 | 1278 |
| RS115 | Gymnogrammitis dareiformis | 3.9 | 31 630 988 | 89.81 | 119 483 | 569 | 449.38 | 1996 | 1220 |
| RS116 | Schizaea dichotoma | 4.5 | 36 668 734 | 89.6 | 67 422 | 1350 | 826.92 | 2035 | 1285 |
| RS119 | Botrychium japonicum | 4.8 | 38 603 000 | 90.28 | 85 236 | 1477 | 846.97 | 1866 | 1283 |
| RS122 | Goniophlebium niponicum | 4.8 | 38 786 214 | 90.82 | 54 152 | 1663 | 951.92 | 2279 | 1300 |
| RS123 | Arthropteris palisotii | 4.4 | 35 646 740 | 91 | 50 700 | 1454 | 891.67 | 2286 | 1311 |
| RS124 | Matteuccia struthiopteris | 4.2 | 34 080 998 | 90.44 | 57 514 | 1345 | 776.52 | 2290 | 1313 |
| RS127 | Salvinia natans | 4.2 | 33 780 056 | 91.17 | 79 393 | 1379 | 767.14 | 1905 | 1173 |
| RS128 | Woodwardia prolifera | 5.1 | 40 967 322 | 91.63 | 69 931 | 1557 | 859.72 | 2328 | 1328 |
| RS14 | Diplazium viridescens | 4.0 | 32 320 416 | 90.46 | 88 236 | 1434 | 780.87 | 2269 | 1310 |
| RS16 | Bolbitis appendiculata | 4.7 | 37 503 336 | 91.66 | 201 426 | 802 | 556.39 | 2226 | 1288 |
| RS17 | Dryopteris pseudocaenopteris | 4.1 | 33 136 196 | 91.23 | 102 751 | 723 | 514.92 | 2236 | 1298 |
| RS18 | Dicranopteris pedata | 4.2 | 33 942 120 | 92.04 | 74 011 | 1193 | 684.09 | 2031 | 1304 |
| RS19 | Haplopteris amboinensis | 4.2 | 42 772 168 | 94.17 | 47 603 | 1713 | 1041.8 | 2249 | 1307 |
| RS21 | Psilotum nudum | 8.5 | 85 199 034 | 93.6 | 66 212 | 1739 | 927.19 | 1741 | 1223 |
| RS24 | Cyclopeltis crenata | 4.6 | 37 158 058 | 91.5 | 29 668 | 600 | 491.82 | 2146 | 1279 |
| RS25 | Asplenium formosae | 4.6 | 46 629 754 | 93.5 | 73 318 | 1722 | 989.84 | 2273 | 1312 |
| RS27 | Lomariopsis spectabilis | 4.1 | 33 233 594 | 91.77 | 98 030 | 1466 | 750.42 | 2225 | 1304 |
| RS28 | Cheiropleuria bicuspis | 5.1 | 41 617 294 | 91.35 | 99 411 | 1435 | 832.82 | 2022 | 1295 |
| RS31 | Plagiogyria japonica | 5.7 | 46 472 760 | 91.92 | 89 532 | 1258 | 733.9 | 2036 | 1222 |
| RS34 | Alsophila podophylla | 4.9 | 48 768 608 | 93.43 | 66 254 | 1580 | 904.62 | 2195 | 1289 |
| RS35 | Histiopteris incisa | 4.3 | 43 115 390 | 93.81 | 61 231 | 1749 | 985.03 | 2319 | 1316 |
| RS36 | Pteris vittata | 4.1 | 41 212 858 | 94.37 | 76 666 | 1868 | 1021.13 | 2296 | 1312 |
| RS37 | Cibotium barometz | 4.1 | 33 263 550 | 91.92 | 85 555 | 1612 | 891.87 | 1790 | 1099 |
| RS38 | Osmunda japonica | 4.1 | 33 485 274 | 92.05 | 58 612 | 1730 | 901.28 | 1732 | 1159 |
| RS39 | Loxogramme chinensis | 3.9 | 31 392 952 | 92.16 | 84 796 | 1065 | 651.88 | 2240 | 1305 |
| RS4 | Microlepia hookeriana | 4.0 | 40 561 422 | 94.49 | 95 951 | 1610 | 874.06 | 2262 | 1301 |
| RS41 | Pteridium aquilinum | 4.6 | 46 157 134 | 93.51 | 55 615 | 1742 | 960.37 | 2321 | 1316 |
| RS42 | Hypolepis punctata | 4.4 | 43 828 154 | 93.56 | 59 717 | 1371 | 833.68 | 2277 | 1308 |
| RS43 | Dicksonia antarctica | 3.9 | 31 210 608 | 91.69 | 56 494 | 1533 | 902.96 | 2045 | 1213 |
| RS45 | Rhachidosorus mesosorus | 4.4 | 35 348 994 | 91.98 | 80 069 | 1541 | 835.92 | 2300 | 1315 |
| RS46 | Drynaria bonii | 4.5 | 36 017 548 | 92.02 | 68 132 | 1077 | 643.93 | 2176 | 1279 |
| RS47 | Platycerium bifurcatum | 4.1 | 33 209 740 | 91.62 | 40 456 | 1097 | 694.56 | 2148 | 1283 |
| RS48 | Angiopteris fokiensis | 4.4 | 35 120 302 | 91.12 | 57 637 | 1629 | 932.57 | 1917 | 1306 |
| RS5 | Diplaziopsis brunoniana | 4.3 | 34 698 846 | 91.35 | 70 184 | 822 | 541.31 | 2040 | 1234 |
| RS50 | Dennstaedtia pilosella | 4.5 | 45 618 446 | 93.63 | 84 813 | 1582 | 831.56 | 2308 | 1313 |
| RS51 | Monachosorum henryi | 4.1 | 41 658 504 | 93.42 | 87 832 | 1465 | 803.17 | 2255 | 1288 |
| RS52 | Acystopteris japonica | 5.5 | 44 662 146 | 91.15 | 57 118 | 1507 | 873.59 | 1222 | 677 |
