Abstract
B cell chronic lymphocytic leukemia (B-CLL), the most common type of leukemia in adults, is still essentially incurable despite the development of novel therapeutic strategies. This reflects the incomplete understanding of the pathophysiology of this disease. A comprehensive proteome analysis of primary human B-CLL cells and B cells from younger as well as elderly healthy donors was performed. For comparison, the chronic B cell leukemia cell line JVM-13 was also included. A principal component analysis comprising 6,945 proteins separated these four groups, placing B cells of aged-matched controls between those of young donors and B-CLL patients, while identifying JVM-13 as poorly related cells. Mass spectrometric proteomics data have been made fully accessible via ProteomeXchange with identifier PXD006570-PXD006572, PXD006576, PXD006578, and PXD006589-PXD006591. Remarkably, B cells from aged controls displayed significant regulation of proteins related to stress management in mitochondria and ROS stress such as DLAT, FIS1, and NDUFAB1, and DNA repair, including RAD9A, MGMT, and XPA. ROS levels were indeed found significantly increased in B cells but not in T cells or monocytes from aged individuals. These alterations may be relevant for tumorigenesis and were observed similarly in B-CLL cells. In B-CLL cells, some remarkable unique features like the loss of tumor suppressor molecules PNN and JARID2, the stress-related serotonin transporter SLC6A4, and high expression of ZNF207, CCDC88A, PIGR and ID3, otherwise associated with stem cell phenotype, were determined. Alterations of metabolic enzymes were another outstanding feature in comparison to normal B cells, indicating increased beta-oxidation of fatty acids and increased consumption of glutamine. Targeted metabolomics assays corroborated these results. The present findings identify a potential proteome signature for immune senescence in addition to previously unrecognized features of B-CLL cells and suggest that aging may be accompanied by cellular reprogramming functionally relevant for predisposing B cells to transform to B-CLL cells.
B cell chronic lymphocytic leukemia (B-CLL)1, the most common type of a non-Hodgkin lymphoma in the Western world, is a disease of the elderly with a median age at diagnosis of 72 years and with approximately twice the incidence in men as in women (1). Several new therapeutic strategies have been developed in recent years; however, while the patient′s survival time could be prolonged and the quality of life improved, an entire cure of the disease is not yet achievable. B-CLL has been intensively studied, especially on the level of genomics and transcriptomics. Nevertheless, several questions remain unanswered, conclusive risk factors for the incidence of the disease could not yet be recognized, and the pathophysiology of the disease is still not fully understood. One of the reasons therefore may be that B-CLL represents a very heterogeneous disorder, associated with a multiplicity of possible genetic alterations (2), which is further strongly dependent on functional changes in the tumor microenvironment (3–5). Genetic as well as environmental factors may both be responsible for the considerably varying disease progression and individual therapeutic response, which are hardly predictable.
Besides genomics and transcriptomics, proteomics is a highly promising approach for characterizing specific features of tumor diseases. We have focused on the investigation of tumor-related pathophysiology using mass-spectrometry-based proteomics (6–9). With regard to B-CLL, proteomics studies have already been successfully conducted (10–13). However, despite the efforts, clear mechanisms explaining the pathogenesis of the disorder have not yet been recognized. The aim of the present study was to further investigate mechanisms that may contribute to the development of B-CLL. To this end, primary human B-CLL cells were analyzed in detail, applying subcellular fractionation as described previously (14). Analyzing normal B lymphocytes of peripheral blood both from young and elderly healthy donors allowed us not only to compare B-CLL cells to age-matched normal B-cells but also to verify if and how aging may be related to B-CLL development. Furthermore, for comparative purposes, the chronic B cell lymphoma cell line JVM-13 was included in the analyses.
In addition, previous studies of our group and others have shown that combining metabolomics with proteomics may contribute to a better understanding of disease pathophysiology (9, 15–17) As metabolic changes seem to play an important role in B-CLL (4, 18, 19), a metabolomics analysis of B-CLL cells in comparison to age-matched normal B lymphocytes was included.
By combining these two omics-type experiments, we could highlight the importance of glutaminolysis in CLL as previously indicated by Koczula et al., which holds great potential to critically influence B-CLL cell survival and therapy response (20). Moreover, many stress-related proteins were identified deregulated in normal B lymphocytes of elderly in comparison to younger healthy donors, suggesting that their regulation may correlate with aging. Most importantly, several of these proteins are related to immunosenescence and potentially promote cell transformation, thus suggesting that aging-related proteome alterations in normal B lymphocytes may predispose for B-CLL development.
EXPERIMENTAL PROCEDURES
Study Cohort and Blood Sampling
Peripheral blood samples were taken from 16 healthy volunteers as well as from 16 B-CLL patients with written consent of each donor and approval of the Ethics Committee of the Rudolfinerhaus Hospital in Vienna. B-CLL patients were diagnosed with B-CLL according to the guidelines provided by the International Workshop on Chronic Lymphocytic Leukemia (21). B cells from B-CLL patients as well as healthy donors were used for several assays, including proteomics, metabolomics, and ROS level measurements, with cells from several donors being used for more than one assay. Blood cell counts of the healthy donors did not show any significantly elevated lymphocyte levels. Treatment regimens for B-CLL patients as well as age and sex distribution for all donors are shown in Supplemental Table S1.
Cell Isolation
Noncoagulated blood from B-CLL patients as well as from healthy donors was collected and diluted 1:2 with PBS buffer. The diluted blood was subsequently overlaid on Ficoll Paque (GE Healthcare, Bio-Sciences AB, Uppsala, Sweden) followed by centrifugation at 720 g for 20 min at room temperature. Peripheral blood mononuclear cells (PBMCs) were collected from the Ficoll Paque-blood plasma interface and washed with PBS.
PBMCs were resuspended in PBS and, after adding magnetic bead-coupled anti-CD19 antibody (Miltenyi Biotec, Bergisch Gladbach, Germany), incubated at 4 °C for 15 min. After another PBS washing step, the cells were resuspended in MACS buffer (1xPBS, 0.5% FBS, 2 mm EDTA) and pipetted onto a preconditioned MACS LS column mounted on a magnetic holder (both Miltenyi Biotec, Bergisch Gladbach, Germany). Cells were washed on the column with MACS buffer, discarding the flow-through. The trapped CD19+ B lymphocytes were eluted by removing the MACS LS column from the magnet and pressing 4 ml of MACS buffer through the column using the column plunger. The B-cell-containing eluate was diluted to 10 ml and the cell number as well as the viability determined using the MOXI Z Mini Automated Cell Counter (ORFLO Technologies, Ketchum, ID, USA). Cells were then pelleted by centrifugation at 590 g for 5 min at 4 °C and supernatants removed.
JVM-13 cells (Leibnitz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) were cultivated in RPMI 1640 medium (Thermo Fisher Scientific, Vienna, Austria) with 10% heat-inactivated FBS (Sigma-Aldrich, St. Louis, MO, USA) and 100 U/ml penicillin/streptomycin (ATCC, Manassas, VA, USA) at 37 °C and 5% CO2 until cell density of 1–2 × 106 cells/ml was reached. Cells were washed with PBS and pelleted by centrifugation at 590 g for 5 min.
ROS Assay
IMDM + GlutaMax (GE Healthcare, Piscataway, NJ, USA) supplemented with 10% FBS (GE Healthcare), 15 μg/ml gentamycin, and 1 μg/ml Amphotericin B (both Sigma Aldrich) was chosen as cultivation medium for PBMCs. PBMCs were isolated as described above and were frozen in culture medium, supplemented with 10% DMSO (Sigma Aldrich), and 20% FBS. PBMCs were thawed and cultivated 12 h before measurement of ROS. For ROS detection, the DCFDA Cellular ROS Detection Assay Kit (Abcam, Cambridge, U.K.) was used. In this assay, 2′,7′ -dichlorofluorescin diacetate, a fluorogenic dye, is deacetylated by cellular esterases and later oxidized by ROS into 2′, 7′-dichlorofluorescein, a highly fluorescent compound that correlates with intracellular ROS activity and can be detected in the FITC channel. For positive controls, cells were prestimulated with 110 μm tert-butyl hydrogen peroxide for 30 min at 37 °C. 2 × 105 cells were stained in culture medium with 20 μm 2′,7′ -dichlorofluorescin diacetate for 30 min at 37 °C and were then immediately transferred on ice. In the following step, cells were incubated with anti-human CD3 (clone OKT3, eFluor450-conjugated) and anti-human CD19 (clone HIB19, APC-H7 conjugated; both eBioscience™, Thermo Fisher Scientific, USA) for 30 min on ice in the dark. Without washing, expression of CD3, CD19, and ROS were quantified on a BD FACS Canto II flow cytometer and analyzed using the FlowJo software (version 10, Tree Star, Ashland OR, USA). Monocytes were identified according to their forward-/side-scatter characteristics.
Cell Lysis and Fractionation for Proteomics Analyses
All working steps during cell lysis and subcellular fractionation were performed on ice. Precipitated B lymphocytes were lysed by applying lysis buffer and mechanical shear stress. Subsequent subcellular fractionation into cytoplasmic and nuclear fractions was performed as described previously (22). In short, cytoplasmic proteins were extracted from the supernatant obtained upon centrifugation of lysed cells, while nuclear proteins were extracted from the remaining pellet using 500 mm NaCl for extraction and dilution with Nonidet P-40 buffer. Both cytoplasmic and nuclear proteins were precipitated with ice-cold ethanol overnight and were dissolved in sample buffer (7.5 m urea, 1.5 m thiourea, 4% CHAPS. 0.05% SDS, 100 mm DTT) after centrifugation. Protein concentrations were assessed by applying a Bradford assay (Bio-Rad-Laboratories, Vienna, Austria).
