Abstract
Introduction
In vivo, cancer cells can utilize tube-like microtracks formed within the extracellular matrix (ECM) of the stroma as ‘highways’ to escape the primary tumor, however very little is known about the molecular mechanisms that govern cell migration through these microtracks. Cell polarization and actin organization are both essential for efficient cell migration and cells are known to migrate very unidirectionally in confined spaces. In this study, we focused on understanding the role of Girdin during unidirectional migration. Girdin is a prometastatic protein known to be involved in cell polarity by directly interacting with the cell polarity protein Par-3 (Partitioning defective-3) and also known as an actin binding protein.
Methods
We utilized a microfabricated platform to recreate these microtracks in vitro using collagen and used siRNA to knockdown Girdin in MDA-MB-231 cells.
Results
Our data indicate that knockdown of Girdin results in decreased cell speed during 3D collagen microtrack migration. Loss of Girdin also results in altered cell morphology and cell orientation. Moreover, Girdin-depletion impairs actin organization and stress fiber formation, which can be restored by upregulating the GTPase RhoA. Activation of RhoA induces actin stress fiber formation, restores elongated migratory cell shape and partial cell migration in 3D collagen microtracks in the absence of Girdin.
Conclusions
Our data suggest that Girdin helps directional migration in collagen microtracks by promoting actin cytoskeletal organization and maintaining morphological cell polarity.
Keywords: Actin cytoskeleton, Cell morphology, Golgi apparatus, Cell orientation, RhoA, Extracellular matrix
Introduction
One of the earliest steps of breast cancer metastasis is cellular migration through the surrounding collagenous stromal extracellular matrix (ECM) after cells dissociate from the primary tumor. The stroma through which cancer cells navigate is a complex network of fiber architectures,11,12 and numerous studies have investigated the mechanisms by which cells move through isotropic collagen networks.8,33,51 However, it is also known that in vivo, metastatic cancer cells can migrate through pre-existing tracks within the collagen matrix.10,11 A subset of cells termed as “leader” cells create invasion paths (known as “microtracks”) by degrading and remodeling the surrounding matrix using proteolytic enzymes. Leader cells first migrate through the stroma leaving tube-like microtracks within the ECM of the stroma and other metastatic “follower” cells utilize these pre-existing microtracks as ‘highways’ to escape the primary tumor without any proteolytic activities.9 – 12 Despite numerous studies, the mechanisms modulating cancer cell migration through the stroma and particularly through these microtracks still remain unclear.
In our earlier studies, we developed a microfabricated platform to recreate three dimensional (3D) collagen microtracks similar to the preformed physiological microtracks found in vivo during cancer metastasis.23 Using our 3D collagen microtrack system, we investigated migration behaviors of invasive and non-invasive breast cancer cells, the roles of cytoskeletal and contractility regulators, and the effects of cell matrix adhesion mechanisms in highly invasive breast cancer cells during migration through the microtracks.2,23,41 Interestingly, our most recent work revealed that vinculin, a well characterized focal adhesion protein, regulates cell directionality by maintaining cell polarity in 3D microtrack migration.41 Given the highly polarized nature of migration within microtracks, we focused here on investigating the role of Girdin, a protein highly expressed in breast cancer cells that has been showed to regulate the establishment of cell polarity, in directing microtrack migration.28,35
One of the most thoroughly characterized protein families involved in determining cell polarity are the components of the partitioning-defective (Par) protein complex, specifically Par-3, Par-6 and atypical protein kinase C (aPKC).13,30,36,44 Girdin has been identified as a novel protein present in the Par protein polarity complex that comprises Par3, Par6 and aPKC, and its role in determining cell polarity has been established due to its direct physical interaction with Par-3.35,43 Girdin (GIRDers of act-IN filaments) is a prometastatic protein, and it binds to actin and regulates migration of breast cancer cells in a PI3K-Akt/PKB signaling pathway dependent way. It is a binding substrate of Akt and an Akt phosphorylation enhancer (APE).6,21,49 Girdin is also termed as a GIV (G -interacting vesicle-associated protein) for its ability to recruit and activate a class of trimeric G proteins (Gi) as a non-receptor Guanine Exchange factor (GEF), which leads to the enhancement of downstream signaling like PI3K-Akt/PKB, Focal Adhesion Kinase (FAK) and small GTPases, resulting in increased cell migration.15,25,26,29 Given its known role in 2D migration, 3D migration, and cell polarity,18,21,35 we are interested in investigating Girdin’s role in directing migration in 3D collagen microtracks that mimic the pre-existing tracks and interstitial spaces found in vivo.
