Abstract
The western honey bee, Apis mellifera, is an enormously influential pollinator in both natural and managed ecosystems. In North America, this species has been introduced numerous times from a variety of different source populations in Europe and Africa. Since then, feral populations have expanded into many different environments across their broad introduced range. Here, we used whole genome sequencing of historical museum specimens and newly collected modern populations from California (USA) to analyze the impact of demography and selection on introduced populations during the past 105 years. We find that populations from both northern and southern California exhibit pronounced genetic changes, but have changed in different ways. In northern populations, honey bees underwent a substantial shift from western European to eastern European ancestry since the 1960s, whereas southern populations are dominated by the introgression of Africanized genomes during the past two decades. Additionally, we identify an isolated island population that has experienced comparatively little change over a large time span. Fine-scale comparison of different populations and time points also revealed SNPs that differ in frequency, highlighting a number of genes that may be important for recent adaptations in these introduced populations.
Keywords: Apis mellifera, population genomics, demography
Introduction
The western honey bee, Apis mellifera, is the world’s most important managed pollinator (Klein et al. 2007) and an iconic home garden visitor. It has been used throughout its native range in Europe, Africa, and western Asia as a source of honey and wax since the Paleolithic era, and the practice of keeping bees in hives likely emerged in Egypt between 3000 and 5000 BCE (Crane 1999). Although beekeeping in the honey bee’s native range has primarily relied on the use of local populations of A. mellifera, beekeepers outside the native range rely on various introduced, and genetically distinct, populations. These populations are now the most important pollinators of agricultural crops (Klein et al. 2007) and understanding temporal and spatial changes in genetic variation within and between these populations is vital to the agricultural industry.
Apis mellifera was first introduced into North America in 1622 (Crane 1999) when populations were brought over by French and English colonists. These populations probably belonged to the M lineage that occurs throughout western Europe (Ruttner’s designations, Ruttner 1988). In 1853, A. mellifera was first introduced to California, when hives from the eastern United States arrived in the city of San Jose (Watkins 1968). Subsequently, in 1859, Italian A. mellifera bees belonging to the C lineage (Ruttner 1988) were imported to North America and were quickly transported to California (Crane 1999). Despite occurring in close geographic proximity in Europe, the M and C lineages represent two separate and genetically distinct radiations of A. mellifera out of Africa into Europe (Ruttner 1988; Whitfield et al. 2006). However, these two major lineages breed freely and there is evidence of admixture in their modern-day contact zone across central Europe (Cridland et al. 2017). These two lineages are the primary contributors to the modern managed honey bee stocks present in California.
In addition to large populations of managed A. mellifera there are also substantial feral populations of bees in California. Feral bees are ultimately derived from a variety of managed stocks and are continuously replenished as managed bees routinely escape and establish new feral colonies. Although managed and feral honey bees share genetic histories, they experience very different sets of selective pressures. Managed honey bee populations experience significant selective pressures for desirable traits (Winston et al. 1983), including honey production (Guzman-Novoa and Page 1999), hygenic behavior, and pathogen resistance (Buchler et al. 2010). At the same time, managed honey bees are shielded from other selective pressures as beekeepers provide supplemental food, shelter, and medication against common pathogens and parasites. Feral bees, however, experience different selective pressures relative to managed honey bees, and thus may provide insight into the processes of adaptation to local conditions. These populations may even serve as a genetic reservoir for future managed stocks (Sheppard and Huttel 1988; Schiff et al. 1994; De la Rúa et al. 2009). Feral bees also contribute to the pollination of agricultural crops, though the amount of that contribution is unclear (Losey and Vaughn 2006).
The introduction of tropical African A. mellifera scutellata (lineage A, Ruttner 1988) to Brazil in 1956 (Kerr 1967; Crane 1999), and its subsequent expansion into California provided an additional, and dissimilar, source of genetic variation to feral Californian A. mellifera populations. These populations have spread rapidly and have quickly altered the genetic landscape of feral population in the southern United States though they have not entirely replaced European-derived feral populations (Pinto et al. 2004). Africanized bees were first documented in southern California in 1994. Since then these populations have introgressed northward, and a 2015 study found evidence of African derived alleles only 40 km south of Sacramento (Kono and Kohn 2015).
California is the seasonal home to nearly half of the managed bee colonies in the continental United States (1,140,000 in January of 2016 [USDA National Agriculture Statistics Service, 2016]). This makes information about changes in the genetic composition of bee populations in California, both feral and managed, of particular interest to bee breeders and the agricultural industry. Apis mellifera populations in California have experienced a number of stresses in the last several decades. First, there are concerns about the spread of Africanized honey bees and the potential of these bees to interbreed with managed colonies, producing offspring with unfavorable traits (i.e., aggressive behavior, lower honey production) (Rinderer et al. 1985). Second, the mortality rate of managed colonies has increased in the United States between 1944 and 2008 (vanEngelsdorp and Meixner 2010). In 2016, the USDA reported a 15% loss of colonies in California between January and March (USDA National Agriculture Statistics Service, 2016). These losses are believed to be influenced by a number of factors that currently threaten managed honey bee populations including pesticides (Brandt et al. 2016), diseases and parasites (vanEngelsdorp and Meixner 2010; Sanchez-Bayo et al. 2016). However, there are indications that genetically diverse hives may be more resistant to disease (Tarpy and Seeley 2006).
Several previous studies have attempted to quantify the contributions of different A. mellifera lineages to the populations in California and the United States generally (Schiff et al. 1994; Magnus and Szalanski 2010; Harpur et al. 2012; Kono and Kohn 2015; Rangel et al. 2016), but previous studies used either mitochondrial markers or small sets of sequenced genes to assess population structure. Here, we present a whole-genome based study of the contributions of European and African honey bee populations to introduced (feral and managed) Californian bees over a 105 year period (1910–2015). Our analyses of how ancestry and genes change through time, across the landscape, and among populations provide insights into adaptive loci and genetic divergence, and are the first steps towards understanding ecologically relevant traits and local adaptation (Luikart et al. 2003; Stinchcombe and Hoekstra 2008; Allendorf et al. 2010). To conduct our analysis, we used paired historical and modern samples from throughout the state, spanning ∼65% of the time that honey bees have existed in California. We hypothesize substantial changes in the contributions of various European and African ancestral lineages to the genetic profile of Californian populations over both spatial and temporal dimensions. In particular, we expect to see an increase in the contribution of African lineages to southern Californian populations over time, including the identification of admixed individuals, paralleling the arrival and spread of Africanized bees. We further hypothesized a reduction in genetic diversity in northern Californian populations in years following Varroa mite outbreaks as feral colonies suffer substantial losses and the expected proportion of feral colonies that are recently derived from managed populations increases. Finally, we expect to identify candidate genes that may be involved in local adaptation to the state of California’s diverse ecological regions.