Sample Preparation for LC-MS Analysis
Both cytoplasmic and nuclear fractions were subjected to an on-filter tryptic digestion described in a previous publication and based on the FASP protocol developed by the Mann group (23, 24). In short, 20 μg of protein extract were applied on a prewetted 3kD molecular weight cutoff filter (Pall Austria Filter GmbH, Vienna, Austria), proteins were reduced with DTT, alkylated with iodoacetamide, and washed with 50 mm ammonium bicarbonate buffer. Digestion of proteins was performed by applying Trypsin/Lys-C Mix (MS grade; Promega Corporation, Madison, WI, USA), followed by a cleanup of the resulting peptide samples on C18 spin columns (Pierce™ C18 Spin Columns, Thermo Fisher Scientific). Finally, after drying via vacuum centrifugation, the samples were stored at −20 °C until further MS analyses.
LC-MS/MS Analysis
Peptides were solubilized in 5 μl of 30% formic acid, further adding four synthetic standard peptides for internal quality control. Dissolved peptide samples were diluted with 40 μl of mobile phase A (97.9% H2O, 2% ACN, 0.1% formic acid), and 10 μl were injected to a Dionex UltiMate 3000 RSLCnano LC system connected to a Q Exactive Orbitrap MS via a nanospray ion source (all Thermo Fisher Scientific, Austria). After trapping and washing the samples on a C18 2 cm × 100 μm precolumn, LC separation was performed on a 50 cm × 75 μm Pepmap100 analytical column (both Thermo Fisher Scientific, Austria). Peptide elution from the precolumn onto the analytical columns was performed by applying a gradient system from 8% to 40% mobile phase B (79.9% ACN, 20% H2O, 0.1% formic acid) at a flow rate of 300 nL/min over a time of 235 min, with a total chromatographic run time of 280 min including the washing and equilibration step. Mass spectrometric resolution on the MS1 level was set to 70,000 (at m/z = 200) with a scan range from 400 to 1,400 m/z. The top eight abundant peptide ions were chosen for fragmentation at 30% normalized collision energy and resulting fragments analyzed in the Orbitrap at a resolution of 17,500 (at m/z = 200).
Proteomics Data Analysis
Raw data were subjected to the freely available software MaxQuant (version 1.5.2.8) utilizing the Andromeda search engine, followed by statistical evaluation with the Perseus software (version 1.5.2.6) (25, 26). For the MaxQuant search, a minimum of two peptide identifications, at least one of them being a unique peptide, was required for valid protein identification. Digestion mode was set to “Specific” choosing Trypsin/P as enzyme specificity. The peptide mass tolerance was set to 50 ppm for the first search and to 25 ppm for the main search. The false discovery rate (FDR) was set to 0.01 both on peptide and protein level, based on the q-value. The database applied for the search was the human Uniprot database (version 09/2014, reviewed entries only) with 20,193 protein entries. Further settings for the search included carbamidomethylation as fixed modification and oxidation of methionine and acetylation of the protein C terminus as variable modifications. Each peptide was allowed to have a maximum of two missed cleavages and two modifications, “Match between runs” was enabled and the alignment window set to 10 min, with the match time window of 5 min.
For statistical data evaluation of the search files obtained from MaxQuant, the Perseus software was used. Reverse sequences and potential contaminants as well as proteins identified only by site were removed. For statistical analysis, data obtained from both biological and technical replicates were used. Label-free quantification values were logarithmized to base 2 and technical replicates averaged. Proteins were filtered for valid values, keeping only proteins that were identified in at least three measurements in at least one sample group (B-CLL, elderly B cells, young B cells, JVM-13). Missing values were then replaced from a normal distribution with a down shift of 1.8 and a width of 0.3 in order to enable t testing and volcano plots. Evaluation of regulatory events between different samples groups was achieved by two-sided t tests using a p value <0.05 (permutation-based FDR correction). For selected proteins, heat maps representing fold changes between sample groups were generated. Heat maps were generated by a custom R (https://www.r-project.org) script plotting the Student's t test difference values obtained from Perseus. Here, we made use of the ColorBrewer Pallettes available at http://cran.r-projects.org/web/packages/RColorBrewer/index.html and gplots available at http://cran.r-projects.org/web/packages/gplots/index.html. Changes marked with asterisks (**) are significant after permutation-based multiparameter testing.
In order to enable upload of mass spectrometric data to a publically available repository, the raw files were also analyzed using Proteome Discoverer 1.4 (Thermo Fisher Scientific, Austria) utilizing Mascot 2.5 (Matrix Science, U.K.). Protein identification was performed by searching raw data against the SwissProt Database (version 11/2015 with 20,193 entries) with a mass tolerance of 50 ppm at the MS1 level and 100 mmu at the MS2 level, allowing for up to two missed cleavages per peptide. Carbamidomethylation was set as fixed peptide modification, methionine oxidation, and protein N-terminal acetylation as variable modifications. Data were submitted to the ProteomeXchange Consortium via the PRIDE partner repository and can be accessed via www.proteomeexchange.org with the identifiers PXD006570 to PXD006572, PXD006576, PXD006578, and PXD006589 to PXD006591 (27) (Supplemental Table S2).
Experimental Design and Statistical Rational
For the proteomics part, peripheral B cells from three elderly and three younger healthy donors as well as nine B-CLL patients were fractionated into cytoplasm and nucleic fraction and analyzed via shotgun proteomics. For comparison, three biological replicates of the chronic B cell leukemia cell line JVM-13 were also analyzed after fractionation. The LC-MS/MS proteomics measurements were carried out in technical duplicates for every sample. All sample groups consisted of both male and female donors. The B-CLL patient group comprised nine donors with different disease stages and treatments (Supplemental Table S1). Principal component analysis was carried out over all samples. Student t tests were applied between data from elderly and younger healthy donors to obtain age-related alterations, while B-CLL-associated deregulations were assessed by t tests between data from B-CLL patients and elderly healthy donors. Targeted metabolomics analyses (see next section) were carried out on whole-cell lysates from five healthy donors and four B-CLL patients at various disease stages, including one donor who was used for both proteomics and metabolomics analyses (Supplemental Table S1). Sample groups again consisted of male and female subjects. Technical replicates were not implemented, as internal standards were applied in every sample in order to correct for sample preparation and measurement variations. Metabolites detected at levels below the lower limit of quantification in the majority of samples were not considered for comparative analyses. For the investigation of regulatory events between B-CLL and healthy donors on the metabolomics level, Student t tests were utilized, including permutation-based multiparameter correction with 250 randomizations and an S0 of 0 at an FDR<0.05. ROS measurements were performed on thawed intact PBMCs from four young and four elderly healthy donors as well as from seven B-CLL patients. PBMCs from every donor were measured with and without fluorescent dye to evaluate fluorescence background levels. As positive controls, two samples per sample group were treated with the ROS-inducer tert-butyl hydrogen peroxide prior to incubation with fluorescent dye. For every sample, B as well as T cells were analyzed using anti-CD3- and anti-CD19-specific antibodies to select T and B cells, respectively. Monocytes were solely separated according to forward and side scattering and present in B-CLL patient samples at very small numbers, which resulted in high variance for corresponding ROS-level measurements. ROS levels in B-CLL patient monocytes can therefore only be seen as a semi-quantitative estimation. Supplemental Fig. S1 exemplifies a representative experiment, including positive and negative controls.
Targeted Metabolomics Experiments
One million B cells were resuspended in lysis buffer (10 mm phosphate buffer in 85% ethanol) and lysed by three freeze–thaw cycles. Targeted metabolomics experiments were conducted using the AbsoluteIDQ p180 kit (Biocrates Life Sciences AG, Innsbruck, Austria), enabling the detection and (semi)quantitation of up to 188 analytes, comprising 40 acylcarnitines, 42 amino acids and biogenic amines, 15 sphingolipids, 90 glycerophospholipids, and the sum of hexoses. Measurements were carried out using LC-MS and flow injection (FIA)-MS analyses on a 4000 QTRAP MS system (AB Sciex, Framingham, MA, USA) coupled to a 1200 RR HPLC system (Agilent, Palo Alto, CA, USA), utilizing the Analyst 1.6.2 software (also AB SCIEX). All required standards, quality controls, and eluents were included in the kit as well as the chromatographic column for the LC-MS/MS analysis part. Phenyl isothiocyanate (Sigma-Aldrich) was purchased separately and used for derivatization of amino acids and biogenic amines according to the kit manual. Preparation of the measurement worklist as well as data evaluation was performed with the software supplied with the kit (MetIDQ, version 5–4-8-DB100-Boron-2607, Biocrates Life Sciences).