To investigate the roles of Girdin in microtrack migration, we utilized a microfabricated in vitro 3D collagen microtrack system.2,23,41 Our findings indicate that depletion of Girdin in MDA-MB-231 cells results in reduced migration speed and changes in cellular morphology within microtracks. Moreover, cells fail to orient their internal machineries in the direction of migration, and they fail to form and organize actin stress fibers, which are essential for promoting directional cell migration.47 Migration can be partially restored in the absence of Girdin by inducing stress fiber formation through the activation of RhoA.
Methods and Materials
Cell Culture and Reagents
Highly migratory breast adenocarcinoma MDA-MB-231 cells (HTB-26, American Type Culture Collection (ATCC), Manassas, VA) were cultured at 37 °C and 5% CO2, and maintained in complete DMEM medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) and 1% penicillin–streptomycin (Life Technologies).
Small Interfering RNA (siRNA) Transfection
MDA-MB-231 cells were transfected with 15 nM of scrambled control siRNA oligos (5′-UUCCUCUCCACGCGCAGUACAUUUA-3′), or 15 nM of Girdin-siRNA oligos (5′-GAAGGAGAGGCAACUGGAUUU-3′)18 using Lipofectamine 2000 (2 μg/ml, Invitrogen). Both siRNAs were purchased from Life Technologies. Girdin siRNA was custom synthesized by Life Technologies. RNA interference-mediated knockdown in transfected cells was confirmed using western blot. Western blot and time-lapse migration assays were performed within 48 to 72 h post transfection.
Western Blotting
siRNA-transfected MDA-MB-231 cells were lysed using preheated (at 90 °C) 2 × Lammeli sample buffer after a quick rinse with ice-cold phosphate buffer saline (PBS) as described previously.20 Cell lysates were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis with a Mini-PROTEAN Tetra System (Bio-Rad, Hercules, CA) and electro-transferred onto a polyvinylidene difluoride (PVF) membrane. Blots were probed using antibodies against Girdin (sc-393757; Santa Cruz Biotechnology Inc., Santa Cruz, CA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, MAB374, Millipore). Anti-mouse horseradish peroxidase (HRP) conjugated secondary antibody was obtained from Rockland (Limerick, PA) to use against primary antibodies. After incubation with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL), blots were exposed and imaged using a FujiFilm ImageQuant LAS-4000.
Three-Dimensional Migration Studies
Three-dimensional (3D) collagen (1.5 mg/mL) uniform isotropic bulk matrices and collagen (3.0 mg/mL) microtracks were prepared using type I collagen extracted from rat tail tendons. As previously described, collagen solutions of desired concentrations were prepared from a 10 mg/mL collagen stock solution by diluting with ice-cold complete media and 1 N NaOH to neutralize the solution to pH 7.0.2,23,41 The final dimensions of the longitudinal collagen microtracks were 1000, 10 and 20 m respectively in the X, Y and Z directions (X = length, Y = width and Z = height). For 3D uniform isotropic bulk matrices studies, siRNA-treated MDA-MB-231 cells (suspended in media at a density of 70,000 cells/ml) were added to the collagen solution and then allowed to polymerize. For 3D collagen microtrack studies, siRNA-treated MDA-MB-231 cells were seeded after collagen was polymerized. In both cases, cells were seeded at a low density (70,000 cells/ml) to obtain isolated cells for single cell migration studies. Where indicated, siRNA-treated cells were incubated in complete medium supplemented with the Rho Activator II (CN03-A; Cytoskeleton, Inc.; Denver, CO).