| RS53 | Monachosorum maximowiczii | 4.8 | 48 497 004 | 93.58 | 101 448 | 1817 | 899.54 | 2257 | 1294 |
| RS54 | Dennstaedtia scabra | 5.1 | 51 360 716 | 93.47 | 92 158 | 1565 | 845.44 | 1818 | 1056 |
| RS56 | Arachniodes nigrospinosa | 5.1 | 50 929 362 | 94.47 | 57 168 | 1623 | 916.1 | 2332 | 1319 |
| RS69 | Cheilanthes chusana | 5.2 | 51 851 066 | 94.18 | 49 449 | 1727 | 1012.63 | 2317 | 1324 |
| RS7 | Elaphoglossum mcclurei | 4.1 | 32 800 248 | 92.31 | 57 330 | 1398 | 846.79 | 2267 | 1299 |
| RS70 | Lomagramma matthewii | 4.4 | 35 218 876 | 91.21 | 65 170 | 1748 | 947.18 | 2258 | 1307 |
| RS71 | Osmolindsaea odorata | 4.6 | 46 808 646 | 94.13 | 113 778 | 1521 | 845.96 | 2257 | 1312 |
| RS72 | Aleuritopteris chrysophylla | 4.8 | 47 955 674 | 94.18 | 61 637 | 1669 | 929.63 | 2307 | 1322 |
| RS77 | Marsilea quadrifolia | 4.3 | 34 724 432 | 91.76 | 65 227 | 1607 | 930.31 | 2188 | 1299 |
| RS8 | Humata repens | 4.5 | 36 606 746 | 91.17 | 68 932 | 1267 | 690.35 | 2264 | 1315 |
| RS81 | Tectaria subpedata | 4.2 | 42 539 482 | 94.43 | 57 384 | 1326 | 797.83 | 2128 | 1242 |
| RS84 | Ophioglossum vulgatum | 4.4 | 35 637 330 | 91.77 | 71 821 | 1226 | 741.62 | 1631 | 1179 |
| RS85 | Nephrolepis cordifolia | 5.0 | 40 063 236 | 90.81 | 55 207 | 1530 | 842.63 | 2302 | 1319 |
| RS86 | Microlepia platyphylla | 4.6 | 46 324 294 | 94 | 74 956 | 1763 | 945.87 | 2267 | 1295 |
| RS88 | Lygodium flexuosum | 4.2 | 34 098 316 | 91.44 | 66 751 | 1514 | 867.82 | 2064 | 1296 |
| RS89 | Hypodematium crenatum | 4.1 | 32 711 798 | 91.58 | 52 813 | 1416 | 852.57 | 2298 | 1319 |
| RS90 | Acrostichum aureum | 5.4 | 43 422 574 | 90.69 | 46 189 | 1729 | 1043.2 | 2303 | 1319 |
| RS91 | Adiantum caudatum | 5.1 | 51 062 204 | 94.23 | 51 145 | 1575 | 950.49 | 2323 | 1327 |
| RS92 | Parahemionitis cordata | 4.1 | 33 309 450 | 91.72 | 47 508 | 1456 | 894.42 | 2306 | 1317 |
| RS93 | Microlepia speluncae | 4.4 | 44 124 842 | 94.55 | 94 980 | 1720 | 917.59 | 2292 | 1308 |
| RS97 | Stenochlaena palustris | 4.7 | 37 887 642 | 91.81 | 58 416 | 1655 | 945.83 | 2300 | 1316 |
| RS98 | Ceratopteris thalictroides | 3.9 | 31 741 082.0 | 91.4 | 74 728 | 1610 | 912.26 | 2231 | 1296 |
The number of ortholog genes used in Matrix 1 and Matrix 2 were shown.
In order to obtain a reliable phylogenetic relationship, we selected 4 species as the outgroup, representing the main lineages of land plants: Amborella trichopoda (representing angiosperms), Picea abies (representing gymnosperms), Selaginella moellendorffii (representing lycophytes), and Physcomitrella patens (representing bryophytes). The translated ORF (protein) sequences of these 4 species were downloaded from Phytozome [29] and used in the following analysis.
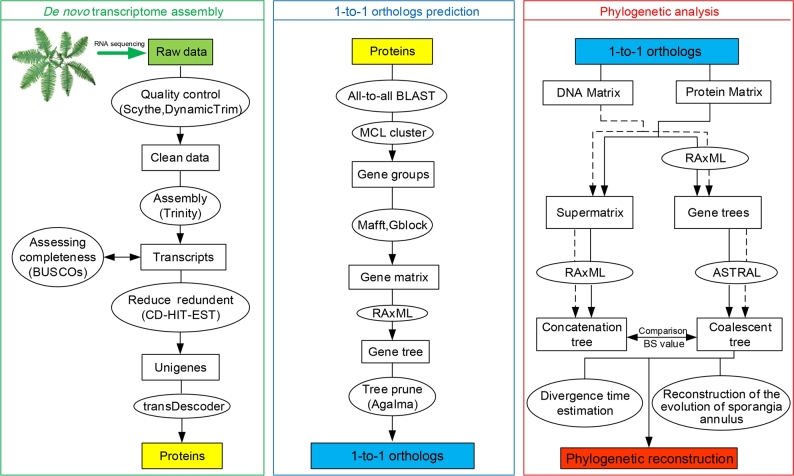
To ensure the consistency of phylogenomic analysis, we used a phylogenetic-based ortholog selection method and obtained 2 subsets of 1-to-1 orthologous genes that differed in gene number and species occupancy rate, named “Matrix 1” and “Matrix 2” [30]. Matrix 1 consists of 2391 genes that are present in at least 52 taxa (that is 75% of the 69 taxa in total), resulting in 2 024 565 nucleotide and 674 855 amino acid positions; the gene and character occupancy were 88% and 85%, respectively. Matrix 2 consists of 1334 genes that are present in at least 62 taxa (that is 90% of the 69 taxa in total), resulting in 1 171 332 nucleotide and 390 444 amino acid positions; the gene and character occupancy reached 94% and 90%, respectively. For each orthologue gene set, coalescent-based and concatenation-based methods were applied separately to both nucleotide and amino acid sequences. A working flow diagram showing the major processes in this study is presented in Fig. 2.