Materials and Methods
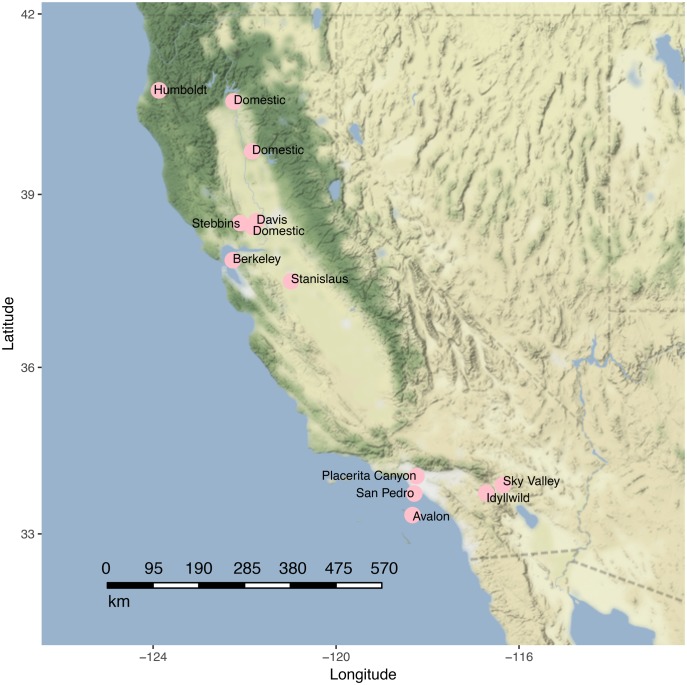
We acquired 29 museum samples of A. mellifera collected in California between 1910 and 2011 (fig. 1 and table 1). In addition, we included a set of A. mellifera individuals from two publicly available data sets (Harpur et al. 2014; Wallberg et al. 2014). We used a SNP data set that we previously generated from these two resources (Cridland et al. 2017) to infer patterns of ancestry in introduced A. mellifera populations from California. This set included samples from Africa (47 individuals), western Europe (39 individuals), eastern Europe (29 individuals), the Middle East (10 individuals), and ten A. cerana individuals from Japan.
Fig. 1.
—Sampling Locations in California. Sky Valley and Idyllwild are collectively referred to as Riverside.
Table 1.
Samples
| Location | Longitude | Latitude | Date | N |
|---|---|---|---|---|
| Avalon, Catalina Island, Los Angeles county | −118.32 | 33.34 | June 1910, September 2014 | 2, 5 |
| UC Berkeley Campus, Alameda county | −122.26 | 37.87 | July 2011 | 1 |
| Arcata, Humboldt county | −124.07 | 40.87 | January 2015 | 6 |
| Blue Lake, Humboldt county | −123.95 | 40.88 | February 1966 | 6 |
| Placerita Canyon Nature Area, Los Angeles county | −118.46 | 34.37 | August 1999, September 2014 | 5, 6 |
| Sky Valley, Riverside county | −116.27 | 33.84 | November 1983, September 2014 | 2, 4 |
| Idyllwild, Riverside county | −116.72 | 33.74 | May 1999, September 2014 | 2, 4 |
| San Pedro, Los Angeles | −118.29 | 33.73 | January 2002 | 2 |
| La Grange, Stanislaus county | −120.46 | 37.66 | September 1976, September 2014 | 2, 6 |
| Stebbins Cold Canyon Reserve, Solano county | −122.08 | 38.3 | March 1996, September 2014 | 5, 5 |
| UC Davis Campus, Yolo county | −121.75 | 38.54 | 1968 and January 2015 | 2, 6 |
| Noble Apiaries, Dixon CA, Yolo county | −121.86 | 38.43 | March 2015 | 3 |
| C.F.Koehnen Apiary, Knight's Landing, Yolo county | −121.72 | 38.8 | March 2015 | 2 |
| Wooten's Golden Queens Apiary, Palo Cedro, Shasta county | −122.23 | 40.61 | March 2015 | 1 |
DNA extraction and library preparation for museum and modern samples were similar. Benchtop, pipettes and extraction forceps were cleaned with bleach and rinsed with distilled water prior to use. Filter tips were used of all steps. Museum samples were handled individually and separately from modern samples. DNA extractions were completed using DNeasy Blood and Tissue kit (Qiagen) following the standard protocol for modern specimens and a reduced elution volume of 130 µl for museum specimens. A vacuum centrifuge concentrator was used to increase DNA concentration for samples with a Qubit reading less than 2.5 ng/µl.
Whole-genome library preparation was completed using Nextera DNA Library Preparation Kit (Illumina) following standard protocol for low plexity index pooling. Library success was confirmed by Bioanalyzer High Sensitivity DNA kit (Agilent Technologies). DNA sequencing of 100 bp single-end reads on Illumina HiSeq2000 was performed at Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley.
We sequenced each of these individuals on between one-sixth and one-third of a lane and generated between 0.38 and 32.7× mean coverage of the genomes. Sequences were aligned to the A. mellifera reference genome version 4.5, available from beebase.org, using Bowtie2 with the very-sensitive-local alignment parameters Langmead and Salzberg (2012). The variation in coverage is largely due to the difficulties in extracting high quality DNA for sequencing from preserved museum specimens and our oldest sequences, especially those from 1910, have the lowest mean coverage (supplementary table 1, Supplementary Material online). Older samples were also more likely to have missing data, though all samples but one had calls for at least half of the SNPs included.
SNP Sets
We generated two sets of SNPs for analyzing the Californian samples. The first SNPs were a previously generated set of SNPs identified in native range African and European bees (Cridland et al. 2017) and was used for all ancestry analyses. We used samtools/vcftools to generate SNP calls for each individual, requiring a quality score of 30 for reads to be included. A minimum coverage of 7× was required to make genotype calls for an individual and we required 2× coverage of an alternate base to make heterozygote calls. The second set was generated from the samples of bees collected in California in 2014–2015 and was used to examine differences between Californian populations. Because many of the historical samples had lower levels of coverage and thus fewer sites where there is genotype information we used the 2014 samples (41 feral + 6 domestic) to identify a set of SNP set for downstream analyses. To include a SNP in the data set we required that we were able to make a genotype call in 40/47 samples from 2014/2015. We then made genotype calls at those positions in all samples using the same set of coverage requirements as above. A total of 3,890,276 SNPs were identified used for downstream analyses.
Ancestry Analyses
We ran ADMIXTURE (Alexander et al. 2009) for the complete set of European, Middle Eastern, African, and Californian individuals for K population values between 2 and 6 to examine population splits at different assumed values. We initially ran this analysis with O group individuals included, but these individuals were never separated by ADMIXTURE from C group individuals. Running the analysis with the O group removed produced the same result with respect to the Californian individuals. We therefore conclude that either the O group does not contribute to the Californian populations or we are not able to distinguish contributions from the O group from contributions from the C group. We found that individuals from the two locations in Riverside County (Sky Valley and Idyllwild) were very similar in each analysis and we combined these two sites into a single population for downstream analyses.
We calculated FST for pairs of populations collected in California in 2014 and the populations from Africa, western Europe and eastern Europe (Harpur et al. 2014, Wallberg et al. 2014) using Dadi (Gutenkunst et al. 2009). To examine the patterns of relatedness between individuals and between major groups we ran a nonparametric multidimensional scaling analysis (nMDS) using the R package ecodist version 1.2.2 and calculated the stress value and R2 value for each analysis. An nMDS analysis represents similarity between individuals by positioning them in multidimensional space. We ran the adonis function from the vegan version 2.3–4 package in R to identify the effect of major groupings on the distance matrix.
We performed formal tests of admixture using ADMIXTOOLS (Patterson et al. 2012). We calculated f3 statistics for all pairs of source populations for each potential target population to identify populations with evidence of admixture, where admixture is signified by a negative f3 statistic. For tests where we identified a negative f3 statistic, we calculated the upper and lower bound on the admixture proportion. We then kept all results for which we found evidence of admixture with an f3 statistic less than −0.01 and a z-score at least four standard deviations from the mean. F4 ratio estimation was performed on the same set of f3 statistics that passed our filters to estimate the proportion of ancestry in the admixed populations.