Confocal Fluorescence and Phase Contrast Microscopy
Confocal fluorescence imaging was performed with a Zeiss LSM700 laser scanning confocal microscope on a Zeiss Axio Observer Z1 inverted stand using a long working distance water immersion C-Apochromat 40×/1.1 NA objective operated by Zen software (version 2010). For filamentous actin imaging, siRNA-treated cells seeded in microtracks were fixed, permeabilized, blocked as previously described8 and stained with Alexa Fluor 568-conjugated phalloidin (Life Technologies) overnight. For nuclei and Golgi apparatus imaging, live cells in microtracks were incubated in complete media for 5 min with a 1:1000 dilution of Hoechst 33342, trihydrochloride trihydrate (Invitrogen) and incubated overnight with Golgi-RFP (Cell Light®, BacMam 2.0, Life Technologies) respectively. Live-cell time-lapse confocal fluorescence imaging of the nucleus and Golgi apparatus was performed in temperature-, humidity- and CO2-controlled incubation chambers. Live cell phase contrast time-lapse images of cells were obtained using a Zeiss observer Z1 m inverted microscope equipped with a Hamamatsu ORCA-ER camera operated by AxioVision software (version 4.8.1.0, Carl Zeiss Microscopy, Thornwood, NY).
Migration Study Analysis
All images were analyzed using ImageJ software (version 1.47 k, National Institutes of Health, Bethesda, MD). For 3D collagen microtrack migration studies, cells were allowed to adhere for 6 h before performing time-lapse imaging at 10× magnification every 20 min for up to 15 h. Cell migration distance was quantified by determining the displacement of the cell from its initial position, and the relative positions of the nucleus and Golgi apparatus were determined by outlining the cell, nucleus and Golgi apparatus and measuring centroid position for each using Image J. Cell migration speed and motility were calculated as previously reported.1,21 For 3D uniform bulk collagen matrices migration studies, time-lapse phase contrast imaging was performed at × 10 magnification every 20 min for 24 h. All imaged cells were analyzed between 12 and 18 h after seeding. Cell trajectories, average displacement and average cell speed were calculated by measuring cell centroid position based on cell outlines as previously described.23 Directional cell persistence during migration in both 3D uniform matrices and 3D collagen microtracks was calculated by measuring the ratio of the sum of displacements between each time point and the total displacement of the cell from the initial to the end time point of the observed time interval. Calculated persistence value ranges between 0 and 1, where a persistence value of 1 indicates migration without changing direction during the observation time.52 Aspect ratio and circularity values of cell shape were generated by analyzing time-lapse phase contrast images of cells in 3D collagen microtracks and 3D bulk uniform matrix using ImageJ software. Aspect ratio (major axis/minor axis or width/height) describes the elongation of a cell and is larger than 1, with increasing values denoting elongated morphology. Circularity (4π × area/perimeter2) describes the roundness of a cell and can range from 0 to 1, with 1 denoting circular cells. All data are represented for 35–45 cells and obtained from a minimum of 3 independent experiments per treatment.
Statistical Analysis
Statistical analysis was carried out using JMP Software (v.11, SAS, Cary, North Carolina). Data are presented as mean ± SD and scatter plots were generated using Microsoft Excel 2016 and Prism 7.03. For migration analysis, statistical significance for comparisons between two groups was determined using two-tailed Student’s t test, and one-way ANOVA was performed to test mean differences for more than two groups with additional post hoc Tukey HSD test to show an overall statistically significant difference in group means. Statistical significance was considered with a p value < 0.05 and < 0.01. All figures are representative of a minimum of 3 replicate studies.