Figure 2:
A working flow diagram showing the major processes of data production and analysis in this study. Three major processes are de novo transcriptome assembly, 1-to-1 orthologs prediction, and phylogenetic analysis. The rectangles represent the main results, and the ellipses represent the main methods and analysis.
Results
Species tree estimated in 69 ferns
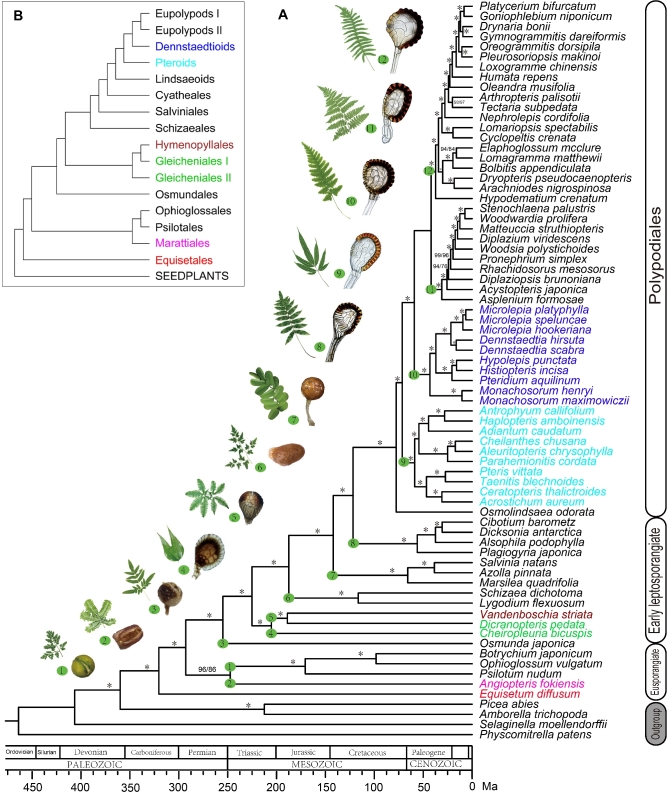
For each combination of reconstruction methods (coalescent-based or concatenation-based) and sequence types (nucleotide or amino acid), Matrix 1 and Matrix 2 [31, 32] always yielded the same topology. In general, the 4 topologies (Fig. 3, Figs S1, S2, S3) from a combination of methods and sequence types are consistent, except for 6 positions (Table 2). Among the topologies, the one estimated by applying a coalescent-based method to the nucleotide sequence (Fig. 3) and the one applying a concatenation-based method (Figure S2) are most congruent.
Figure 3:
Phylogeny of ferns reconstructed by coalescent-based method using nucleotide sequence with divergence times calculated. Support values for the main phylogeny (A) calculated from Matrix 1/Matrix 2 are listed as percentages. *Indicates 100%/100%. Representative leave(s), sporangium, and the corresponding lineage are labeled with a same number. Simplified topology (B) shows the main linages as in Fig. 1. Species in phylogeny (A) and the corresponding lineage in topology (B) are shown in the same color.
Table 2:
Inconsistent topologies using different methods and sequences
| Coalescent-based method | Concatenation-based method | |||
|---|---|---|---|---|
| Site | Nucleotide | Amino acid | Nucleotide | Amino acid |
| A | (Anfo,(Pnu,(Ovu,Bja))) | (Anfo,(Pnu,(Ovu,Bja))) | ((Pnu,(Ovu,Bja)),(Anfo,a)) | ((Pnu,(Ovu,Bja)),(Anfo,a)) |
| B | (Cbi,(Dpe,Vst)) | (Cbi,(Dpe,Vst)) | (Cbi,(Dpe,Vst)) | ((Dpe,Vst),(Cbi,a)) |
| C | (Asfo,(Aja,(Dbr,a))) | (Asfo,(Aja,(Dbr,a))) | (Asfo,(Aja,(Dbr,a))) | (Asfo,((Aja,Dbr),a)) |
| D | (Dvi,(Mst,(Spa,Wpr))) | ((Dvi,Mst),(Spa,Wpr)) | (Dvi,(Mst,(Spa,Wpr))) | (Dvi,(Mst,(Spa,Wpr))) |
| E | (Bap,(Emc,Lma)) | (Emc,(Bap,Lma)) | (Bap,(Emc,Lma)) | (Emc,(Bap,Lma)) |
| F | (Nco,((Tsu,Apa),a)) | (Nco,(Tsu,(Apa,a))) | (Nco,((Tsu,Apa),a)) | (Nco,((Tsu,Apa),a)) |
(A) Anfo: Angiopteris fokiensis, Pnu: Psilotum nudum, Ovu: Ophioglossum vulgatum, Bja: Botrychium japonicum; (B) Cbi: Cheiropleuria bicuspis, Dpe: Dicranopteris pedata, Vst: Vandenboschia striata; (C) Asfo: Asplenium formosae, Aja: Acystopteris japonica, Dbr: Diplaziopsis brunoniana; (D) Dvi: Diplazium viridescens, Mst: Matteuccia struthiopteris, Spa: Stenochlaena palustris, Wpr: Woodwardia prolifera; (E) Bap: Bolbitis appendiculata, Emc: Elaphoglossum mcclurei, Lma: Lomagramma matthewii; (F) Nco: Nephrolepis cordifolia, Tsu: Tectaria subpedata, Apa: Arthropteris palisotii.
aIndicates other sampled species within this lineage. Topologies consistent with the one yielded from coalescent-based methods and nucleotide sequences are shown in bold.