Geographic Differentiation
We calculated FST per site for pairs of populations between African, western European, and eastern European groups and Californian 2014 populations. For these tests we combined the southern Californian populations of Placerita and Riverside into a single population and the northern Californian populations of Davis, Humboldt, Stanislaus, and Stebbins into a single population. We required genotype information for at least 80% of individuals for each population in a given population comparison to include that site in the analysis. We then calculated the 95th percentile of FST.
We identified all sites for which an alternate SNP was only ever seen in one of the ancestral populations. We then compared the SNPs found in only one ancestral population at intermediate to high frequency, 10% or greater, to the frequencies of those SNPs in the 2014 northern and southern Californian populations and used a Fisher’s Exact Test to examine the proportions of SNPs between each ancestral population and the southern versus northern Californian populations. We identified for each site in each population comparison if the site was in an exonic region and, if so, if the SNP difference produces a synonymous or nonsynonymous amino acid.
For each site identified in an exon we also identified which amino acid it coded for and the alternate amino acid encoded for by the alternate base.
For each pair of populations for the modern Californian populations as well as for the Humboldt 1966 versus 2015 populations we calculated a mid-P value based on a 2 by 2 contingency table for the number of reference and alternate allele calls for the two populations (epitools R-package). We then performed a q-value correction for the set of p-values using the R package qvalue.
Gene Ontology Analysis
We used the DAVID version 6.8 Functional Annotation tool (Huang et al. 2009a, 2009b) to identify enriched gene ontology terms. We used the medium stringency for gene classification in DAVID and used an enrichment score of 2.5 as the minimum score for identifying enriched clusters and the Benjamini corrected P value (Benjamini and Hochberg 1995) to identify significantly enriched terms within clusters.
Results
We called SNPs in a set of museum specimens and modern populations of A. mellifera collected throughout the state of California between 1910 and 2015 (fig. 1). We acquired museum specimens for a number of sites in northern and southern California as well as from Avalon, Catalina Island off the southern coast of California. Sampling bees from the same sites in 2014–2015 generated a modern collection. In addition, we included domestic drones from three different bee breeders in northern California. To identify patterns of variation in Californian populations with respect to changes in contributions from ancestral populations we used a previously curated set of SNPs generated from native range, African and European, bees for inferring the demographic history of A. mellifera (Cridland et al. 2017).
Californian Populations Derive Ancestry from Multiple Native Range Honey Bee Lineages
We tested the hypothesis that Californian A. mellifera derive ancestry from some combination of A. mellifera populations from eastern Europe (C lineage), western Europe (M lineage), and Africa (A lineage). We calculated mean π (nucleotide diversity) along 10 kb windows for each or our modern populations (table 2). The African population had the highest level of diversity followed by the southern Californian populations. The domesticated drone grouping showed the lowest nucleotide diversity in California. However, it exhibited similar levels of variation to that observed in the eastern and western European populations. We find that northern Californian populations have diversity levels intermediate to eastern and western European populations.
Table 2.
Mean π in Californian Populations
| Population | Location | Mean π |
|---|---|---|
| Avalon | Island | 7.14E-05 |
| Davis | Northern California | 7.62E-05 |
| Humboldt | Northern California | 6.43E-05 |
| Placerita Canyon | Southern California | 1.05E-04 |
| Riverside | Southern California | 1.14E-04 |
| Stanislaus | Northern California | 7.54E-05 |
| Stebbins | Northern California | 7.46E-05 |
| Domestic | Northern California | 6.38E-05 |
| A | Africa | 1.63E-04 |
| C | Central Europe | 5.17E-05 |
| M | Western Europe | 8.38E-05 |
Native Range Populations Are Differentially Represented across California
Modern populations of bees from northern California, southern California, and Avalon were genetically distinct from each other, and each population was most similar to a different native range population. To quantify genetic differentiation across populations from distinct geographic regions across California, we calculated FST between pairs of modern populations using Dadi (Gutenkunst et al. 2009). The southern Californian populations (Riverside County and Placerita Canyon) were more similar to the African population than they were to either the central or the western European populations (supplementary table 1, Supplementary Material online). In contrast, the northern Californian populations and the domesticated individuals were most similar to the eastern European population. Avalon was most similar to the western European population. Within California, we found that modern populations from the Central Valley (Stanislaus, Stebbins, and Davis) were most similar both to each other and to the domesticated populations, also from the Central Valley (supplementary table 2, Supplementary Material online). The southern Californian populations from Riverside County and Placerita Canyon were most similar to each other.
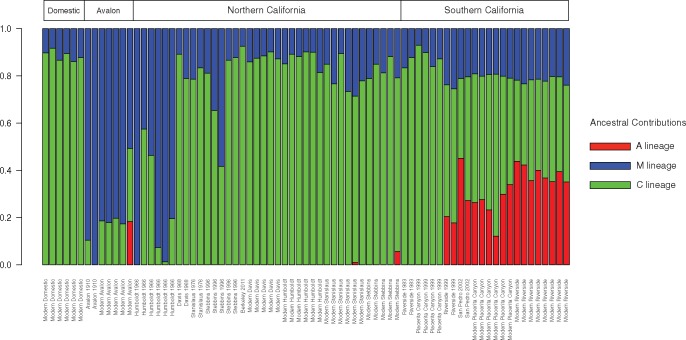
We tested the hypotheses that Californian bees exhibit admixture with between 2 and 5 source populations using the program ADMIXTURE (Alexander et al. 2009). In the following analysis, we also included two outgroup populations: A. cerana, a sister species, and Y lineage A. mellifera, which are not thought to have contributed to the genetic makeup of Californian bees (Magnus and Szalanski 2010). The samples from the four native range populations (African [A], western European [M], eastern European [C], and Middle Eastern [Y]) plus A. cerana clustered into five distinct lineages, as expected and as previously reported (Harpur et al. 2014; Wallberg et al. 2014; Cridland et al. 2017) (fig. 2 and supplementary fig. 1, Supplementary Material online). Comparing the native range bee population to introduced Californian populations showed that the latter derive ancestry from the African, eastern, and western European populations, which is in line with our expectations based on historical data documenting the initial introduction of honey bees to California.
Fig. 2.
—ADMIXTURE results for K = 5 for Californian bees.
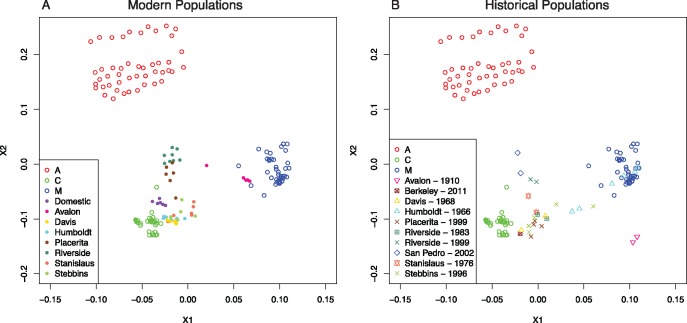
We examined the relatedness between the modern individuals in California and the African, eastern European, and western European population representatives by creating a distance matrix from the genotype data and performing a nonmetric Multidimensional Scaling Analysis (nMDS, stress = 0.071, R2 = 0.989). All Californian individuals were placed intermediate to the native range individuals (fig. 4A) and southern Californian individuals were placed closer to African individuals than northern Californian or Avalon individuals.
Fig. 4.
—Non-metric Multidimensional Scaling Analyses for ancestral population representatives, modern, and historical Californian individuals. (A) Modern populations. (B) museum collected.