Results
Girdin Depletion Impairs Cell Migration in 3D Collagen Matrices and Microtracks
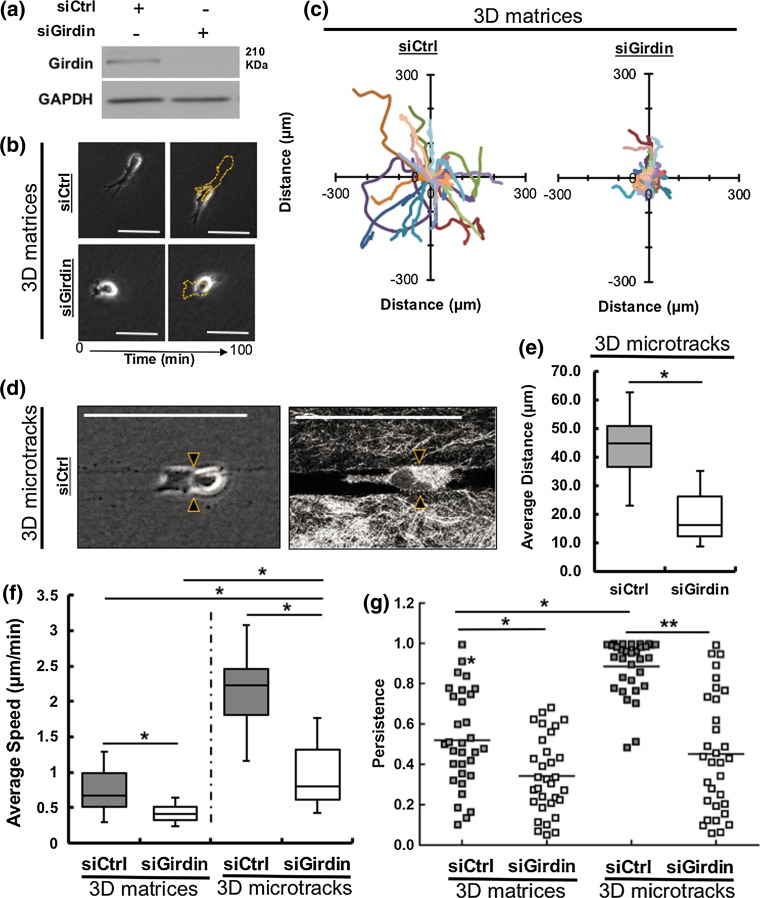
In previous work, we demonstrated the molecular mechanisms governing migration through 3D isotropic collagen matrices are different than cell migration mechanisms in 3D collagen microtracks.2,41 Cancer cells employ context-specific mechanisms to migrate.2,41 While migrating through 3D collagen microtracks, cells encounter little resistance and do not require significant traction generation, matrix remodeling, or cell body deformation.2 Therefore, we sought to test the effects of Girdin depletion in migration through two different 3D collagen systems: bulk matrices and tube-like microtracks. We confirmed the depletion of Girdin protein in Girdin siRNA-treated MDA-MB-231 cell using western blot (Fig. 1a). In 3D isotropic bulk collagen matrices, Girdin siRNA-treated cells travelled less far compared to control (scrambled) siRNA-treated cells over a period of 100 min (Fig. 1b), and cell trajectories indicate that Girdin knockdown cells moved slower (Fig. 1c). In 3D collagen microtracks with an average width of 10 microns (Fig. 1d), cells travelled significantly shorter distances in the absence of Girdin (Fig. 1e). Average cell migration speed was significantly less in Girdin siRNA-treated cells compared to control siRNA-treated cells in both 3D collagen matrices and microtracks. Interestingly, the average migration speed of Girdin-depleted cancer cells in 3D collagen microtracks was significantly higher than both Girdin-depleted cells and control cells migrating through 3D collagen isotropic matrices (Fig. 1f). In addition, Girdin siRNA treated cells’ directional migration persistence was significantly lower compared to control cells, in both 3D uniform bulk matrices and 3D microtracks. In 3D microtracks, control siRNA treated cells showed higher persistence than control cells in 3D matrices, and knockdown of Girdin results in less persistent cells. Altogether, Girdin knockdown cells became significantly less persistent compared to control cells in 3D microtracks (p < 0.01) than Girdin knockdown cells compared to control cells (p < 0.05) in 3D isotropic collagen matrices.
Figure 1.
Girdin regulates migration through 3D isotropic bulk collagen matrices and microtracks. (a) Western blot confirming the knockdown of Girdin in siRNA-treated MDA-MB-231 cells; (b) Time-lapse phase contrast images of control (scrambled) siRNA-treated (top), and Girdin siRNA-treated (bottom) MDA-MB-231 cells showing migration distance travelled by siRNA-treated cells in a 100 min time interval. Yellow dashed outlines indicate the initial positions of cells; c Rose plots showing trajectories of control and Girdin siRNA-treated cells in 3D collagen matrices, # of cells presented = 15; (d) Phase contrast (left) and confocal reflectance (right) images of a control siRNA-treated MDA-MB-231 cell (double arrowheaded) migrating through a 3D in vitro microfabricated collagen microtracks, (e) Average travelled distance of control and Girdin siRNA-treated MDA-MB-231 cells in microtracks over a period of more than 6 h. (f) Comparison of average migration speeds and (g) directional persistence of control and Girdin siRNA-treated MDA-MB-231 cells in 3D collagen isotropic matrices and 3D collagen microtracks. # cells per treatment = 32–45, scale bars 25 µm, *p value < 0.05, **p value < 0.01.