Reconstruction of the evolutionary history of sporangial annulus
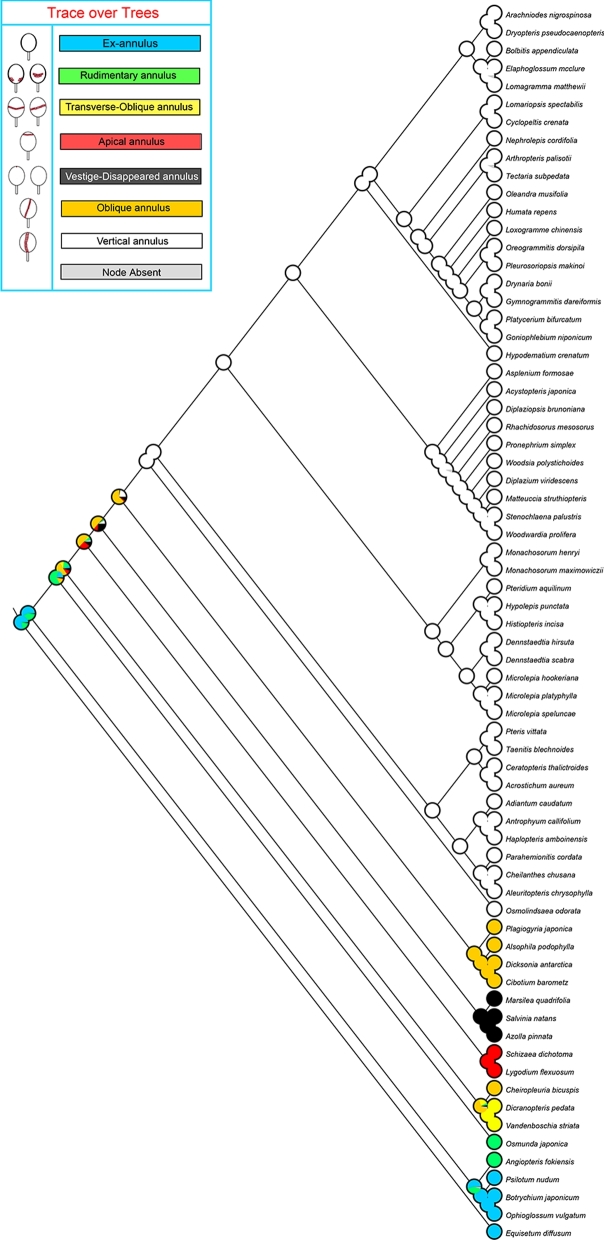
Our reconstruction of the evolution of sporangial annulus (Fig. 4) showed that ex-annulus sporangia are inferred to be the ancestral state (proportional likelihood [PL] = 1), and the rest of annulus states are likely derived from ex-annulus sporangia. Vertical annulus is suggested as synapomorphy for all polypod ferns (PL > 0.99). Both oblique annulus and rudimentary annulus have experienced parallel evolution.
Figure 4:
Reconstruction of the evolutionary history of sporangial annulus in ferns. Sampled species with 7 types of sporangial annulus are shown in different colours. For each ancient node, percentage of character state of sporangial annulus is shown.
Discussion
Comparison of topologies estimated by various methods
By comparing topologies estimated by coalescent-based and concatenation-based methods using both nucleotide and amino acid sequences (Table 2), we found that the topologies yielded from coalescent-based and concatenation-based methods using nucleotide sequences are mostly consistent, except for the position of Angiopteris fokiensis. Topologies yielded from coalescent-based methods using nucleotide sequences and amino acid sequences showed 3 positions of inconsistency, all of which belong to eupolypods. As eupolypods have experienced rapid evolutionary radiation in Cenozoic [7] and nucleotide sequences usually provide more information to reconstruct relationships at a shallow phylogenetic scale, we consider the topology yielded from nucleotide sequences to be more reliable. However, the inconsistent positions among topologies often show relatively lower supporting values, and are often the controversial nodes from past studies based on different genes; we suggest that such inconsistency might be caused partially by ILS and reticulate evolution.
Relationships of eusporangiate ferns
Which clade is sister to the remaining taxa in ferns is a long-debated question (Fig. 1). Our results strongly supported that Equisetales (horsetails) are the sister group to all other monilophytes. This topology confirmed the results reported by Rai and Graham [12] and Kuo et al. [33] based on plastid genes, and it was accepted by the PPG I [3] in 2016. Distinct from most fern phylogeny based on molecular evidence (Fig. 1), our results based on a coalescent method revealed that Psilotales (whisk ferns), Ophioglossales (moonworts), and Marattiales (king ferns) form a monophyletic clade as ([Psilotales, Ophioglossales], Marattiales), which is sister to leptosporangiate ferns. The monophyletic origin of Psilotales, Ophioglossales, and Marattiales, which belong to eusporangiate ferns, is supported by the structure of sporangia. Being different from the leptosporangiate type, sporangia of eusporangiate ferns have no sporangiophore; they are thick in wall and large in volume, produce large amounts of spores, and have no sporangial annulus or only have a few enlarged parenchyma cells. The incongruence between the results based on coalescent and concatenation methods may be caused by strong ILS effect, which is a main pitfall when using the concatenation method [21].