We performed formal tests of admixture using ADMIXTOOLS (Patterson et al. 2012) to estimate contributions of native range populations to the Californian individuals (table 3). In the modern Davis, Humboldt, and Stanislaus populations, we find evidence of admixture between the Western European (M) group the domestic individuals and the eastern European (C) group as indicated by negative f3 scores and large negative Z-scores. In the modern southern Californian populations, we observed evidence of admixture between the African (A) group and the domesticated populations and western European populations (table 3).
Table 3.
Admixture in Modern Californian Populations
| Source 1 | Source 2 | Target | f_3 | std.err | Z | SNPs | αL | αU |
|---|---|---|---|---|---|---|---|---|
| C | M | Davis | −0.052211 | 0.001976 | −26.424 | 31070 | 0.769 | 0.841 |
| M | Domestic | Davis | −0.014021 | 0.003084 | −4.546 | 25749 | 0.037 | 0.379 |
| C | M | Humboldt | −0.025731 | 0.002279 | −11.289 | 30161 | 0.804 | 0.867 |
| A | Domestic | Placerita | −0.038386 | 0.00093 | −41.287 | 42501 | 0.435 | 0.493 |
| A | M | Placerita | −0.018291 | 0.001474 | −12.405 | 51054 | 0.284 | 0.708 |
| A | M | Riverside | −0.024526 | 0.001092 | −22.468 | 59331 | 0.481 | 0.712 |
| A | Domestic | Riverside | −0.024418 | 0.000992 | −24.612 | 51768 | 0.591 | 0.603 |
| C | M | Riverside | −0.010761 | 0.001425 | −7.554 | 56143 | 0.445 | 0.883 |
| C | M | Stanislaus | −0.065307 | 0.001788 | −36.535 | 31404 | 0.689 | 0.789 |
| A | Domestic | Stanislaus | −0.013122 | 0.001643 | −7.985 | 25962 | 0.092 | 0.237 |
For source population pairs where we found evidence of admixture, we calculated the F4 ratio (Patterson et al. 2012) to estimate ancestry proportions for the source populations. We observed that the proportion of the European M group in modern northern Californian populations ranges from around 53.5% ± 8.3% to 71.4% ± 9.2% depending upon the other source population considered (table 4). In modern southern Californian populations we found that the contribution of African populations was between 37.6% ± 7.3% and 73.3% ± 3.9% depending on the other source population considered. The variation in percentages is likely a reflection of the populations in question having more than two ancestral populations contributing to the target population.
Table 4.
Estimated Admixture Proportions in Modern Californian Populations
| Outgroup | Ougroup 2 | Source 1 | Source 2 | Target | alpha | std.err | Z Score |
|---|---|---|---|---|---|---|---|
| Cerana | Y | M | C | Davis | 0.645738 | 0.088711 | 7.279 |
| Cerana | Y | Domestic | M | Davis | 0.59859 | 0.122403 | 4.89 |
| Cerana | Y | M | C | Humboldt | 0.534668 | 0.083113 | 6.433 |
| Cerana | Y | A | Domestic | Placerita | 0.577514 | 0.039873 | 14.484 |
| Cerana | Y | A | M | Placerita | 0.376642 | 0.073008 | 5.159 |
| Cerana | Y | A | M | Riverside | 0.603464 | 0.060694 | 9.943 |
| Cerana | Y | A | Domestic | Riverside | 0.732793 | 0.03965 | 18.482 |
| Cerana | Y | M | C | Stanislaus | 0.714388 | 0.091494 | 7.808 |
| Cerana | Y | Domestic | A | Stanislaus | 0.826711 | 0.038753 | 21.333 |
Patterns of Differentiation between Modern Californian and Native Range Populations
To investigate the patterns of differentiation between modern California and native range populations, we conducted an FST analysis across all sites between the modern Californian population and the African, eastern European, and western European populations. For these population comparisons we grouped the modern Humboldt, Stanislaus, Stebbins, and Davis populations into a single, northern Californian population. Similarly, we grouped the modern Placerita Canyon and the Riverside populations into a single mainland southern California population. We found that the southern Californian population was the least differentiated from the African lineage population, 95th percentile (supplementary table 2, Supplementary Material online). For northern California the FST differences were lowest for the eastern European lineage. Avalon was most differentiated from the eastern European lineage.
Shared Patterns of High Frequency Alternate SNPs in Modern Populations
We examined nonreference SNPs that were at high frequency (≥10%) in one or more native range populations and also at high frequency in one or more of the Californian populations (table 5). Alleles that are observed at high frequency in both a Californian and a native range population can be interpreted as alleles in California that are most likely to have originated from the corresponding native range population(s). In general, we observed a higher proportion of SNPs at high frequency in both the African population and the southern Californian population than in the African population and the northern Californian population. We also observed a higher proportion of high frequency SNPs in both the European (C) population and southern California than in European (C) population and southern California.
Table 5.
High FrequencySNPs
| Ancestral Population | Northern California (%) | Southern California (%) | Avalon (%) |
|---|---|---|---|
| All Shared High Frequency SNPs | |||
| A | 2.43 | 4.82 | 3.04 |
| C | 10.76 | 7.33 | 8.73 |
| M | 10.50 | 9.55 | 13.78 |
| Unique Shared High Frequency SNPs | |||
| A | 0.05 | 1.29 | 0.48 |
| C | 0.95 | 0.47 | 0.33 |
| M | 0.38 | 0.39 | 0.85 |
We then restricted our high frequency native range SNPs to the set of SNPs we only observe in one of the native range lineages. We found a greater proportion of SNPs that are both unique and at high frequency in the African ancestral population and in southern California than in Africa and northern California (Fisher’s Exact Test [FET], = ∼0) (table 5). Similarly, the southern Californian population also had a greater proportion of high frequency African SNPs than the Avalon population (FET, P = ∼0). We detected a higher proportion of eastern European SNPs in northern California than in southern California (FET, P = 7.62809e-26). However, we did not observe any difference in the proportions of western European SNPs between northern and southern California, but we observed a higher proportion of western European SNPs in Avalon than we found in either northern California (FET, 9.803935e-43) or southern California (FET, 6.480419e-45).
To determine whether enriched clusters of genes exhibit SNPs at high frequency in both native range populations and within a particular Californian population, we performed a GO analysis using DAVID version 6.8 (Huang et al. 2009a, 2009b). We found enriched clusters of high frequency SNPs only in the Africa-southern California comparison (supplementary table 3, Supplementary Material online). These included a number of clusters that are associated with developmental processes such as growth factor binding protein and calcium-binding.
Temporal Patterns of Native Range Contributions
We observed two main patterns of temporal variation in our ADMIXTURE results (fig. 2). The oldest honey bee samples from northern California (Humbolt 1966; Stebbins 1996) appear to have extensive western European (M) ancestry. Over time, however, these populations appear to have undergone a dramatic genetic shift, as this western European ancestry was largely supplanted by contributions from the eastern European (C) lineage. Moreover, in contrast to southern Californian populations (below), modern populations in northern California show very little introgression of African (A) alleles. Two individuals, one from Stebbins and one from Stanislaus County show very small contributions from Africa, but this is not seen in any other samples from northern California, either historical or modern (fig. 2).