Girdin Knockdown Alters Cell Shape During Migration Through Microtracks
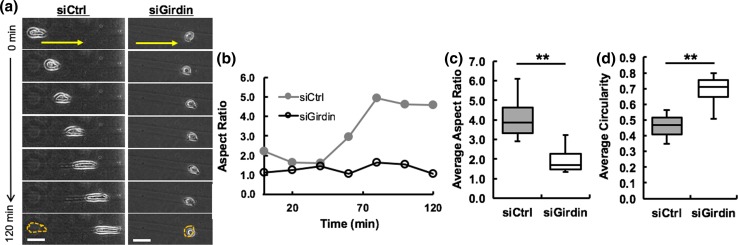
Cell morphology and migration are closely interrelated cell behaviors.3 The cell body must modify its shape to migrate. During migration, a cell becomes elongated as it extends the leading edge to attach to the surrounding ECM, and contracts at the rear end to pull the entire cell body forward.11 Time-lapse phase contrast migration imaging demonstrates that control siRNA-treated cells elongate their cell body during migration through microtracks, whereas Girdin knockdown cells were unable to elongate their cell body (Fig. 2a). Plotting aspect ratio against time further demonstrates that control cells exhibit dynamic shape changes as they move, whereas the aspect ratio remained unchanged for Girdin knockdown cells (Fig. 2b). The average aspect ratio of control siRNA-treated cells was significantly higher than Girdin knockdown cells (Fig. 2c) and correspondingly the average circularity was significantly lower for control cells compared to Girdin knockdown cells (Fig. 2d). These data indicate that Girdin helps to maintain cell elongation and direct migration in 3D collagen microtracks.
Figure 2.
Knockdown of Girdin results in morphological differences. (a) Time-lapse phase contrast images of a single control and Girdin siRNA-treated cell in a microtrack over a period of 120 min, yellow arrows and dashed outlines indicate the direction of migration and initial positions of cells at the beginning of migration, respectively; (b) Plotted aspect ratio of the same single control and Girdin siRNA-treated cells (showed in 3C) migrating through microtracks in a total time period of 120 at 20 min intervals; c Average aspect ratio and (d) Average circularity of control and Girdin knockdown MDA-MB-231 cells in microtracks. Scale bars 25 µm, # of cells per treatment = 35, **p value < 0.01.
Absence of Girdin Impacts Cell Orientation
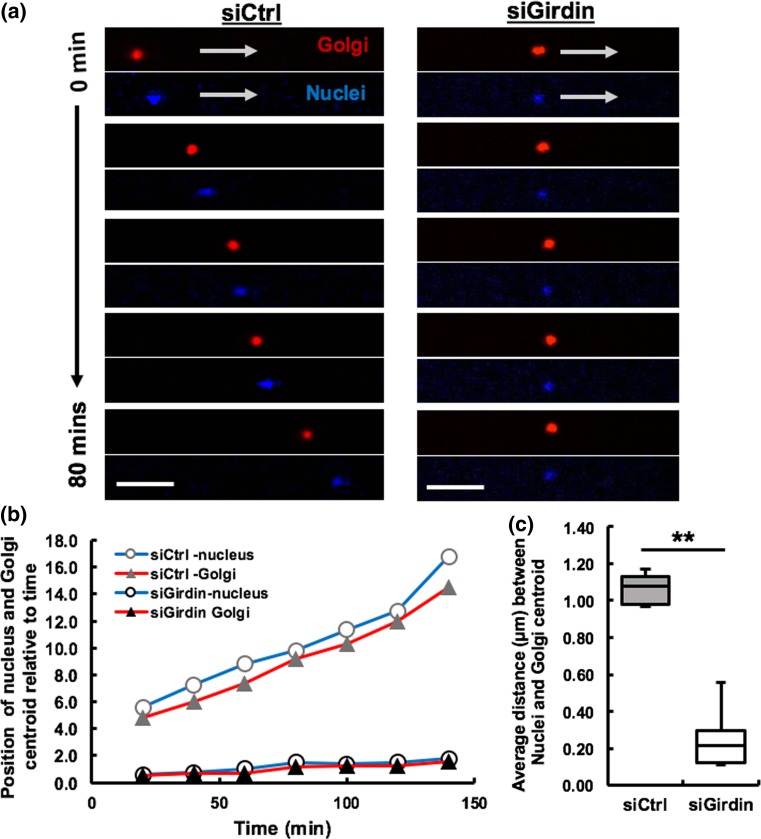
Previous studies performed using an in vitro wound healing assay model showed that Girdin is involved in determining cell polarity and orienting cells towards the direction of migration.35 To test Girdin’s ability to polarize and orient cells within microtracks, we tracked the position of the Golgi apparatus in control and Girdin siRNA-treated MDA-MB-231 cells using a fluorescent Golgi tracker. The Golgi were consistently positioned behind the nuclei in control cells migrating through the microtracks, but the Golgi were not positioned behind the nuclei in the absence of Girdin (Fig. 3a). In control siRNA-treated cells, the Golgi centroid was located further away from the nuclei centroid as the cell was able to polarize its cell body, whereas the Golgi centroid was located in very close proximity of the nuclei centroid in the absence of Girdin (Fig. 3b). The average distance between the nuclei and the Golgi centroid was significantly greater within the control cells compared to the Girdin knockdown cells (Fig. 3c). Altogether, our results indicate that Girdin enables cell elongation and affects the location of the Golgi relative to the nucleus.