Relationship of early leptosporangiates
Within early leptosporangiates, our results revealed a new monophyletic clade in which Gleicheniaceae (forking ferns) is sister to Hymenophyllaceae (filmy ferns), which is different from the mainstream [3, 10, 12–14, 34]. Similar but still different from the topology ([Dipteridaceae, Matoniaceae], Gleicheniaceae], Hymenophyllaceae) reported by Pryer et al. in 2004 [5], in our results, Cheiropleuria, which belongs to Dipteridaceae and was formerly placed in Gleicheniales [2, 5, 12, 26, 35, 36], is sister to the monophyletic clade of (Gleicheniaceae, Hymenophyllaceae).
This new relationship is supported by sporangia character. Early leptosporangiates [36] are characterized by diverse sporangia and annulus. However, both Gleicheniaceae and Hymenophyllaceae have spherical sporangia with transverse-oblique annulus, as well as a short sporangial stalk connecting to a prominent receptacle [37]. On the other hand, flattened sporangia with slightly oblique annulus are found in Cheiropleuria. Moreover, long sporangial stalk and inapparent receptacle are common in Cheiropleuria, Dipteris, and Matonia. We suggest that Dipteridaceae, probably together with its sister lineage Matoniaceae [5, 12], may be sister to the clade of (Gleicheniaceae, Hymenophyllaceae). According to our results, Gleicheniales, which is comprised of Dipteridaceae, Matoniaceae, and Gleicheniaceae [26], is no longer a monophyletic lineage, but a paraphyletic one.
Relationships within polypod ferns
Polypods include more than 80% of living ferns, and their phylogeny remains somewhat controversial and elusive [26, 35, 36]. Our results strongly supported that Dennstaedtiaceae instead of Pteridaceae is sister to eupolypods. This pattern confirmed the topology suggested recently by Rothfels et al. based on 25 low-copy nuclear genes [14] and Lu et al. based on plastid genes [13], as well as the PPG I system [3]. According to our results, the relationships of Pteridaceae [34, 36, 38] and Dennstaedtiaceae [36] are also well resolved. Notably, Monachosorum is sister to the rest of the members in Dennstaedtiaceae, rather than being sister to the lineage of Pteridium, Hypolepis, and Histiopteris [36].
Our results showed that eupolypods are divided into 2 major lineages, eupolypods I and eupolypods II, in agreement with the consensus opinion [3]. Within eupolypods II, our results supported that Aspleniaceae is the sister group to the rest of the members, which is different from the current viewpoint [26, 36, 39]. Within eupolypods I, our result strongly supported that Lomariopsidaceae and Nephrolepidaceae form a paraphyletic group, rather than a monophyletic clade based on plastid genes [10, 26, 36].
Our new topology confirmed the morphology-based hypothesis that Dennstaedtiaceae with 2 indusial, rather than Pteridaceae with 1 false indusium, is more closely related to eupolypod ferns [40]. In Pteridaceae, the unstable structure of spherical sporangia, including variable annulus and short sporangial stalk, indicates that these characters of sporangia are relatively original and are close to those with oblique annulus in early leptosporangiates [23]. We also noticed that the characters of spherical sporangia with slightly oblique annulus in Monachosorum should be more ancestral than the flattened sporangia with typical vertical annulus in other genera of Dennstaedtiaceae. For distinguishing eupolypods I and eupolypods II, the number and shape of the vascular bundles at the base of petiole have been demonstrated to be of a powerful diagnostic character [36, 39].
The evolution of sporangial annulus in ferns
By observing the character of sporangial annulus of abundant samples in each fern group and combining these characters with our well-resolved backbone phylogeny (Fig. 3), we reconstructed the evolutionary history of sporangial annulus in ferns (Fig. 4). According to the results, we infer that ex-annulus sporangia, as in Equisetaceae, Psilotaceae, and Ophioglossaceae, is the ancestral state in ferns; rudimentary multiseriate annulus, which is inverse U-shaped in Marattiaceae and U-shaped in Osmundaceae; equatorial transverse-oblique uniseriate annulus, as in Gleicheniaceae and Hymenophyllaceae; oblique annulus as in Cyatheales (tree ferns); and vertical annulus as synapomorphy in polypods have been derived from the ex-annulus state. Both apical annulus, as in Lygodium and Schizaea, and vestige or disappeared annulus, as in Salviniales (aquatic ferns), are likely to be specialized in parallel from oblique annulus. Inconsistent with Bower's hypothesis [23], our results showed that sporangia with apical annulus as in Schizaeales are no longer the ancestral type in ferns but a specialized one. Correspondingly, the oldest fossils of Schizaeaceae are now believed to appear in the Jurassic period (201–145 MY BP) rather than formerly thought Carboniferous period (359–252 MY BP) [41].
Conclusion
Our results confirmed that Equisetales is sister to all the other monilophytes and that Dennstaedtiaceae is sister to eupolypods, which have been reported previously. Moreover, our results revealed some new relationships, such as that eusporangiate ferns, except Equisetales, may form a monophyletic clade as ([Psilotaceae, Ophioglossaceae], Marattiaceae), while Gleicheniaceae and Hymenophyllaceae form a monophyletic clade, which is sister to Dipteridaceae, and that Aspleniaceae is sister to the rest of the groups in eupolypods II. Most of these results are supported by sporangia characters, and a new evolutionary route of sporangial annulus in ferns is suggested.
Potential implications
Here, we present a robust fern phylogeny yielded from a large-scale phylogenomic analysis based on a high-quality RNA-seq dataset covering 69 fern species. This backbone phylogeny in ferns sets a foundation for further studies in biology and evolution in ferns and therefore in plants, especially when fern genomes are not available.