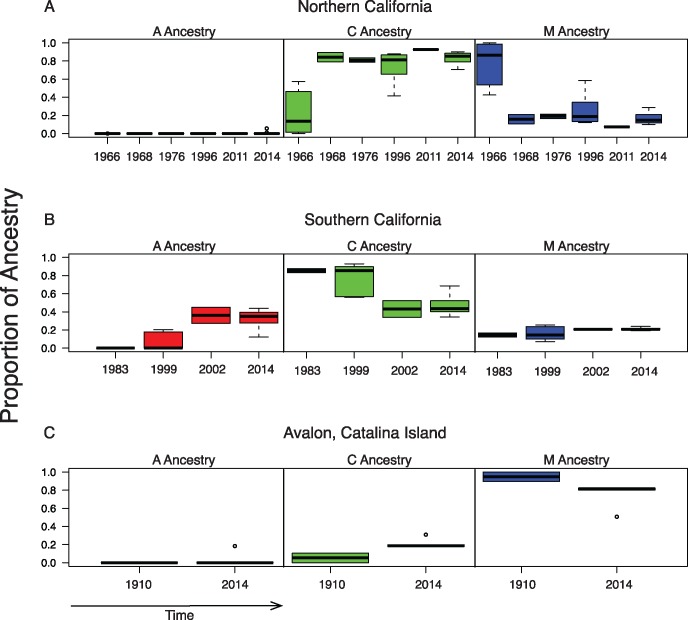
We grouped individual bees based on their location into the categories of northern California (Davis, Humboldt, Stanislaus, and Stebbins), southern California (Placerita, Riverside, and San Pedro) and Avalon. We then generated boxplots for each regional population for each time period for which we have data (fig. 3). We find that there is a decline in mean western European ancestry in northern California over time and at the same time an increase in the mean eastern European ancestry in these individuals. In southern California, we find an increase in the mean African ancestry over time along with a corresponding increase in mean western European ancestry. The Avalon population appears fairly consistent over time with modest changes in the mean contributions of western and eastern European populations.
Fig. 3.
—Boxplots of ADMIXTURE estimates for regional populations over time.
These changes are also reflected in the nMDS analysis. We find that the modern southern Californian populations are most similar to the African individuals (fig. 4A) than the historical southern Californian populations (fig. 4B). Additionally, we find that modern Avalon individuals are positioned closer to the western European individuals than to the eastern European individuals (fig. 4A).
The more recent arrival and spread of Africanized bees (descended from A lineage A. mellifera scutellata) in southern California left a clear signature in our genomic data. The earliest specimens in our data set from this region, the Riverside individuals from 1983, were collected prior to the arrival of Africanized bees and, accordingly, showed no evidence of African ancestry (fig. 2). Individuals collected 5 years after the arrival of Africanized bees (Riverside 1999), however, begin to show evidence of African introgression with 33% of individuals deriving some of their ancestry from the African lineage. Then, southern Californian specimens collected more recently (in Riverside and Placerita Canyon in 2014) showed a substantially greater contribution from Africa in all individuals examined with 100% of individuals collected after 2002 deriving some of their ancestry from the African lineage (fig. 2 and table 3).
Although also located in southern California, the individuals from Avalon exhibited lower introgression from African-derived alleles, likely due to their isolation on Santa Catalina Island, 22 miles from the mainland. We observed that the modern Avalon population received a relative higher contribution from western Europe than did Riverside and Placerita Canyon in 2014. Moreover, only one individual from Avalon, out of the five individuals we sampled, shows any evidence of African ancestry.
Genetic Diversity within California
In addition to the set of SNPs used for our ancestry analysis, we identified an additional set of 3,890,279 SNPs in California that we used to examine both temporal and spatial population changes within California. The southern populations appear to be more genetically diverse (mean π is 1.09 e−4 vs. 7.02 e−5 in northern California), likely due to the recent contribution from African-derived alleles.
Geographic Patterns of Variation
We identified all sites that exhibited differentiation between all modern Californian population pairs. For each comparison, we specified a false discovery rate of 1% and identified SNPs as belonging to intronic or exonic regions within this subset. In all of our population comparisons, we found that there were significantly fewer SNPs in exonic regions compared with the number expected if SNPs were distributed randomly throughout the genome (supplementary table 4, Supplementary Material online). In contrast to exonic SNPs, we find more SNPs in intronic regions than expected in most of our comparisons (supplementary table 4, Supplementary Material online). There are more differences between the Avalon population and the mainland populations than between the all mainland populations, further supporting the idea that the Avalon population has remained genetically isolated from the mainland for a relatively long period of time.
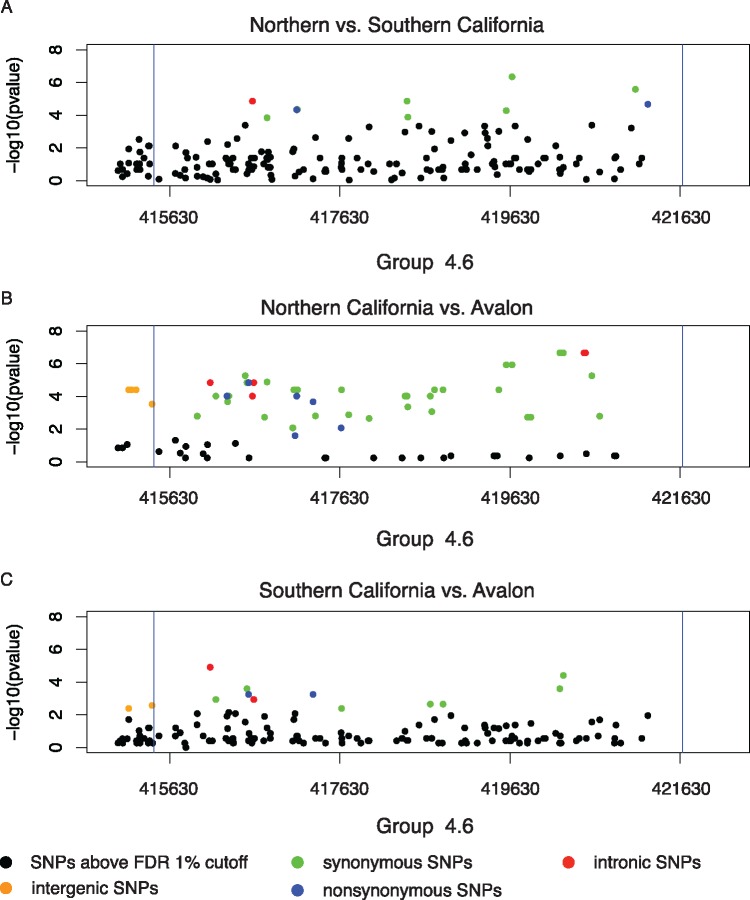
We examined the genomic location of differences and found that, between northern and southern California, there are peaks of SNPs in genic regions that pass the FDR cutoff. We found a total of 208 genes with nonsynonymous SNPs within this subset of the data. Vitellogenin, for example, is found in one of these peaks on chromosome 4 with nine synonymous SNPs and one nonsynonymous SNP that pass the FDR cutoff (fig. 5). Although most genes have only one nonsynonymous SNP, there were four genes with five or more. In the gene zormin, which is involved in actin binding, we found 16 nonsynonymous SNPs at the 1% FDR level. We found ten SNPs in cubilin, which is involved in nephrocyte filtration in Drosophila, nine SNPs LOC100578512; hydrocephalus-inducing protein-like, and five in LOC725833, which is an uncharacterized gene.
Fig. 5.
—Differentiated SNPs in Vitellogenin in modern population comparisons. Vg is a minus strand gene and there are no SNPs observed in the 500 bp upstream of the 5′ end.