RESULTS
High-Resolution Mass-Spectrometry-Based Proteomics Yields Comprehensive Protein Profiles
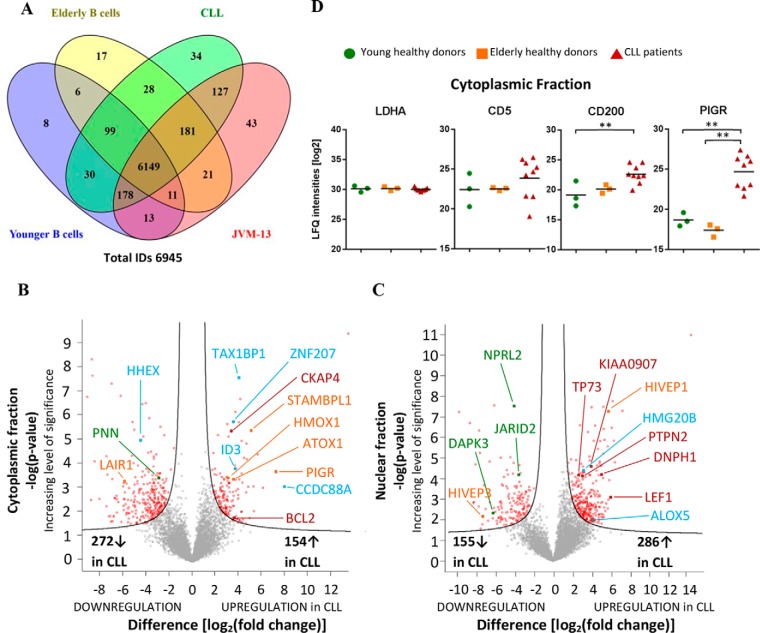
High-resolution Orbitrap-MS-based proteomic analyses of the cytoplasmic and nuclear fractions of B lymphocytes from six healthy donors, including three young donors (aged 25–50 years), three elderly donors (aged 71–83 years), and nine B-CLL patients, including Rai stages I-IV aged 67–88 years (Supplemental Table S1) and three biological replicates of JVM-13 samples resulted in the identification of 6,945 protein groups (Fig. 1A, Supplemental Table S3 and S4). Subcellular fractionation improved the chances to identify low-abundant proteins, such as transcription factors, and thus increased the proteome coverage as described (14). Protein identifications contained at least two distinct peptides with one proteotypic peptide and fulfilled an FDR<0.01 at both peptide and protein level. The B cell marker molecules CD19, CD20, CD21, CD38, CD40; the SLAM receptors characteristic for B cells CD84, CD150, CD229, CD319, and CD352; the B-cell-specific transcription factors PAX5, EBF1, and POU2F2; the B-CLL markers CD5, CD22, CD23/FCER2, and CD200; and other known leukemia-associated proteins, such as BCL2 (28), TCF3 (29) and LAIR1 (30), were all successfully identified (Supplemental Tables S3 and S4). These observations may serve to qualify both the cell preparation as well as the proteome analysis protocol. Actually, the present data also suggest novel candidate marker molecules such as EBAG9 (Fig. 3) and PIGR (Figs. 1B and 1D) robustly identified in all CLL cells but hardly detectable in normal B cells or peripheral blood mononuclear cells (24). Fig. 1D demonstrates that the differential expression of PIGR was found more pronounced in comparison to the known CLL marker molecules CD5 and CD200, while the household protein LDHA shows rather little variation between the groups and the individual samples. When comparing B cells from B-CLL patient with age-matched controls based on label-free quantitation using the MaxQuant software (25), 426 and 441 proteins in cytoplasmic and nuclear fractions, respectively, were found to be significantly altered (FDR<0.05) (Figs. 1B and 1C). However, no relevant alterations were found when comparing low-stage patients (Rai stage 0 and I) with high-risk patients (Rai stage III and IV). A complete list of all identified proteins including abundance differences is provided in Supplemental Table S3.
Fig. 1.
Data-dependent proteome analyses. (A) Venn diagram (96) of proteins identified in young and elderly normal B cells, CLL cells, and JVM13 cell line, indicating high qualitative ID overlap between sample groups. Results from cytoplasmic and nuclear fractions were combined. (B, C) Volcano plots of t tests between CLL cells and age-matched normal B cells for cytoplasmic and nuclear fractions, respectively. Permutation-based multiparameter correction was applied with an FDR of 0.05. Red data points indicate significant regulations, with up-regulations on the right and down-regulations on the left side of the volcano plot. Proteins associated with inflammatory response are highlighted in orange, while proteins relevant for stemness and differentiation are shown in blue color. Tumor proteins and tumor suppressors are depicted in red and green, respectively. (D) Levels of a household protein LDHA serving as loading control, two common CLL-associated proteins (CD5 and CD200) as well as of a novel potential CLL marker protein (PIGR) determined in the cytoplasm of young and elderly normal B cells as well as in primary CLL cells.
Fig. 3.
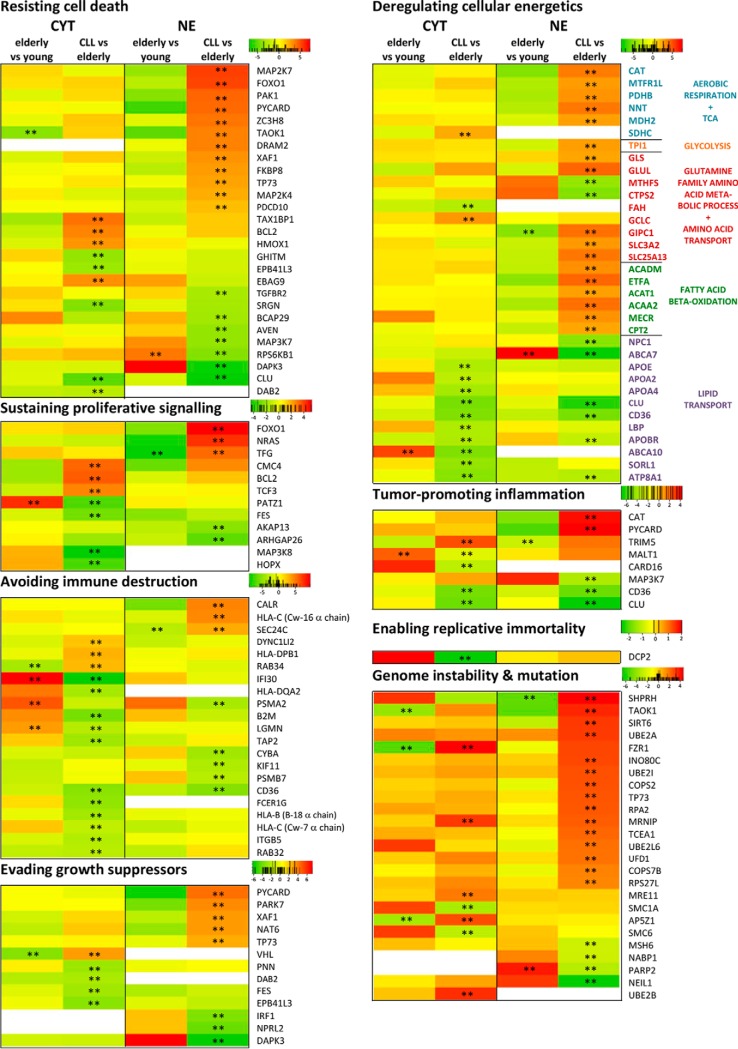
Heat maps for proteins related to cancer hallmarks (54) relevant for CLL. Heat maps are shown for proteins that were significantly regulated (FDR<0.05, indicated by **) in at least one fraction between CLL samples and samples from age-matched healthy donors. Red color indicates up-regulation, green color down-regulation, and white color absence of the protein in the respective fraction. Numbers given in the color scales represent fold changes in the form of 2x. Proteins were associated with the respective cancer hallmark term based on Uniprot GO terminology and/or keywords. So, to assign proteins to the hallmarks “Resisting cell death,” “Sustained proliferative signaling,” and “Evading growth suppression,” the Uniprot keywords “Apoptosis,” “Proto-oncogene,” and “Tumor suppressor” were utilized, respectively. To associate proteins to the hallmark “Deregulating cellular energetics,” a combined list of the GO terms “Aerobic respiration,” “Glycolysis,” “Fatty acid beta-oxidation,” “TCA cycle,” “Glutamine family amino acid metabolic process,” “Lipid transport,” “Glucose transport,” and “Amino acid transport” was used. Proteins involved in “Avoiding immune destruction” were determined using the GO terms “Antigen processing and presentation.” The Uniprot GO terms “positive regulation of NF-kappaB transcription factor activity,” “DNA repair,” and “negative regulation of telomere maintenance via telomerase” were utilized for the cancer hallmarks “Tumor-promoting inflammation,” “Genome instability and mutation,” and “Enabling replicative immortality,” respectively.
Aging-Associated Proteome Alterations of B cells May Predispose for B-CLL
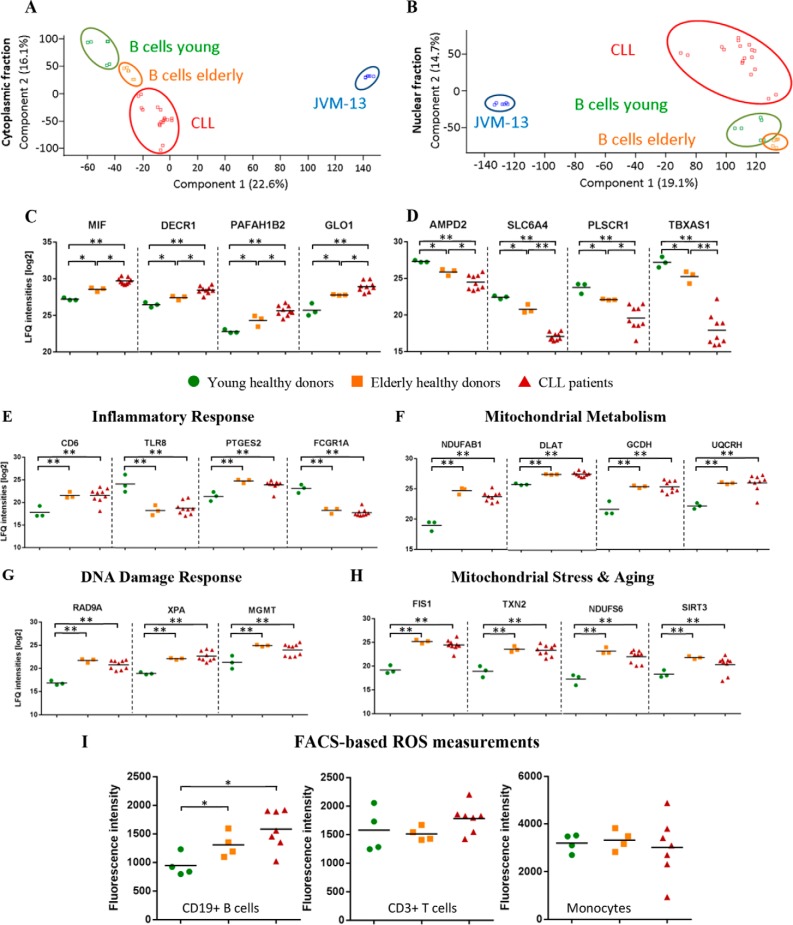
Principal component analyses of the data as depicted in Figs. 2A and 2B clearly demonstrate the successful separation of four different sample cohorts. Particularly striking was the order of appearance from B cell samples from young donors to the elderly donors and then the CLL cell cohort in the cytoplasmic fraction, hinting to age-associated proteome alterations functionally related to CLL. In the nuclear fraction, young and elderly donors were hardly separated. JVM-13 cells (Supplemental Table S4), used as model for B-CLL (31), were found to be rather poorly related to both primary B cells and B-CLL cells and were therefore excluded from further analysis.