Methods
De novo transcriptome assembly
For each paired-end library, we first removed the Illumina adapter of raw reads using Scythe (Scythe, RRID: SCR_011844) [42] and trimmed the poor-quality bases using DynamicTrim Perl script of the SolexQA package with default parameters [43]. Next, de novo transcriptome assembly of each species was conducted using the Trinity package, version trinityrnaseq_r20140413 (Trinity, RRID: SCR_013048) with default parameters [44]. To discard the duplicated sequences, the obtained contigs were clustered using CD-HIT-EST v4.6.1 (CD-HIT, RRID: SCR_007105) to generate nonredundant contigs. All contigs longer than 200 bp in length were used for downstream analysis. We used TransDescoder, a program in the Trinity package, to identify the candidate coding sequences (CDS) from the contigs with default criteria. Finally, the translated protein sequences of CDS were searched by BLASTP against the nonredundant protein database in NCBI with an e-value threshold of 1e-5. These BLASTP hit sequences were used for further analysis.
Orthology assignment, alignment, and alignment masking
For orthology assignment for the 69 sample assemblies together with the 4 outgroup species, a phylogenetic-based clustering method described previously [16] was used. In short, an all-vs-all BLAST search of amino acid sequence was performed across different species; the BLAST results were clustered using MCL [45] software with the parameters ‘-I 2–tf ΄gq(20)΄.’ Optimization of the inflation parameter (I) was conducted as described previously [46], and the default value 2.0 was selected ultimately. As the de novo assembly by Trinity produces many sequences with high similarity, which contain both paralogs and isoforms [47], when a clustered gene family contains too many sequences (e.g., more than 10), the risk of contamination of isoforms rises, along with the computational infeasibility. Hence, when a species had more than 10 sequences in a gene family, we removed all sequences in this gene family of this species. Then, groups with at least 35 (50%) fern species were aligned using the einsi command, implemented in MAFFT (MAFFT, RRID: SCR_011811) [48], and trimmed by Gblocks with default parameters [49]. Next, for each group, a homologous gene tree was built with RAxML software, version 8.0.20 (RAxML, RRID: SCR_006086), by implementing the maximum likelihood method (ML) [50]. To infer orthologous genes, we used treeprune in the Agalma package [51] to mask the monophyletic sequences. We pruned the paralogous subtrees from the homologous gene trees until only 1 monophyletic subtree was retained. Next, the resulting orthologous gene trees were further filtered by the criteria that each species should be represented by only 1 sequence, and the resulting subset genes were referred to “1-to-1 orthologs,” which were largely free of gene duplication. Then, we extracted both the CDS (nucleotide sequence) and translated amino acid sequence from each orthologous gene group, followed by aligning with MAFFT and trimming with Gblocks. The alignment with coding and corresponding translated sequences longer than 150 bp (or 50 amino acids) in length were kept for further analysis.
Basic Universal Single Copy Orthologs analysis
The Basic Universal Single Copy Orthologs (BUSCO, RRID: SCR_015008), which employs a core set of orthologs conservative in eukaryotic species to determine the gene coverage of each assembly [52], was employed to assess the completeness of the transcriptome assembly we obtained (Table S2) [53]. A total of 303 BUSCOs were employed to blast against by translated amino acid of the assemblies using BLASTP. Then the numbers of complete and partially matched genes from each assembly were counted. Out of the 69 samples in total, the gene coverage of 65 samples (94.2%) exceeded 82%, with at least 251 complete genes identified. Unexpectedly, among our total assemblies, 1 sample (Aleuritopteris chrysophylla, named RS_72) presented an extremely low gene coverage degree, in which only 72 (23.8%) complete housekeeping genes were found (Supplementary Table S2). However, when the sample was deleted from the matrix used to construct the backbone of the phylogenetic tree, the topology remained unchanged, indicating that the lower completeness in this sample did not affect our results (data not shown).
Phylogenetic analysis
The coalescent-based species trees were reconstructed by ASTRAL v4.10.4 [54], carried out by 100 replicates of multilocus bootstrapping [55]. Each gene tree was constructed with the PROTCATJTTF model by RAxML v8.2.4 (RAxML, RRID: SCR_006086) [50], performed using 100 random replicates to calculate bootstrap value. For the concatenation analysis, we preformed the ML for each matrix using RAxML software (version 8.0.20). Branch support was evaluated using 100 bootstrap replicates. We used the “GTR + Γ4 + I” model for DNA matrices, and the JTTF model for the corresponding protein matrices, selected by “ProtienModelselection.pl” [56]. To estimate the divergence times, we used the concatenated alignment of orthologs, calibrated with the ages of 2 fossils (Archaeocalamites Senftenbergia: 354 MY, Grammatopteris: 280 MY) [6, 57] as the minimum ages of monilophytes and leptosporangiate ferns, respectively, and a maximum age constraint of 500 MY for land plants in a Bayesian relaxed clock method using MCMCTREE [58] on the coalescent-based species tree.
Reconstruction of the evolution of sporangial annulus
Characters of sporangial annulus of the sampled species were observed using a polarized light microscope (Axio Scope.A1, ZEISS) after the fresh and mature sporangia were treated with sodium hypochlorite (NaClO) solution. The evolution of sporangial annulus was reconstructed with the likelihood method, implemented in Mesquite v2.7.5 [59]. All character states (i.e., vertical annulus, oblique annulus, rudimentary annulus, ex-annulus, apical annulus, transverse annulus, and vestigial annulus) were treated as unordered and equally weighted. To reconstruct character evolution, a maximum likelihood approach using Markov k-state 1 parameter model [60] was applied. To account for phylogenetic uncertainty, the “Trace-characters-over-trees” command was used to calculate the ancestral states at each node, including probabilities in the context of likelihood reconstructions. To carry out these analyses, characters were plotted onto 100 trees that were sampled in the ML analyses of the combined dataset using RAxML v7. The results were finally summarized as percentage of changes of character states on a given branch among all 100 trees utilizing the option of “Average-frequencies-across-trees.”
Availability of data and materials
Raw reads of RNA-Seq for 69 fern species were deposited in GenBank under Bioproject accession number PRJNA281136. Transcriptome datasets, alignments, phylogenetic trees, BUSCO results and other supporting data are available via the GigaScience repository, GigaDB [61].