We also examined the subset of genes with nonsynonymous SNPs in the comparisons between modern Avalon and the mainland populations. Within this differentiated set, 2,064 genes exhibited differentiation between Avalon and northern California and 906 genes showed differentiation between Avalon and southern California. A total of 807 genes showed differentiation in both comparisons. Odorant receptor (OR) genes occur in the list of differentiated genes with six OR genes differentiated between northern California and Avalon and two OR genes differentiated between southern California and Avalon. A further four OR genes are differentiated in both comparisons. Vitellogenin is differentiated in both population comparisons (fig. 4) and its receptor, yolkless, is differentiated between Avalon and northern California. These genes may represent local adaptation between populations within California and are good candidates for further study.
Temporal Patterns
We compared population pairs for each of the seven locations for which we had both historical and a modern samples. The temporal difference between sampling times ranged from 15 to 104 years. We found that genetic differentiation increased with increasing time between sampling dates at a given location (regression R2 = 0.5649, P = 0.01184) (supplementary table 5, Supplementary Material online). However, there are more differences between modern northern and modern southern Californian populations (FST = 0.0193) than between northern Californian populations from the 1960s and 1970s (combined) and the modern populations (mean FST = 0.0084). The mean FST between southern Californian populations from 1999 and 2014 was 0.0148.
We compared all sites where there was a difference between the population from Humboldt County in 1966 and in 2014, the only historical to modern comparison where we have even sample sizes and high quality sequence for all individuals. We found that there are fewer SNPs at a FDR of 1% in exonic regions and more in intronic regions than expected. We found 306 genes with nonsynonymous SNPs in the Humboldt comparison, nine of which have five or more nonsynonymous SNPs. Most of these genes were uncharacterized, but hemolectin, which is involved in clotting, and LOC551016, which is described as midasin-like, are in this set. The overall set of genes with nonsynonymous SNPs is very different between the Humboldt and the modern Californian comparison with only 29 genes in both comparisons, and 456 that are in only one comparison. Of the genes that do occur in both comparisons, one is yolkless, a vitellogenin receptor.
Discussion
Here, we document patterns of genetic diversity of Apis mellifera populations over temporal and spatial scales in the state of California. We document substantial genetic diversity in the A. mellifera populations that are likely the result of 1) changes in beekeeping practices over time, 2) the introduction of Africanized bees to California in 1994, 3) the introduction of the Varroa mite to California in 1987, and 4) the life history of feral A. mellifera and potential local adaptation of feral populations.
Modern Populations
Populations examined in this study were collected away from agricultural areas, but cannot be confirmed to be feral bees. Examining the patterns of genetic differences between populations can indicate the relatedness of these individuals to managed stocks. Regardless, these bees serve as indicators of the genetic diversity of A. mellifera found in regions throughout California. We identified three main geographically and genetically distinct groups of honey bee populations in California. First, there are populations of northern Californian bees that closely resemble managed stocks. Prior to the introduction of Varroa mites, feral colonies appear to be generally stable over time, but the demographics of feral bees can still change relatively rapidly. The density of feral colonies has been measured in Ithaca, NY was found to change slowly as established colonies are likely to survive through the winter (∼79% survival; Seeley 1978), which is similar to the survival of feral populations in Arizona (∼80%; Taber 1979), where winters are milder. Baum et al. (2005), who examined feral A. mellifera colonies in Texas between 1993 and 2000, found that, although colony turnover was 5–30% per year, over that time Africanized colonies increased from an initial colony discovered in 1993 to 80.3% of colonies Africanized in 2000 (Baum et al. 2005). This baseline rate of turnover gives us a good idea of what we should expect in terms of the genetic relationships between feral populations and nearby managed bees if we assume that managed populations continue to generate new feral colonies and feral colonies are not experiencing any additional pressures such as Varroa mites or African introgression.
The introduction of the Varroa mite has substantially altered the population dynamics of feral colonies. After being first discovered in California in 1987, Varroa reduced the feral colony population of California to less than one fifth its previous size by 1994 (Kraus and Page 1995). We expect that this substantial reduction in the feral bee population would result in reduced genetic diversity of modern feral bees both by wiping out most feral colonies that existed prior to Varroa, and also increasing the turnover of feral colonies by reducing the mean feral colony survival time, as was shown in Kraus and Page (1995). These predictions match well with the patterns we documented in northern California. Historical populations exhibited a shift over time, concurrent with changing beekeeping preferences from M lineage to C lineage populations (Crane 1999), and resembling more closely domesticated populations, derived mostly from European (C) lineages with some European (M) ancestry as well. Many of these populations were taken from the area at the northern end of California’s Central Valley, an area with high mite infestation rates (Kraus and Page 1995).
Our analysis based on southern Californian bees revealed that these populations derive substantial genetic ancestry from African lineages and this influx of new alleles result in populations that are much more genetically diverse than northern Californian bees. This diversity is consistent with observations in Texas of hybrid swarms that were found in the contact zone of African and European derived bees, post Africanization (Pinto et al. 2005). The southern Californian mainland populations appear to be primarily influenced by different factors than northern Californian populations, though the timing of the Varroa mite outbreak and the introduction of Africanized bees into the United States may have played a role in favoring Africanized bee spread (Pinto et al. 2004). The introduction of Africanized bees to California in 1994 provide a source of genetic variation that is currently lacking in northern California, though Africanized bees appear to be spreading northwards (Kono and Kohn 2015) and our analyses support this conclusion.
We find that African populations contribute between 37.6% and 73.3% of the ancestry of modern southern Californian populations. In addition, because of concerns of Africanization, modern beekeepers in southern California have altered breeding practices to reduce or eliminate the introduction of Africanized alleles into domesticated lineages. This makes it very likely that the southern Californian bees are genuine feral bees. Compared with northern Californian populations, modern southern Californian populations also appear to have more substantial genetic contributions from western European (M) lineages, a pattern which has been observed previously (Whitfield et al. 2006; Rangel et al. 2016). This may be due to a decrease in gene flow between managed and feral populations in southern Californian, selective pressures favoring western European alleles, or hitchhiking of M alleles with African spread. Further studies are required to determine what processes may be at work here. This produces a population with high diversity and contributions from both western and eastern European bees as well as Africanized strains and is similar to the population dynamics of feral bees in the Yucatan in between 1993 and 1998 as Africanization of the local populations occurred (Clarke et al. 2002). Our results support the hypothesis that populations from southern California are experiencing rapid ongoing changes, possibly due to local adaptation and admixture from multiple lineages.
The increased genetic diversity we observed in mainland southern Californian populations bears directly on some of the current issues facing A. mellifera populations in California. Genetic diversity has been shown to be an important factor in colony health, with genetically diverse colonies exhibiting higher disease resistance (Tarpy and Seeley 2006), increased thermoregulation capacity (Jones et al. 2004), increased diversity of worker responses to environmental stimuli (Page et al. 1995), and higher productivity and fitness (Mattila and Seeley 2007). Managed bee colonies have recently been found to have higher levels of genetic variation than honey bee populations in the eastern or western Europe (Harpur et al. 2012). However, those studies were based on colonies that exhibited much lower genetic diversity than the Africanized populations we documented here. We find that within California, Africanized southern Californian populations exhibit more genetic diversity than managed or modern northern Californian populations, but the managed and modern northern Californian populations show similar levels of genetic diversity both with respect to each other and to western and eastern European bees.