Fig. 2.
Aging may predispose for CLL. (A, B) Principal component analysis of (A) cytoplasmic and (B) nuclear fractions of all four sample groups; a clear distinction between B cells from young donors, elderly donors, and CLL patients can be observed in the principal component analysis of the cytoplasmic fractions. (C, D) Selected proteins up-regulated (C) or down-regulated (D) in the cytoplasmic fractions of aged B cells from elderly donors and even more so in B cells from CLL patients are depicted. (E-H) Cytoplasmic proteins deregulated in elderly donors but hardly further altered in CLL. These proteins are related to (E) inflammatory response, (F) mitochondrial metabolism, (G) DNA damage response, and (H) mitochondrial stress and aging. (I) FACS-based ROS analyses of B cells, T cells, and monocytes obtained from young and elderly healthy donors as well as CLL patients. Statistical significance for displayed regulations was assessed by Student's t test with; *, p ≤ 0.05 and **, FDR≤0.05 applying multiparameter correction. Lines indicate mean label-free quantification values.
In order to interpret the clear trend observed in the principal component analysis for the cytoplasmic fraction, proteins successively up- or down-regulated in this fraction when comparing young and aged B cells with B-CLL cells were investigated in more detail. Up-regulation of MIF, DECR1, PAFAH1B2 and GLO1 (Fig. 2C) point to stress adaptation responses associated with chronic inflammation (32), altered metabolism (33), and signaling (34) as well as increased detoxification (35). Similar aging-related processes may as well account for the down-regulated proteins AMPD2, SLC6A4, PLSCR1 and TBXAS1 (Fig. 2D). Loss of function of the cellular energy homoeostasis protein AMPD2 was found associated with neurodegeneration (36), whereas down-regulation of the sodium-dependent serotonin transporter SLC6A4 was described as marker of life adversity exposure clearly associated with massive stress (37). The down-regulation of PLSCR1 may most plausibly inhibit inflammatory responses resulting from acute cell stress as described in case of hypoxia and reoxygenation (38). TBXAS1 has been described to be expressed at significantly reduced levels in high-grade breast tumors and tumor patients with poor prognosis (39).
Remarkably, several proteins related to cancer-associated processes such as inflammatory response (Fig. 2E), altered mitochondrial metabolism (Fig. 2F), DNA damage response (Fig. 2G), and mitochondrial stress and aging (Fig. 2H) were found significantly deregulated in the aged but healthy individuals, showing no further significant changes in the B-CLL cells. We hypothesized that the mitochondrial alterations might cause increased ROS levels eventually accounting for increased DNA damage and consequent DNA damage responses. FACS-based ROS assays indeed demonstrated significantly elevated ROS levels in B cells from elderly donors. This finding was specific for B cells and not observed in case of T cells and monocytes (Fig. 2I).
Proteins found to be significantly regulated between B cells from young and elderly donors were further submitted to the oPOSSUM software (version 3.0) (40). This allowed the detection of overrepresented conserved transcription factor binding sites in the corresponding sets of genes. Age-related protein up-regulation was thus suggested to be most apparently related to ZNF143 and GABPA (Supplemental Fig. S5). This is compatible with our previous interpretation, as ZNF143 is regulated through ROS production (41), and GABPA is involved in mitochondrial biogenesis (42). While these transcription factors were positively identified by mass spectrometry, no significant alteration was observed (Supplemental Table S3). Other proteins found significantly down-regulated in aged donors may relate to STAT3, which was indeed found fourfold down-regulated (p < 0.033) (Supplemental Fig. S5). This is in accordance with the observation that age-associated impairment of B cell functions correlate with reduced STAT3 activity, affecting especially naïve B cells (43, 44).
These data suggest age-associated mitochondrial stress associated with increased ROS levels causing DNA damage and thus indicate molecular mechanisms how aging may predispose for CLL.
B-CLL Proteome and the Hallmarks of Cancer
Several proteins previously associated with various tumor types, such as TP73 (45), CKAP4 (46), PTPN2 (TC-PTP) (Fig. 1C) (47), DNPH1 (48), and KIAA0907 (49) were found significantly up-regulated in B-CLL (Figs. 1B and 1C). Furthermore, the tumor suppressor molecules JARID2 (50), NPRL2 (51), and PNN (52) were found significantly down-regulated (Figs. 1B and 1C). In order to learn more about the potential meaning of B-CLL-associated proteome alterations, proteins known to relate to hallmarks of cancer according to Hanahan and Weinberg (53) were evaluated separately (Fig. 3, Supplemental Table S6). Fig. 3 depicts heat maps of all proteins found to be significantly deregulated in B-CLL and listed in one of eight hallmarks according to gene ontology (GO) terms. In contrast to protein regulations associated with aging, which were found especially in the cytoplasmic fraction (Fig. 2), protein regulations specific for CLL were observed in both the cytoplasmic and nuclear fraction. In CLL, important regulatory effects were observed for proteins related to the GO terms “genome instability and mutation,” “avoiding immune destruction,” “resisting cell death,” and “deregulation of cellular energetics.” Rather little changes of proliferation markers MKI67 and PCNA as well as the growth factors EGF, IGF2, TGFB1, FGF2, CTGF, and HDGF as shown in Supplemental Table S3 are in accordance with the known quiescence of peripheral B-CLL cells (54).
Remarkably, two known age-associated proteins, CD21 (complement receptor type 2, CR2) (39) and CXCR5 (40), were found to be significantly up-regulated in the aged donors but partially brought back to normal levels in the B-CLL patients (Supplemental Table S3, CYT). This may relate to some kind of dedifferentiation and an apparent gain of a stem cell phenotype as indicated by up-regulated cell differentiation inhibitors ID3 (55), HMG20B (56), and TAX1BP1 (57) and other proteins characteristic for stem cells, including ZNF207 (58), CCDC88A (59), and ALOX5 (60) (Figs. 1B and 1C). This was accompanied by the significant deregulation of several kappaB motif binding transcription factors, i.e. up-regulation of NF-kappaB promoters HIVEP1 (61) and STAMBPL1 (62) and down-regulation of the NF-kappaB inhibitor HIVEP3 (63) (Fig. 1C).
Using again the oPOSSUM software (40), beside an apparent induction of p53 target genes mainly a loss of functionality of transcription factors in B-CLL cells relative to B cells was indicated (Supplemental Table S5). Targets of the transcription factors involved in B cell differentiation, apoptosis induction and inhibiting stem cell formation such as FOXA1 (64), FOXI1, FOXD3, and ELF5 (65, 66) (Supplemental Table S5) were most apparently affected. The repression of FOXD3 has been reported to be an early event in B-CLL, which leads to epigenetic silencing of specific target genes in mature B-CLL cells (67).
Metabolomic Analyses of B-CLL Cells Corroborate Alterations of Metabolically Relevant Proteins
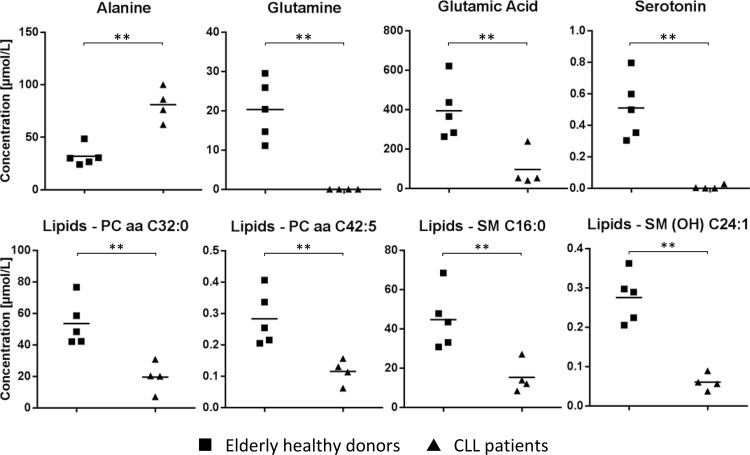
In order to support conclusions based on the observed alterations of proteins involved in metabolic functions between B-CLL cells and B cells from aged healthy donors, a metabolome screening assay covering 180 molecules was performed using primary B cells from four different B-CLL patients aged 68–83 years and five healthy elderly adults aged 54–63 years (Supplemental Table S1). While the concentration of most molecules was not affected, significant differences were observed in case of alanine, glutamine, glutamic acid, and serotonin as well as several phosphatidylcholines and sphingomyelins (Fig. 4). The apparent exhaustion of glutamine and glutamic acid accompanied by the increase of the glutaminolysis side product alanine indicated increased glutaminolysis in B-CLL cells as suggested by increased levels of GLS and GLUL (Fig. 3). The observed loss of serotonin was compatible with the massive down-regulation of the serotonin transporter SLC6A4 (Fig. 2D).