Abbreviations
BUSCOs: Basic Universal Single-Copy Orthologs; ILS: incomplete lineage sorting; ML: maximum likelihood; MY: million years; PPG: Pteridophyte Phylogeny Group; RNA-Seq: transcriptome sequencing.
Additional files
Additional file 1: Tables S1 and S2 and Figures S1–S3.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was funded by Shanghai Landscaping and City Appearance Administrative Bureau of China, Scientific Research Grants (G142433, G152420, and F112422), and the National Natural Science Foundation of China (31370234).
Author contributions
Y.H.Y., H. Shen, and D.M.J. conceived and designed the study. M.L., J.P.S., D.M.J., R.W., and L.L. implemented the data analyses. Y.H.Y., H. Shen, H.J.W., X.L.Z., H. Shang, and Y.F.G. collected the specimens. H. Shen, R.Z., and Y.F.G. prepared the specimens for sequencing. X.L.Z. provided the anatomical data. D.M.J., H. Shen, Y.H.Y., J.P.S., M.L., R.W., H. Shang, X.L.Z., and X.C.Z. interpreted the results and wrote the manuscript.
Supplementary Material
31 Jul 2017 Reviewed
06 Sep 2017 Reviewed
01 Aug 2017 Reviewed
03 Aug 2017 Reviewed
Acknowledgements
We thank Prof. Yong-Hong Hu, Prof. Jin-Shuang Ma, Prof. Zhao-Qing Chu, and Dr. Jun Yang from Shanghai Chenshan Botanical Garden of China, as well as Prof. Fu-Wu Xing from South China Botanical Garden of CAS, for helpful comments and suggestions. We thank Prof. Paul G. Wolf from Utah State University, Prof. Yin-Long Qiu from the University of Michigan, and Dr. Jin-Long Zhang from Kadoorie Farm and Botanic Garden for providing important suggestions regarding the research method. We appreciate helpful comments and suggestions from 3 reviewers of previous versions of this manuscript.
References
- 1. Duff RJ, Nickrent DL. Phylogenetic relationships of land plants using mitochondrial small-subunit rDNA sequences. Am J Bot 1999;86(3):372–86. [PubMed] [Google Scholar]
- 2. Pryer KM, Schneider H, Smith AR et al. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 2001;409(6820):618–22. [DOI] [PubMed] [Google Scholar]
- 3. The Pteridophyte Phylogeny Group A community-derived classification for extant lycophytes and ferns. J Syst Evol 2016;54(6):563–603. [Google Scholar]
- 4. Qiu Y-L, Li L, Wang B et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci U S A 2006;103(42):15511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pryer KM, Schuettpelz E, Wolf PG et al. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am J Bot 2004;91(10):1582–98. [DOI] [PubMed] [Google Scholar]
- 6. Schneider H, Schuettpelz E, Pryer KM et al. Ferns diversified in the shadow of angiosperms. Nature 2004;428(6982):553–7. [DOI] [PubMed] [Google Scholar]
- 7. Schuettpelz E, Pryer KM. Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc Natl Acad Sci U S A 2009;106(27):11200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang LB, Zhang L, Dong SY et al. Molecular circumscription and major evolutionary lineages of the fern genus Dryopteris (Dryopteridaceae). BMC Evol Biol 2012;12(1):180–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu H-M, Zhang X-C, Wang W et al. Molecular phylogeny of the fern family dryopteridaceae inferred from chloroplast rbc L and atp B genes. Int J Plant Sci 2007;168(9):1311–23. [Google Scholar]
- 10. Liu H-M. Embracing the pteridophyte classification of Ren-Chang Ching using a generic phylogeny of Chinese ferns and lycophytes. J Syst Evol 2016;54(4):307–35. [Google Scholar]
- 11. Schneider H, Smith AR, Pryer KM. Is morphology really at odds with molecules in estimating fern phylogeny? Syst Bot 2009;34(3):455–75. [Google Scholar]
- 12. Rai HS, Graham SW. Utility of a large, multigene plastid data set in inferring higher-order relationships in ferns and relatives (monilophytes). Am J Bot 2010;97(9):1444–56. [DOI] [PubMed] [Google Scholar]
- 13. Lu J-M, Zhang N, Du X-Y et al. Chloroplast phylogenomics resolves key relationships in ferns. J Syst Evol 2015;53(5):448–57. [Google Scholar]
- 14. Rothfels CJ, Li F-W, Sigel EM et al. The evolutionary history of ferns inferred from 25 low-copy nuclear genes. Am J Bot 2015;102(7):1089–107. [DOI] [PubMed] [Google Scholar]
- 15. Hittinger CT, Johnston M, Tossberg JT et al. Leveraging skewed transcript abundance by RNA-Seq to increase the genomic depth of the tree of life. Proc Natl Acad Sci U S A 2010;107(4):1476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith S, Wilson N, Goetz F et al. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 2011;480(7377):364–7. [DOI] [PubMed] [Google Scholar]
- 17. Misof B, Liu S, Meusemann K et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014;346(6210):763–7. [DOI] [PubMed] [Google Scholar]
- 18. Wen J, Xiong Z, Nie Z-L et al. Transcriptome sequences resolve deep relationships of the grape family. PLoS One 2013;8(9):e74394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng L, Zhang Q, Sun R et al. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun 2014;5:4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wickett NJ, Mirarab S, Nam N et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A 2014;111(45):E4859–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mirarab S, Bayzid MS, Boussau B et al. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 2014;346(6215):1250463. [DOI] [PubMed] [Google Scholar]
- 22. Noblin X, Rojas N, Westbrook J et al. The fern sporangium: a unique catapult. Science. 2012;335(6074):1322-. [DOI] [PubMed] [Google Scholar]
- 23. Bower FO, eds. The Ferns (Filicales): Treated Comparatively with a View to Their Natural Classification. 1–3 London: Cambridge University Press; 1923,1926,1928. [Google Scholar]
- 24. Pichi-Sermolli REG. Historical review of the higher classification of the Filicopsida. In: Jermy AC, Crabb JA, Thomas BA, eds. Phylogeny and Classification of the Ferns. London: Botanical Journal of the Linnean Society; 1973, 11–40. [Google Scholar]
- 25. Smith AR. Non-molecular phylogenetic hypotheses for ferns. Am Fern J 1995;85(4):104–22. [Google Scholar]
- 26. Smith AR, Pryer KM, Schuettpelz E et al. A classification for extant ferns. Taxon 2006;55:705–31. [Google Scholar]
- 27. Raw reads https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA281136. Accessed 5 July 2017.