Africanized bees have been found to be more resistant to Varroa mites, likely due to a higher grooming rate and shorter period between egg laying and capping of the larvae (Camazine 1986; Moritz and Mautz 1990; Emsen et al. 2012), Moreover, it has been hypothesized that Africanization of Californian honey bees may assist feral A. mellifera populations in surviving Varroa by introducing these beneficial behaviors into southern Californian populations (Kraus and Page 1995). Environmental variables may also play an important role in facilitating mite infestations. Medina-Flores et al. (2014) observed an association between drier conditions and lower infestation rates in Mexico. Our study is congruent with this observation, and may help explain the observed patterns of higher genetic diversity in southern California where environmental conditions are similar. Further studies should investigate the relative role of genetic background and environmental conditions with respect to honey bee resistance Varroa and other health threats.
Our analysis uncovered the existence of a third modern population in Catalina Island, which is located off the coast of California near Los Angeles. This population is genetically distinct from the mainland populations and bears more resemblance to the western European (M) lineages and the Humboldt County 1966 population. This population is likely to have been on the island for some time, possibly since 1890, when A. mellifera was introduced on nearby Santa Cruz Island (Wenner and Thorp 1994) and there are no managed bees on the island. Additionally, this population is likely to be free of Varroa mites. The Varroa mite was never found on Santa Cruz Island until it was introduced in an effort to biologically control honey bees (Wenner and Thorp 1994) and is therefore also unlikely to have made it to Santa Catalina Island. Moreover, the genetic differences between the mainland population and Santa Catalina Island indicate that there is little to no gene flow between the populations, and thus little opportunity for the transfer of mites. This population provides an interesting look into past honey bee populations and has likely undergone adaptation to the local environment.
Temporal Changes
The temporal changes we observed were dramatically different between southern California and northern California. The first Africanized bees to reach California in 1994 have, over the past couple of decades, contributed substantially to the population structure. In contrast, historical populations of A. mellifera prior to the 1970s in northern California differ from more modern northern Californian populations primarily due to a reduction in the contribution from western European (M) lineages and a concomitant increase in the contribution from eastern European (C) lineages. Modern northern Californian populations in the Central Valley (Davis, Stanislaus, and Stebbins), are also very similar to modern domesticated bees, FST = 0.059–0.065, which likely reflects frequent dispersal of domesticated individuals into feral populations as escaped queens and drones establish new colonies. It is difficult to assess the precise timing of this change from more western European (M) like bees to primarily eastern European (C) like bees, but we suspect that it was driven by the reduction of the feral population in the mid 1990s and changes in breeding practices over that time period.
Differentiated Genes
In addition to the broad-scale changes in the genetic composition of modern honey bee populations of California we identified a substantial number of genes that are differentiated between populations. Some genes, like vitellogenin, show differentiation between multiple population comparisons. These genes constitute a candidate sets for both local adaptation to the various environmental conditions along California, and future studies may investigate their potential role in resistance to Varroa or other diseases faced by honey bee populations. We also find some interesting candidates in the comparison of the 1966 and modern Humboldt population, though these populations are composed of only a few individuals each and the differentiation observed may be due to sampling. The gene spatzle 2, a toll receptor, is highly differentiated between the Humboldt 1966 and Humboldt 2015 populations. It was found to be downregulated in high Varroa treatment groups in a previous study of the effects of deformed wing virus and Varroa on honey bees (Ryabov et al. 2014). Other functions such as neuronal development, neuronal sensitivity, and olfaction have been implicated in studies of the genetic basis of tolerance to Varroa (Zakar et al. 2014).
Conclusions
Our study provides an in-depth examination of temporal and spatial changes in honey bee populations in California. We identified substantial genetic differences across these two dimensions in populations that are likely the result of both biotic and abiotic factors. Our analysis uncovers the potential genetic signatures that beekeeping practices and the introduction of Varroa have had on the changes experienced by bee populations over time and suggests that southern Californian populations may harbor higher genetic variation that could be harnessed to improve the health of A. mellifera and its important role in the agriculture of California.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank Dr. Michael Mesler for help in collecting bees. We would also like to thank C.F. Koehnen and Sons, Phil Hofland, and Glenda Wooten for providing drones. This work was supported by the Berkeley Initiative for Global Change Biology; the Office of the Vice Chancellor for Research at UC Berkeley; the Gordon and Betty Moore Foundation [grant number GBMF2983], the Vincent Coates Genome Sequencing Facility; National Institutes of Health, S10 Instrumentation Grants [grant numbers S10RR029668 and S10RR027303]; the USDA National Institute of Food and Agriculture, Hatch Project [grant number CA-B-INS-0087-H].
Literature Cited
- Allendorf FW, Hohenlohe PA, Luikart G.. 2010. Genomics and the future of conservation genetics. Nat Rev Genet. 1110:697–709.http://dx.doi.org/10.1038/nrg2844 [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 199:1655–1664.http://dx.doi.org/10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum KA, Rubink WL, Pinto MA, Coulson RN.. 2005. Spatial and temporal distribution and nest site characteristics of feral honey bee (Hymenoptera: Apidae) colonies in a coastal prairie landscape. Environ Entomol. 343:610–618.http://dx.doi.org/10.1603/0046-225X-34.3.610 [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 1:289–300. [Google Scholar]
- Brandt A, Gorenflo A, Siede R, Meixner M, Büchler R.. 2016. The neonicotinoids thiacloprid, imidacloprid, and clothianidin affect the immunocompetence of honey bees (Apis mellifera L.). J Ins Phys 86:40–47. [DOI] [PubMed] [Google Scholar]
- Buchler R, Berg S, Le Conte Y.. 2010. Breeding for resistance to Varroa destructor in Europe. Apidologie 413:393–408.http://dx.doi.org/10.1051/apido/2010011 [Google Scholar]
- Camazine S. 1986. Differential reproduction of the mite Varroa jacobsoni (Mesostigmata: Varroidae), on Africanized and European bees (Hymenoptera: Apidae). Ann Entomol Soc Am. 795:801–803.http://dx.doi.org/10.1093/aesa/79.5.801 [Google Scholar]
- Clarke KE, Rinderer TE, Franck P, Quezada-Euan JG, Oldroyd BP.. 2002. The Africanization of honeybees (Apis mellifera L.) of the Yucatan: a study of a massive hybridization event across time. Evolution 56:1462–1474. [DOI] [PubMed] [Google Scholar]
- Crane E. (1999) The world history of beekeeping and honey hunting. UK: Tayor and Francis. p. 1–682.