Fig. 4.
Metabolite analyses. Comparative visualization of selected metabolite species measured via Biocrates p180 MetIDQ kit. Asterisks (**) indicate significance after Student's t test (p value <0.05) including multiparameter correction. Lines indicate mean concentration values.
DISCUSSION
Normal B Cells from Elderly Donors Display Features of “Inflamm-Aging” and Immunosenescence
The necessity to include age-matched control samples for the analysis of B-CLL-associated proteome alterations provided us with somewhat unexpected results of this study, a distinct proteome signature of B cells from elderly but healthy donors when compared with younger donors. As demonstrated in Fig. 2C, MIF was found increased in aged cells and further increased in B-CLL. MIF is a well-known marker molecule for chronic inflammation and has been functionally linked to the pathogenesis of various forms of cancer (68), including B-CLL (69). Similarly, GLO1 confers a stress-inducible enzymatic defense against dicarbonyl stress associated with aging and was found associated with multidrug-resistant tumors (70). As PLSCR1 mediates inflammation-induced phosphatidylserine exposure and subsequent apoptosis induction (38), the observed down-regulation (Fig. 2D) may again relate to chronic inflammation and reduced cell death. The general down-regulation of SLC6A4 in response to various forms of stress is very well documented (37); in rats, a physical restraint stress has been demonstrated to down-regulate SLC6A4 in lymphocytes (71). Additional age-associated proteome alterations in B cells confirmed the association with inflammation (Figs. 2E–2H). T cell differentiation antigen CD6 and PTGES2 potentially conferring positive cell survival signals (72, 73) were found up-regulated, whereas the down-regulation of TLR8 and FCGR1A may reduce immunogenicity (74, 75). The mitochondrial stress management and DNA repair proteins (Figs. 2F–2H) are highly reminiscent to activation-induced molecular mechanisms described in activated T cells resulting in the formation of ROS (76). The observed age-related increase in ROS-formation supported this interpretation and was rather specific for B cells and not observed in T cells and monocytes of the same donors (Fig. 2I). The important contribution of ROS for the formation and specific properties of incurable tumors was reviewed thoroughly by Watson (77). Three key mechanisms associated with aging, i.e. altered mitochondrial function, altered metabolic activity and increased DNA repair (78) were found deregulated in aged B cells. Consequently, we suggest that the age-associated proteome alterations presented in Figs. 2C-2H may be representing a proteome signature of inflamm-aging (79) and immunosenescence (80) and may mediate an increased risk for the formation of B-CLL.
Environmental Conditions Are Drivers of B-CLL Development
Chronic inflammation has been described as most important cause for immunosenescence, which is characterized by an “aged stem cell” phenotype with less replicative power and increased genomic instability (81). Actually, the present proteome analysis results suggest that B-CLL cells underwent some kind of selection toward genome instability, avoiding immune destruction, resisting apoptosis and deregulation of cellular energetics, and less toward uncontrolled proliferation and cell cycle. Various proteins characteristic for stem cells as well as differentiation inhibitors directly suggest stem-cell-like properties or stemness of B-CLL cells. The contribution of inflammatory processes to establish an immunosenescence phenotype in B cells from aged cells (Fig. 2) seems thus to proceed further in B-CLL cells. The inflammation-associated proteins PIGR (82) were found up-regulated (Fig. 1D), while the inflammation suppressor molecule LAIR1 (83) was consistently down-regulated (Fig. 1B), in line with published data (84). Moreover, PIGR deregulation seems to be sufficiently robust to potentially serve as additive diagnostic tool for B-CLL and eventually innovative therapeutic target. The expression of this transforming protein (82) in B-CLL cells not only indicates an adaptive response but may directly contribute to tumor promotion and thus represents a potential functional link between inflammatory signaling and increased risk for B-CLL formation. Furthermore, chronic inflammation may account for the presently observed down-regulation of tumor suppressors (85) as well as the up-regulation of HMOX1 and ATOX1 (Fig. 1B) indicating a further resistance to radical stress. It is remarkable that the present proteome profiles revealed a rather consistent protein pattern across the individual donors as demonstrated in Fig. 1D, despite B-CLL is associated with a rather heterogeneous genotype (86).
The alteration of enzymes and metabolites related to glutaminolysis (Figs. 3 and 4) is also compatible with stem-cell-like properties of CLL cells in an inflammatory background (87). Glutaminolysis is a well-recognized feature of cancer cells as glutamine may serve as fuel for fatty acid synthesis, protein synthesis, and ROS control (15). The metabolic control of immune cell differentiation and activation is a recent hot topic in immunology (89). If the metabolic environment played such an important role as suggested by these recent data, the poor relation of JVM-13 cells compared with the primary cells (Fig. 2) will be a direct consequence of cell culturing conditions. As these cells are cultured using FBS, they are kept in a metabolic environment extremely different from the in vivo situation in elderly individuals. The adaptation of the JVM-13 cells to the fetal environment may thus account for the observed alterations of the proteome profiles.
Reprogrammed Mitochondrial Function May Precede B-CLL Development
Mitochondria are the center of cellular stress control managed by a complex mito-nuclear communication system (89) and have been suggested to play a causative role in tumorigenesis (90, 91). We suggest that deregulated mitochondrial functions are resulting from chronic inflammation and may precede the formation of B-CLL cells. It is well described that ROS released from deregulated mitochondria cause DNA damage and thus promote mutations, as also indicated by the induction of DNA damage repair proteins demonstrated in Fig. 5A. The molecular signature of immunosenescence may represent a status description for a biological system at risk to positively select for transforming mutations to gain stem cell properties (Fig. 5B).
Fig. 5.
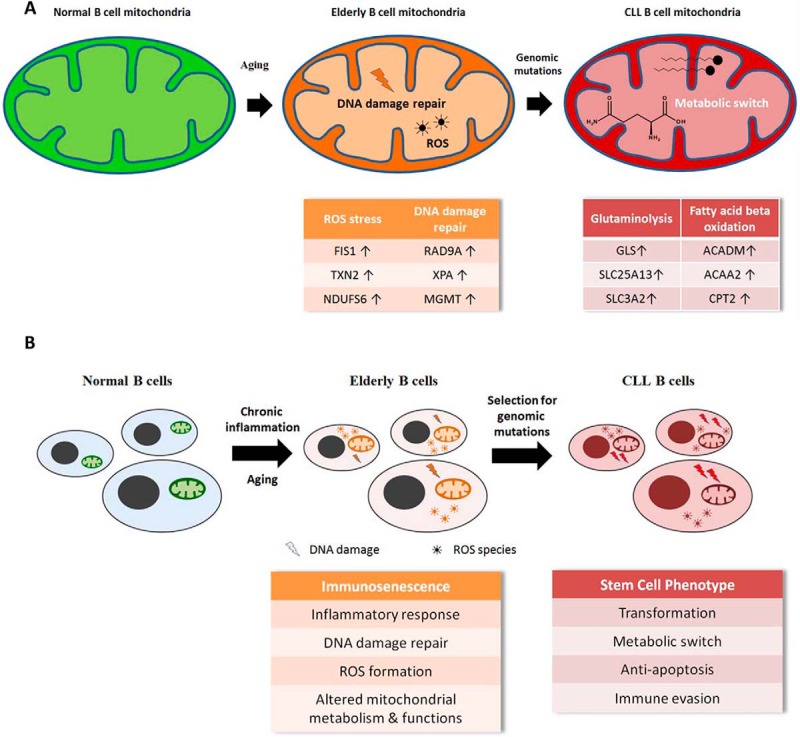
CLL tumorigenesis model. Proposed model of metabolic and proteomic alterations associated with development of CLL at the mitochondrial (A) and at the cellular level (B).
Individual Systemic Parameters and the Microenvironment Are Relevant for CLL
This line of interpretation also implies a strong regulatory function of systemic parameters such as the individual metabolome, as demonstrated recently by us with regard to ovarian cancer (16) and tumor-associated cachexia (9). In addition, the tumor microenvironment in the bone marrow was demonstrated by us to support CLL cells in order to become less sensitive with regard to apoptosis (92). The significant alteration of proteins regulating immune evasion such as EBAG9 (93) (Fig. 3) indicate the importance of adapting to conditions defined by the host. Actually, none of the deregulated tumor marker molecules, apoptosis regulators, tumor suppressor proteins, and other molecular players described in the present study was found to correlate with Rai stages. This observation suggests that disease progression may relate to individual systemic parameters and the tumor microenvironment rather than tumor cell properties. The term anakoinosis was coined in order to describe a novel therapeutic concept based on the reprogramming of tumor cells to reestablish apoptosis competence by modulating the microenvironment (94). It remains to be established whether such therapeutic concept, already proven to be successful in case of therapy-refractory Hodgkin lymphoma (95), may as well be beneficial for the treatment of B-CLL.
CONCLUSIONS
The present proteome profiling study provides evidence for an age-related reprogramming of normal B cells potentially predisposing for B-CLL by mitochondrial changes causing ROS stress and increased mutation rates and a metabolic pressure to differentiate into more long-lived cells. A rather homogenous protein expression pattern of B-CLL cells indicating a gain of stem cell properties with distinct metabolic features indicates previously unrecognized properties of B-CLL cells and may support the development of novel therapeutic strategies. Considering the treatment of the conditions supporting the formation of tumor cells in addition to the direct treatment of tumor cells will perhaps become an option.