- 28. Transcriptome datasets https://figshare.com/s/0f773861b6813f97ff63. Acessed 5 July 2017.
- 29. Phytozome http://phytozome.jgi.doe.gov/. Accessed 5 July 2017.
- 30. Alignments https://figshare.com/s/f835735cb66911ff1ffd. Accessed 5 July 2017.
- 31. Datasets of coalescent-based species tree https://figshare.com/s/e5e70c2fd3990e5176d8. Accessed 5 July 2017.
- 32. Datasets of concatenation based phylogenetic tree https://figshare.com/s/8af236b660f61078e40b. Accessed 5 July 2017.
- 33. Kuo LY, Li FW, Chiu WL et al. First insights into fern matK phylogeny. Mol Phylogenet Evol 2011;59:556–66. [DOI] [PubMed] [Google Scholar]
- 34. Schneider H. Evolutionary morphology of ferns (monilophytes). Annu Plant Rev 2013;45:115–40. [Google Scholar]
- 35. Christenhusz MJM, Chase M. Trends and concepts in fern classification. Ann Bot 2014;113:571–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schuettpelz E, Pryer KM. Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes. Taxon 2007;56:1037–50. [Google Scholar]
- 37. Bierhorst DW. Morphology of Vascular Plants. New York: Macmillan; 1971. [Google Scholar]
- 38. Schuettpelz E, Schneider H, Huiet L et al. A molecular phylogeny of the fern family Pteridaceae: assessing overall relationships and the affinities of previously unsampled genera. Mol Phylogenet Evol 2007;44:1172–85. [DOI] [PubMed] [Google Scholar]
- 39. Rothfels CJ, Sundue MA, Kuo L-Y et al. A revised family-level classification for eupolypod II ferns (Polypodiidae: Polypodiales). Taxon 2012;61:515–33. [Google Scholar]
- 40. Mickel JT. The classification and phylogenetic position of the Dennstadtieaceae. In: Jeremy AC, Crabbe JA, Thomas BA, eds. The Phylogeny and Classification of the Ferns. London: Academic Press for The Linnean Society of London; 1973:135–44. [Google Scholar]
- 41. Taylor TN, Taylor EL, Krings M. Paleobotany: The Biology and Evolution of Fossil Plants. 2nd ed San Diego: Academic Press; 2009. [Google Scholar]
- 42. Scythe https://github.com/ucdavis-bioinformatics/scythe. Accessed 5 July 2017.
- 43. Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 2010;11:485 doi:10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grabherr MG, Haas BJ, Yassour M et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 2011;29:644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Dongen S. A cluster algorithm for graphs. Technical Report INS-R0010, National Research Institute for Mathematics and Computer Science in the Netherlands. 2000. http://micans.org/mcl/index.html?sec_thesisetc. Accessed 5 July 2017.
- 46. Hejnol A, Obst M, Stamatakis A et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Royal Soc B Biol Sci 2009;276:4261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Haas BJ, Papanicolaou A, Yassour M et al. De novo transcript sequence reconstruction from RNA-seq 600 using the Trinity platform for reference generation and analysis. Nat Protoc 2013;8(8):1494–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 2007;56:564–77. [DOI] [PubMed] [Google Scholar]
- 50. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dunn CW, Howison M, Zapata F. Agalma: an automated phylogenomics workflow. BMC Bioinformatics 2013;14:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Simao FA, Waterhouse RM, Ioannidis P et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015;31:3210–2. [DOI] [PubMed] [Google Scholar]
- 53. BUSCO results https://figshare.com/s/bf999173d04b4c311d46. Accessed 5 July 2017.
- 54. Mirarab S, Reaz R, Bayzid MS et al. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 2014;30:i541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Seo TK. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Mol Biol Evol 2008;25:960–71. [DOI] [PubMed] [Google Scholar]
- 56. ProtienModelselection.pl https://github.com/stamatak/standard-RAxML/. Accessed 5 July 2017.
- 57. Rößler R, Galtier J. First Grammatopteris tree ferns from the Southern Hemisphere – new insights in the evolution of the Osmundaceae from the Permian of Brazil. Rev Palaeobot Palynol 2002;121:205–30. [Google Scholar]
- 58. dos Reis M, Yang Z. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol 2011;28:2161–72. [DOI] [PubMed] [Google Scholar]
- 59. Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2011. http://mesquiteproject.org. Accessed 5 July 2017.
- 60. Lewis PO, Olmstead R. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol 2001;50:913–25. [DOI] [PubMed] [Google Scholar]
- 61. Shen H, Jin D, Shu J et al. Supporting data for “Large scale phylogenomic analysis resolves a backbone phylogeny in ferns.” GigaScience Database 2017. http://dx.doi.org/10.5524/100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
31 Jul 2017 Reviewed
06 Sep 2017 Reviewed
01 Aug 2017 Reviewed
03 Aug 2017 Reviewed
Data Availability Statement
Raw reads of RNA-Seq for 69 fern species were deposited in GenBank under Bioproject accession number PRJNA281136. Transcriptome datasets, alignments, phylogenetic trees, BUSCO results and other supporting data are available via the GigaScience repository, GigaDB [61].