- Cridland JM, Tsutsui ND, Ramirez SR.. 2017. The complex demographic history and evolutionary origin of the western honey bee, Apis mellifera. Genome Biol Evol. doi: 10.1093/gbe/evx009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Rúa P, Jaffé R, Dall'Olio R, Muñoz I, Serrano J.. 2009. Biodiversity, conservation and current threats to European honeybees. Apidologie 403:263–284. [Google Scholar]
- Emsen B, Petukhova T, Guzman-Novoa E.. 2012. Factors Limiting the Growth of Varroa destructor Populations in Selected Honey Bee (Apis mellifera L.) Colonies. J Anim Vet Adv. 11:4519–4525. [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD.. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP data. PLoS Genet. 510:e1000695.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Novoa E, Page RE.. 1999. Selective breeding of honey bees (Hymenoptera: Apidae) in Africanized areas. J Econ Entomol. 923:521–525.http://dx.doi.org/10.1093/jee/92.3.521 [Google Scholar]
- Harpur BA, et al. 2014. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc Natl Acad Sci U S A. 1117:2614–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpur BA, MinaeI S, Kent CF, Zayed A.. 2012. Management increases genetic diversity of honey bees via admixture. Mol Ecol. 2118:4414–4421.http://dx.doi.org/10.1111/j.1365-294X.2012.05614.x [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009a. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009b. Bioinformatics enrichment tools: paths towards the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Myerscough MR, Graham S, Oldroyd BP.. 2004. Honey bee nest thermoregulation: diversity promotes stability. Science 3055682:402–404.http://dx.doi.org/10.1126/science.1096340 [DOI] [PubMed] [Google Scholar]
- Kerr EW. 1967. The history of the introduction of African bees in Brazil. South Afric Bee J. 39:33–35. [Google Scholar]
- Klein AM, et al. 2007. Importance of pollinators in changing landscapes for world crops. Proc Biol Sci. 2741608:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono Y, Kohn JR.. 2015. Range and frequency of Africanized honey bees in Calfornia (USA). PLoS One 109:e0137407.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus B, Page RE Jr.. 1995. Effect of Varroa jacobsoni (Mesostigmata: Varroidae) on feral Apis mellifera (Hymenopter: Apidae) in California. Environ Entomol. 246:1473–1480.http://dx.doi.org/10.1093/ee/24.6.1473 [Google Scholar]
- Langmead B, Salzberg S.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 94:357–359.http://dx.doi.org/10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, 1000 Genome Project Data Processing Subgroup, et al. 2009. The sequence alignement/map (SAM) format and SAMtools. Bioinformatics 2516:2078–2079.http://dx.doi.org/10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losey JE, Vaughn M.. 2006. The economic value of ecological services provided by insects. BioScience 56:311–323.http://dx.doi.org/10.1641/0006-3568(2006)56[311:TEVOES]2.0.CO;2 [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P.. 2003. The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet. 412:981–994. [DOI] [PubMed] [Google Scholar]
- Magnus R, Szalanski AL.. 2010. Genetic evidence for honey bees (Apis mellifera L.) of middle eastern lineage in the United States. Sociobiology 55:285–296. [Google Scholar]
- Mattila HR, Seeley TD.. 2007. Genetic diversity in honey bee colonies enhances productivity and fitness. Science 3175836:362–364.http://dx.doi.org/10.1126/science.1143046 [DOI] [PubMed] [Google Scholar]
- Moritz RFA, Mautz D.. 1990. Development of Varroa jacobsoni in colonies of Apis mellifera capensis and Apis mellifera carnica. Apidologie 211:53–58.http://dx.doi.org/10.1051/apido:19900107 [Google Scholar]
- Medina-Flores CA, Guzmán-Novoa E, Hamiduzzaman MM, Aréchiga-Flores CF, López-Carlos MA.. 2014. Africanized honey bees (Apis mellifera) have low infestation levels of the mite Varroa destructor in different ecological regions in Mexico. Genet Mol Res. 133:7282–7293. [DOI] [PubMed] [Google Scholar]
- Page RE, Robinson GE, Fondrk MK, Nasr ME.. 1995. Effects of worker genotypic diversity on honey bee colony development and behavior (Apis mellifera L.). Behav Ecol Sociobiol. 366:387..http://dx.doi.org/10.1007/BF00177334 [Google Scholar]
- Patterson N, et al. 2012. Ancient admixture in human history. Genetics. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AM, Rubink WL, Coulson RN, Patton JC, Johnston JS.. 2004. Temporal pattern of Africanization in a feral honeybee population from Texas inferred from mitochondrial DNA. Evolution 585:1047–1055. [DOI] [PubMed] [Google Scholar]
- Pinto AM, Rubink WL, Patton JC, Coulson RN, Johnston JS.. 2005. Africanization in the United States. Genetics 1704:1653–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel J, et al. 2016. Africanization of a feral honey bee (Apis mellifera) population in south Texas: does a decade make a difference?. Ecol Evol. 67:2158–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinderer TE, Collins AM, Tucker KW.. 1985. Honey production and underlying nectar harvesting activities of Africanized and European honeybees. J Apicult Res. 243:161–167.http://dx.doi.org/10.1080/00218839.1985.11100666 [Google Scholar]
- Ruttner F. (1988) The western honeybee Apis mellifera L. classification and natural history. Biography and taxonomy of honeybees. Berlin Heidelberg GmbH: Springer-Verlag. P. 163–175.
- Ryabov EV, et al. 2014. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathogn. doi: 10.1371/journal.ppat.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Bayo F, et al. 2016. Are bee diseases linked to pesticides?—A brief review. Environ Inter. 89–90:7–11. [DOI] [PubMed] [Google Scholar]
- Schiff NM, Sheppard WS, Loper GM, Shimanuki H.. 1994. Genetic diversity of feral honey bee (Hymenoptera: Apidae) populations in the southern United States. Ann Entomol Soc Am. 876:842–848.http://dx.doi.org/10.1093/aesa/87.6.842 [Google Scholar]
- Seeley TD. 1978. Life history strategy of the honey bee, Apis mellifera. Oecologia 321:109–118.http://dx.doi.org/10.1007/BF00344695 [DOI] [PubMed] [Google Scholar]
- Sheppard WS, Huettel MD.. 1988. Biochemical genetic markers, intraspecific variation, and population genetics of the honey bee, Apis mellifera. Environ Entomol. 221:183–189. [Google Scholar]
- Stinchcombe JR, Hoekstra HE.. 2008. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity 1002:158–170.http://dx.doi.org/10.1038/sj.hdy.6800937 [DOI] [PubMed] [Google Scholar]
- Taber S., III 1979. A population of feral honey bee colonies. Am Bee J. 119:842–847. [Google Scholar]
- Tarpy DR, et al. 2010. Mating frequencies of Africanized honey bees in the south western USA. J Apicult Res. 494:302–310.http://dx.doi.org/10.3896/IBRA.1.49.4.02 [Google Scholar]
- Tarpy DR, Seeley TD.. 2006. Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs monoandrous queens. Naturwissenschaften 934:195..http://dx.doi.org/10.1007/s00114-006-0091-4 [DOI] [PubMed] [Google Scholar]
- US Department of Agriculture, National Agricultural Statistics Service. 2016. Honey Bee Colonies (US Department of Agriculture, Washington, DC). Available from: http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do? documentID=1943; last accessed February 2017.
- vanEngelsdorp D, Meixner MD.. 2010. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J Invertebr Pathol. 103:S80–S95. [DOI] [PubMed] [Google Scholar]
- Wallberg, A, et al. 2014. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet. 46:1081–1088. [DOI] [PubMed] [Google Scholar]
- Watkins LH. 1968. California’s first honey bees. Am Bee J. 108:190–191. [Google Scholar]
- Wenner AM, Thorp RW.. 1994. Removal of feral honey bee (Apis mellifera) colonies from Santa Cruz Island. In: Halvorson WL, Meander GL, editors. The Fourth California Islands Symposium: update on the status of resources. Santa Barbara (CA): Santa Barbara Museum of Natural History. p. 513–522.
- Winston ML, Taylor OR, Otis GW.. 1983. Some differences between temperate European and tropical African and South American honeybees. Bee World 641:12–21.http://dx.doi.org/10.1080/0005772X.1983.11097902 [Google Scholar]
- Whitfield CW, et al. 2006. Thrice Out of Africa: Ancient and Recent Expansions of the Honey Bee, Apis mellifera. Science 3145799:642–645.http://dx.doi.org/10.1126/science.1132772 [DOI] [PubMed] [Google Scholar]
- Zakar E, Javor A, Kusza S.. 2014. Genetic basis of tolerance to Varroa destructor in honey bees (Apis mellifera L.). Insect Soc. 613:207–215.http://dx.doi.org/10.1007/s00040-014-0347-5 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.