DATA AVAILABILITY
Data were submitted to the ProteomeXchange Consortium via the PRIDE partner repository and can be accessed via www.proteomeexchange.org with the identifiers PXD006570 to PXD006572, PXD006576, PXD006578, and PXD006589 to PXD006591 (27) (Supplemental Table S2).
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support by the Fellinger Krebsforschung and the Faculty of Chemistry of the University of Vienna. We thank Günter Walder for his help with the cell culture part of the experiments.
Footnotes
* This work was supported by the Fellinger Krebsforschung.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- B-CLL
- B cell chronic lymphocytic leukemia
- CD19
- cluster of differentiation 19
- FDR
- false discovery rate
- IDs
- identifications
- MACS
- magnetic-activated cell sorting
- PBMCs
- peripheral blood mononuclear cells
- ROS
- reactive oxygen species.
REFERENCES
- 1. Zenz T., Mertens D., Küppers R., Döhner H., and Stilgenbauer S. (2010) From pathogenesis to treatment of chronic lymphocytic leukaemia. Nature Rev. Cancer 10, 37–50 [DOI] [PubMed] [Google Scholar]
- 2. Ghamlouch H., Nguyen-Khac F., and Bernard O. A. (2017) Chronic lymphocytic leukaemia genomics and the precision medicine era. Br. J. Haematol. 178, 852–870 [DOI] [PubMed] [Google Scholar]
- 3. Shehata M., Schnabl S., Demirtas D., Hilgarth M., Hubmann R., Ponath E., Badrnya S., Lehner C., Hoelbl A., Duechler M., Gaiger A., Zielinski C., Schwarzmeier J. D., and Jaeger U. (2010) Reconstitution of PTEN activity by CK2 inhibitors and interference with the PI3-K/Akt cascade counteract the antiapoptotic effect of human stromal cells in chronic lymphocytic leukemia. Blood 116, 2513–2521 [DOI] [PubMed] [Google Scholar]
- 4. Jitschin R., Braun M., Qorraj M., Saul D., Le Blanc K., Zenz T., and Mougiakakos D. (2015) Stromal cell-mediated glycolytic switch in CLL cells involves Notch-c-Myc signaling. Blood 125, 3432–3436 [DOI] [PubMed] [Google Scholar]
- 5. Frenzel L. P., Reinhardt H. C., and Pallasch C. P. (2016) Concepts of chronic lymphocytic leukemia pathogenesis: DNA damage response and tumor microenvironment. Oncol. Res. Treatment 39, 9–16 [DOI] [PubMed] [Google Scholar]
- 6. Groessl M., Slany A., Bileck A., Gloessmann K., Kreutz D., Jaeger W., Pfeiler G., and Gerner C. (2014) Proteome profiling of breast cancer biopsies reveals a wound healing signature of cancer-associated fibroblasts. J. Proteome Res. 13, 4773–4782 [DOI] [PubMed] [Google Scholar]
- 7. Paulitschke V., Gruber S., Hofstätter E., Haudek-Prinz V., Klepeisz P., Schicher N., Jonak C., Petzelbauer P., Pehamberger H., Gerner C., and Kunstfeld R. (2012) Proteome analysis identified the PPARgamma ligand 15d-PGJ2 as a novel drug inhibiting melanoma progression and interfering with tumor-stroma interaction. PloS One 7, e46103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Slany A., Haudek-Prinz V., Meshcheryakova A., Bileck A., Lamm W., Zielinski C., Gerner C., and Drach J. (2014) Extracellular matrix remodeling by bone marrow fibroblast-like cells correlates with disease progression in multiple myeloma. J. Proteome Res. 13, 844–854 [DOI] [PubMed] [Google Scholar]
- 9. Muqaku B., Eisinger M., Meier S. M., Tahir A., Pukrop T., Haferkamp S., Slany A., Reichle A., and Gerner C. (2017) Multi-omics analysis of serum samples demonstrates reprogramming of organ functions via systemic calcium mobilization and platelet activation in metastatic melanoma. Mol. Cell. Proteomics 16, 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barnidge D. R., Tschumper R. C., Jelinek D. F., Muddiman D. C., and Kay N. E. (2005) Protein expression profiling of CLL B cells using replicate off-line strong cation exchange chromatography and LC-MS/MS. J. Chromatography B 819, 33–39 [DOI] [PubMed] [Google Scholar]
- 11. Boyd R. S., Adam P. J., Patel S., Loader J. A., Berry J., Redpath N. T., Poyser H. R., Fletcher G. C., Burgess N. A., Stamps A. C., Hudson L., Smith P., Griffiths M., Willis T. G., Karran E. L., Oscier D. G., Catovsky D., Terrett J. A., and Dyer M. J. (2003) Proteomic analysis of the cell-surface membrane in chronic lymphocytic leukemia: Identification of two novel proteins, BCNP1 and MIG2B. Leukemia 17, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 12. Gez S., Crossett B., and Christopherson R. I. (2007) Differentially expressed cytosolic proteins in human leukemia and lymphoma cell lines correlate with lineages and functions. Biochim. Biophys. Acta 1774, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 13. Thurgood L. A., Chataway T. K., Lower K. M., and Kuss B. J. (2017) From genome to proteome: Looking beyond DNA and RNA in chronic lymphocytic leukemia. J. Proteomics 155, 73–84 [DOI] [PubMed] [Google Scholar]
- 14. Bileck A., Mayer R. L., Kreutz D., Weiss T., Taschner-Mandl S., Meier S. M., Slany A., and Gerner C. (2017) Evaluation of inflammation-related signaling events covering phosphorylation and nuclear translocation of proteins based on mass spectrometry data. J. Proteomics 152, 161–171 [DOI] [PubMed] [Google Scholar]
- 15. Altman B. J., Stine Z. E., and Dang C. V. (2016) From Krebs to clinic: Glutamine metabolism to cancer therapy. Nature Rev. Cancer 16, 619–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bachmayr-Heyda A., Aust S., Auer K., Meier S. M., Schmetterer K. G., Dekan S., Gerner C., and Pils D. (2017) Integrative systemic and local metabolomics with impact on survival in high-grade serous ovarian cancer. Clin. Cancer Res. 23, 2081–2092 [DOI] [PubMed] [Google Scholar]
- 17. Tahir A., Bileck A., Muqaku B., Niederstaetter L., Kreutz D., Mayer R. L., Wolrab D., Meier S. M., Slany A., and Gerner C. (2017) Combined proteome and eicosanoid profiling approach for revealing implications of human fibroblasts in chronic inflammation. Analytical Chem. 89, 1945–1954 [DOI] [PubMed] [Google Scholar]
- 18. Heintel D., Kienle D., Shehata M., Kröber A., Kroemer E., Schwarzinger I., Mitteregger D., Le T., Gleiss A., Mannhalter C., Chott A., Schwarzmeier J., Fonatsch C., Gaiger A., Döhner H., Stilgenbauer S., and Jäger U. (2005) High expression of lipoprotein lipase in poor risk B-cell chronic lymphocytic leukemia. Leukemia 19, 1216–1223 [DOI] [PubMed] [Google Scholar]
- 19. Rozovski U., Hazan-Halevy I., Barzilai M., Keating M. J., and Estrov Z. (2016) Metabolism pathways in chronic lymphocytic leukemia. Leukemia Lymphoma 57, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koczula K. M., Ludwig C., Hayden R., Cronin L., Pratt G., Parry H., Tennant D., Drayson M., Bunce C. M., Khanim F. L., and Günther U. L. (2016) Metabolic plasticity in CLL: Adaptation to the hypoxic niche. Leukemia 30, 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hallek M., Cheson B. D., Catovsky D., Caligaris-Cappio F., Dighiero G., Döhner H., Hillmen P., Keating M. J., Montserrat E., Rai K. R., and Kipps T. J. (2008) Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 111, 5446–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haudek-Prinz V. J., Klepeisz P., Slany A., Griss J., Meshcheryakova A., Paulitschke V., Mitulovic G., Stöckl J., and Gerner C. (2012) Proteome signatures of inflammatory activated primary human peripheral blood mononuclear cells. J. Proteomics 76, 150–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nature Meth. 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 24. Bileck A., Kreutz D., Muqaku B., Slany A., and Gerner C. (2014) Comprehensive assessment of proteins regulated by dexamethasone reveals novel effects in primary human peripheral blood mononuclear cells. J. Proteome Res. 13, 5989–6000 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 26. Cox J., and Mann M. (2012) 1D and 2D annotation enrichment: A statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 16, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Majid A., Tsoulakis O., Walewska R., Gesk S., Siebert R., Kennedy D. B., and Dyer M. J. (2008) BCL2 expression in chronic lymphocytic leukemia: Lack of association with the BCL2 938A>C promoter single nucleotide polymorphism. Blood 111, 874–877 [DOI] [PubMed] [Google Scholar]
- 29. Kardava L., Yang Q., St Leger A., Foon K. A., Lentzsch S., Vallejo A. N., Milcarek C., and Borghesi L. (2011) The B lineage transcription factor E2A regulates apoptosis in chronic lymphocytic leukemia (CLL) cells. Int. Immunol. 23, 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang X., Lu Z., Cui C., Deng M., Fan Y., Dong B., Han X., Xie F., Tyner J. W., Coligan J. E., Collins R. H., Xiao X., You M. J., and Zhang C. C. (2015) The ITIM-containing receptor LAIR1 is essential for acute myeloid leukaemia development. Nature Cell Biol. 17, 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konig A., Menzel T., Lynen S., Wrazel L., Rosen A., Al-Katib A., Raveche E., and Gabrilove J. L. (1997) Basic fibroblast growth factor (bFGF) upregulates the expression of bcl-2 in B cell chronic lymphocytic leukemia cell lines resulting in delaying apoptosis. Leukemia 11, 258–265 [DOI] [PubMed] [Google Scholar]
- 32. Das R., Koo M. S., Kim B. H., Jacob S. T., Subbian S., Yao J., Leng L., Levy R., Murchison C., Burman W. J., Moore C. C., Scheld W. M., David J. R., Kaplan G., MacMicking J. D., and Bucala R. (2013) Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, E2997–E3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Young C. D., and Anderson S. M. (2008) Sugar and fat - that's where it's at: Metabolic changes in tumors. Breast Cancer Res. 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kohnz R. A., Mulvihill M. M., Chang J. W., Hsu K. L., Sorrentino A., Cravatt B. F., Bandyopadhyay S., Goga A., and Nomura D. K. (2015) Activity-based protein profiling of oncogene-driven changes in metabolism reveals broad dysregulation of PAFAH1B2 and 1B3 in cancer. ACS Chem. Biol. 10, 1624–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rabbani N., Xue M., Weickert M. O., and Thornalley P. J. (2017) Multiple roles of glyoxalase 1-mediated suppression of methylglyoxal glycation in cancer biology—Involvement in tumour suppression, tumour growth, multidrug resistance and target for chemotherapy. Semin. Cancer Biol. In press [DOI] [PubMed] [Google Scholar]
- 36. Akizu N., Cantagrel V., Schroth J., Cai N., Vaux K., McCloskey D., Naviaux R. K., Van Vleet J., Fenstermaker A. G., Silhavy J. L., Scheliga J. S., Toyama K., Morisaki H., Sonmez F. M., Celep F., Oraby A., Zaki M. S., Al-Baradie R., Faqeih E. A., Saleh M. A., Spencer E., Rosti R. O., Scott E., Nickerson E., Gabriel S., Morisaki T., Holmes E. W., and Gleeson J. G. (2013) AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell 154, 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Provenzi L., Giorda R., Beri S., and Montirosso R. (2016) SLC6A4 methylation as an epigenetic marker of life adversity exposures in humans: A systematic review of literature. Neurosci. Biobehav. Rev. 71, 7–20 [DOI] [PubMed] [Google Scholar]
- 38. Slone E. A., Pope M. R., and Fleming S. D. (2015) Phospholipid scramblase 1 is required for beta2-glycoprotein I binding in hypoxia and reoxygenation-induced endothelial inflammation. J. Leukocyte Biol. 98, 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Watkins G., Douglas-Jones A., Mansel R. E., and Jiang W. G. (2005) Expression of thromboxane synthase, TBXAS1 and the thromboxane A2 receptor, TBXA2R, in human breast cancer. Intl. Sem. Surgical Oncol. 2, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ho Sui S. J., Mortimer J. R., Arenillas D. J., Brumm J., Walsh C. J., Kennedy B. P., and Wasserman W. W. (2005) oPOSSUM: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 33, 3154–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paek A. R., and You H. J. (2011) GAIP-interacting protein, C-terminus is involved in the induction of zinc-finger protein 143 in response to insulin-like growth factor-1 in colon cancer cells. Molecules Cells 32, 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Z. F., Drumea K., Mott S., Wang J., and Rosmarin A. G. (2014) GABP transcription factor (nuclear respiratory factor 2) is required for mitochondrial biogenesis. Mol. Cell Biol. 34, 3194–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Duggal N. A., Upton J., Phillips A. C., Sapey E., and Lord J. M. (2013) An age-related numerical and functional deficit in CD19(+) CD24(hi) CD38(hi) B cells is associated with an increase in systemic autoimmunity. Aging Cell 12, 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deenick E. K., Avery D. T., Chan A., Berglund L. J., Ives M. L., Moens L., Stoddard J. L., Bustamante J., Boisson-Dupuis S., Tsumura M., Kobayashi M., Arkwright P. D., Averbuch D., Engelhard D., Roesler J., Peake J., Wong M., Adelstein S., Choo S., Smart J. M., French M. A., Fulcher D. A., Cook M. C., Picard C., Durandy A., Klein C., Holland S. M., Uzel G., Casanova J. L., Ma C. S., and Tangye S. G. (2013) Naive and memory human B cells have distinct requirements for STAT3 activation to differentiate into antibody-secreting plasma cells. J. Experiment. Med. 210, 2739–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inoue K., and Fry E. A. (2014) Alterations of p63 and p73 in human cancers. Subcell. Biochem. 85, 17–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kikuchi A., Fumoto K., and Kimura H. (2017) The Dickkopf1-CKAP4 axis creates a novel signaling pathway and may represent a molecular target for cancer therapy. Br. J. Pharmacol. 174, 4651–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pike K. A., and Tremblay M. L. (2016) TC-PTP and PTP1B: Regulating JAK-STAT signaling, controlling lymphoid malignancies. Cytokine 82, 52–57 [DOI] [PubMed] [Google Scholar]
- 48. Amiable C., Paoletti J., Haouz A., Padilla A., Labesse G., Kaminski P. A., and Pochet S. (2014) 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: Synthesis, structural studies and cytotoxic activities. Eur. J. Med. Chem. 85, 418–437 [DOI] [PubMed] [Google Scholar]
- 49. Mei Y. P., Liao J. P., Shen J., Yu L., Liu B. L., Liu L., Li R. Y., Ji L., Dorsey S. G., Jiang Z. R., Katz R. L., Wang J. Y., and Jiang F. (2012) Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 31, 2794–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Su C. L., Deng T. R., Shang Z., and Xiao Y. (2015) JARID2 inhibits leukemia cell proliferation by regulating CCND1 expression. Int. J. Hematol. 102, 76–85 [DOI] [PubMed] [Google Scholar]
- 51. Peng Y., Dai H., Wang E., Lin C. C., Mo W., Peng G., and Lin S. Y. (2015) TUSC4 functions as a tumor suppressor by regulating BRCA1 stability. Cancer Res. 75, 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pehlivan D., Gunduz E., Gunduz M., Nagatsuka H., Beder L. B., Cengiz B., Rivera R. S., Fukushima K., Palanduz S., Ozturk S., Yamanaka N., and Shimizu K. (2008) Loss of heterozygosity at chromosome 14q is associated with poor prognosis in head and neck squamous cell carcinomas. J. Cancer Res. Clin. Oncol. 134, 1267–1276 [DOI] [PubMed] [Google Scholar]
- 53. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 54. Bueso-Ramos C. E., Ferrajoli A., Medeiros L. J., Keating M. J., and Estrov Z. (2004) Aberrant morphology, proliferation, and apoptosis of B-cell chronic lymphocytic leukemia cells. Hematology 9, 279–286 [DOI] [PubMed] [Google Scholar]
- 55. Miao T., Symonds A. L. J., Singh R., Symonds J. D., Ogbe A., Omodho B., Zhu B., Li S., and Wang P. (2017) Egr2 and 3 control adaptive immune responses by temporally uncoupling expansion from T cell differentiation. J. Experiment. Med. 214, 1787–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Esteghamat F., van Dijk T. B., Braun H., Dekker S., van der Linden R., Hou J., Fanis P., Demmers J., van I. W., Ozgür Z., Horos R., Pourfarzad F., von Lindern M., and Philipsen S. (2011) The DNA binding factor Hmg20b is a repressor of erythroid differentiation. Haematologica 96, 1252–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matsushita N., Suzuki M., Ikebe E., Nagashima S., Inatome R., Asano K., Tanaka M., Matsushita M., Kondo E., Iha H., and Yanagi S. (2016) Regulation of B cell differentiation by the ubiquitin-binding protein TAX1BP1. Sci. Rep. 6, 31266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Toh C. X., Chan J. W., Chong Z. S., Wang H. F., Guo H. C., Satapathy S., Ma D., Goh G. Y., Khattar E., Yang L., Tergaonkar V., Chang Y. T., Collins J. J., Daley G. Q., Wee K. B., Farran C. A., Li H., Lim Y. P., Bard F. A., and Loh Y. H. (2016) RNAi reveals phase-specific global regulators of human somatic cell reprogramming. Cell Rep. 15, 2597–2607 [DOI] [PubMed] [Google Scholar]
- 59. Liu C., Xue H., Lu Y., and Chi B. (2012) Stem cell gene Girdin: A potential early liver metastasis predictor of colorectal cancer. Mol. Biol. Rep. 39, 8717–8722 [DOI] [PubMed] [Google Scholar]
- 60. Hu Y., and Li S. (2016) Survival regulation of leukemia stem cells. Cell Mol. Life Sci. 73, 1039–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu B., Mitchell G. A., and Richter A. (2009) Cirhin up-regulates a canonical NF-kappaB element through strong interaction with Cirip/HIVEP1. Exp. Cell Res. 315, 3086–3098 [DOI] [PubMed] [Google Scholar]
- 62. Lavorgna A., and Harhaj E. W. (2012) An RNA interference screen identifies the deubiquitinase STAMBPL1 as a critical regulator of human T-cell leukemia virus type 1 tax nuclear export and NF-kappaB activation. J. Virol. 86, 3357–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hicar M. D., Liu Y., Allen C. E., and Wu L. C. (2001) Structure of the human zinc finger protein HIVEP3: Molecular cloning, expression, exon-intron structure, and comparison with paralogous genes HIVEP1 and HIVEP2. Genomics 71, 89–100 [DOI] [PubMed] [Google Scholar]
- 64. Yamaguchi N., Nakayama Y., and Yamaguchi N. (2017) Down-regulation of Forkhead box protein A1 (FOXA1) leads to cancer stem cell-like properties in tamoxifen-resistant breast cancer cells through induction of interleukin-6. J. Biol. Chem. 292, 8136–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yamamizu K., Piao Y., Sharov A. A., Zsiros V., Yu H., Nakazawa K., Schlessinger D., and Ko M. S. (2013) Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Rep. 1, 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ma K., Araki K., Ichwan S. J., Suganuma T., Tamamori-Adachi M., and Ikeda M. A. (2003) E2FBP1/DRIL1, an AT-rich interaction domain-family transcription factor, is regulated by p53. Mol. Cancer Res. 1, 438–444 [PubMed] [Google Scholar]
- 67. Chen S. S., Raval A., Johnson A. J., Hertlein E., Liu T. H., Jin V. X., Sherman M. H., Liu S. J., Dawson D. W., Williams K. E., Lanasa M., Liyanarachchi S., Lin T. S., Marcucci G., Pekarsky Y., Davuluri R., Croce C. M., Guttridge D. C., Teitell M. A., Byrd J. C., and Plass C. (2009) Epigenetic changes during disease progression in a murine model of human chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 106, 13433–13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. O'Reilly C., Doroudian M., Mawhinney L., and Donnelly S. C. (2016) Targeting MIF in cancer: Therapeutic strategies, current developments, and future opportunities. Med. Res. Rev. 36, 440–460 [DOI] [PubMed] [Google Scholar]
- 69. Reinart N., Nguyen P. H., Boucas J., Rosen N., Kvasnicka H. M., Heukamp L., Rudolph C., Ristovska V., Velmans T., Mueller C., Reiners K. S., von Strandmann E. P., Krause G., Montesinos-Rongen M., Schlegelberger B., Herling M., Hallek M., and Fingerle-Rowson G. (2013) Delayed development of chronic lymphocytic leukemia in the absence of macrophage migration inhibitory factor. Blood 121, 812–821 [DOI] [PubMed] [Google Scholar]
- 70. Rabbani N., Xue M., and Thornalley P. J. (2016) Methylglyoxal-induced dicarbonyl stress in aging and disease: First steps towards glyoxalase 1-based treatments. Clin. Sci. 130, 1677–1696 [DOI] [PubMed] [Google Scholar]
- 71. Medina-Martel M., Urbina M., Fazzino F., and Lima L. (2013) Serotonin transporter in lymphocytes of rats exposed to physical restraint stress. Neuroimmunomodulation 20, 361–367 [DOI] [PubMed] [Google Scholar]
- 72. Consuegra-Fernández M., Martínez-Florensa M., Aranda F., de Salort J., Armiger-Borràs N., Lozano T., Casares N., Lasarte J. J., Engel P., and Lozano F. (2017) Relevance of CD6-mediated interactions in the regulation of peripheral T-cell responses and tolerance. Front Immunol. 8, 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ke J., Yang Y., Che Q., Jiang F., Wang H., Chen Z., Zhu M., Tong H., Zhang H., Yan X., Wang X., Wang F., Liu Y., Dai C., and Wan X. (2016) Prostaglandin E2 (PGE2) promotes proliferation and invasion by enhancing SUMO-1 activity via EP4 receptor in endometrial cancer. Tumour Biol. 37, 12203–12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang Y., Wang Y., Li L., Chen F., and Zhang P. (2017) Activation of the Toll-like receptor 8 pathway increases the immunogenicity of mesenchymal stem cells from umbilical cord. Mol. Med. Rep. 16, 2061–2068 [DOI] [PubMed] [Google Scholar]
- 75. Mancardi D. A., Albanesi M., Jönsson F., Iannascoli B., Van Rooijen N., Kang X., England P., Daëron M., and Bruhns P. (2013) The high-affinity human IgG receptor FcgammaRI (CD64) promotes IgG-mediated inflammation, anaphylaxis, and antitumor immunotherapy. Blood 121, 1563–1573 [DOI] [PubMed] [Google Scholar]
- 76. Kamiński M. M., Sauer S. W., Kamiński M., Opp S., Ruppert T., Grigaravičius P., Grudnik P., Gröne H. J., Krammer P. H., and Gülow K. (2012) T cell activation is driven by an ADP-dependent glucokinase linking enhanced glycolysis with mitochondrial reactive oxygen species generation. Cell Rep. 2, 1300–1315 [DOI] [PubMed] [Google Scholar]
- 77. Watson J. (2013) Oxidants, antioxidants and the current incurability of metastatic cancers. Open Biol. 3, 120144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kubben N., and Misteli T. (2017) Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 18, 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bulati M., Caruso C., and Colonna-Romano G. (2017) From lymphopoiesis to plasma cells differentiation, the age-related modifications of B cell compartment are influenced by “inflamm-ageing.” Ageing Res. Rev. 36, 125–136 [DOI] [PubMed] [Google Scholar]
- 80. Masters A. R., Haynes L., Su D. M., and Palmer D. B. (2017) Immune senescence: Significance of the stromal microenvironment. Clin. Exp. Immunol. 187, 6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Skamagki M., Correia C., Yeung P., Baslan T., Beck S., Zhang C., Ross C. A., Dang L., Liu Z., Giunta S., Chang T. P., Wang J., Ananthanarayanan A., Bohndorf M., Bosbach B., Adjaye J., Funabiki H., Kim J., Lowe S., Collins J. J., Lu C. W., Li H., Zhao R., and Kim K. (2017) ZSCAN10 expression corrects the genomic instability of iPSCs from aged donors. Nature Cell Biol. 19, 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yue X., Ai J., Xu Y., Chen Y., Huang M., Yang X., Hu B., Zhang H., He C., Tang W., Peng X., Dong L., Wang H., Fan J., Ding J., and Geng M. (2017) Polymeric immunoglobulin receptor promotes tumor growth in hepatocellular carcinoma. Hepatology, 65, 1948–1962 [DOI] [PubMed] [Google Scholar]
- 83. Meyaard L. (2008) The inhibitory collagen receptor LAIR-1 (CD305). J. Leukocyte Biol. 83, 799–803 [DOI] [PubMed] [Google Scholar]
- 84. Perbellini O., Falisi E., Giaretta I., Boscaro E., Novella E., Facco M., Fortuna S., Finotto S., Amati E., Maniscalco F., Montaldi A., Alghisi A., Aprili F., Bonaldi L., Paolini R., Scupoli M. T., Trentin L., Ambrosetti A., Semenzato G., Pizzolo G., Rodeghiero F., and Visco C. (2014) Clinical significance of LAIR1 (CD305) as assessed by flow cytometry in a prospective series of patients with chronic lymphocytic leukemia. Haematologica 99, 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Walter B., Schrettenbrunner I., Vogelhuber M., Grassinger J., Bross K., Wilke J., Suedhoff T., Berand A., Wieland W. F., Rogenhofer S., Andreesen R., and Reichle A. (2012) Pioglitazone, etoricoxib, interferon-alpha, and metronomic capecitabine for metastatic renal cell carcinoma: Final results of a prospective phase II trial. Med. Oncol. 29, 799–805 [DOI] [PubMed] [Google Scholar]
- 86. Ghamlouch H., Nguyen-Khac F., and Bernard O. A. (2017) Chronic lymphocytic leukaemia genomics and the precision medicine era. Br. J. Haematol. 178, 852–870 [DOI] [PubMed] [Google Scholar]
- 87. Oburoglu L., Tardito S., Fritz V., de Barros S. C., Merida P., Craveiro M., Mamede J., Cretenet G., Mongellaz C., An X., Klysz D., Touhami J., Boyer-Clavel M., Battini J. L., Dardalhon V., Zimmermann V. S., Mohandas N., Gottlieb E., Sitbon M., Kinet S., and Taylor N. (2014) Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell 15, 169–184 [DOI] [PubMed] [Google Scholar]
- 88. Buck M. D., Sowell R. T., Kaech S. M., and Pearce E. L. (2017) Metabolic instruction of immunity. Cell 169, 570–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Quirós P. M., Mottis A., and Auwerx J. (2016) Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17, 213–226 [DOI] [PubMed] [Google Scholar]
- 90. Warburg O. (1956) On respiratory impairment in cancer cells. Science 124, 269–270 [PubMed] [Google Scholar]
- 91. Seyfried T. N. (2015) Cancer as a mitochondrial metabolic disease. Front. Cell Dev. Biol. 3, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Plander M., Ugocsai P., Seegers S., Orsó E., Reichle A., Schmitz G., Hofstädter F., and Brockhoff G. (2011) Chronic lymphocytic leukemia cells induce anti-apoptotic effects of bone marrow stroma. Ann. Hematol. 90, 1381–1390 [DOI] [PubMed] [Google Scholar]
- 93. Miyazaki T., Ikeda K., Horie-Inoue K., Kondo T., Takahashi S., and Inoue S. (2014) EBAG9 modulates host immune defense against tumor formation and metastasis by regulating cytotoxic activity of T lymphocytes. Oncogenesis 3, e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hart C., Vogelhuber M., Wolff D., Klobuch S., Ghibelli L., Foell J., Corbacioglu S., Rehe K., Haegeman G., Thomas S., Herr W., and Reichle A. (2015) Anakoinosis: Communicative reprogramming of tumor systems—For rescuing from chemorefractory neoplasia. Cancer Microenviron. 8, 75–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ugocsai P., Wolff D., Menhart K., Hellwig D., Holler E., Herr W., and Reichle A. (2016) Biomodulatory metronomic therapy induces PET-negative remission in chemo- and brentuximab-refractory Hodgkin lymphoma. Br. J. Haematol. 172, 290–293 [DOI] [PubMed] [Google Scholar]
- 96. Oliveros J. C. (2007–2015) Venny. An interactive tool for comparing lists with Venn's diagrams. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were submitted to the ProteomeXchange Consortium via the PRIDE partner repository and can be accessed via www.proteomeexchange.org with the identifiers PXD006570 to PXD006572, PXD006576, PXD006578, and PXD006589 to PXD006591 (27) (Supplemental Table S2).