Figure 3.
Girdin facilitates migration by orienting cell in the direction of migration. (a) Time-lapse confocal fluorescence imaging of nucleus (blue) and the Golgi apparatus (red) positions of migrating single control siRNA-treated (left) and single Girdin siRNA-treated (right) MDA-MB-231 cell over a period of 80 min in 3D microtracks, white arrows indicating the direction of movement, scale bars 25 µm; (b) Centroid positions of nucleus and the Golgi in a representable control and Girdin knockdown cell relative to time; (c) Average distance between Nuclei and Golgi centroids in control cells is greater compared to Girdin knockdown cells, # of cells per treatment = 12, **p value < 0.01.
Activation of RhoA Partially Restores Migration in the Absence of Girdin
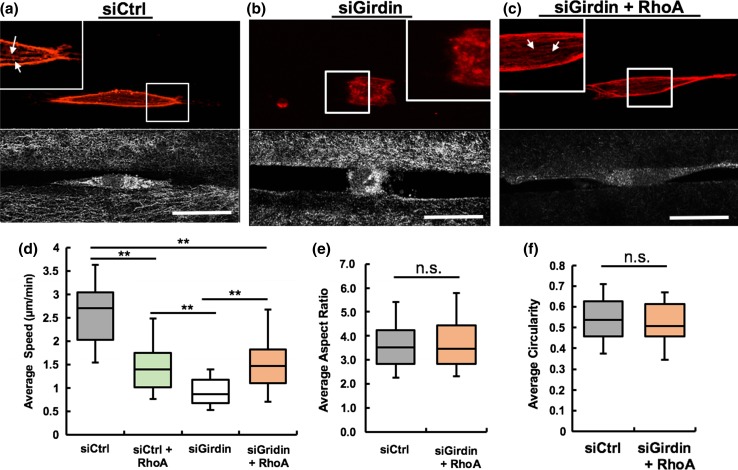
Girdin is known to be an actin binding protein, and depletion of Girdin has been showed to disrupt F- actin rearrangement.18,50 Therefore, we sought to focus on investigating the organization of actin filaments in siRNA-treated MDA-MB-231 cells during migration through 3D collagen microtracks. Actin polymerization was impaired in Girdin-depleted cells, and the cells exhibited rugged boundaries compared to control cells, in which cortical actin at the periphery of the cells and actin stress fibers within the cells was clearly observed (Figs. 4a, 4b). To determine role of RhoA in the absence of Girdin, we incubated Girdin knockdown cells with RhoA activator and visualized formation of stress fibers and cortical actin in the cells (Fig. 4c). RhoA is a small GTPase that can directly promote actin stress fiber formation.40 Interestingly, Girdin knockdown cells treated with the RhoA activator migrated significantly faster compared to untreated Girdin knockdown cells, but their average speed was significantly lower compared to control siRNA-treated cells (Fig. 4d). Interestingly, RhoA activated control siRNA treated cells also migrated with significant reduced speed compared to untreated control cells, but higher speed compared to Girdin knockdown cells (Fig. 4d). The average aspect ratio and circularity of treated cells were similar to control cells (Fig. 4e, 4f). Overall, these data indicate that Girdin knockdown impairs actin polymerization and impedes cell migration ability, and these effects can be ameliorated by activating RhoA.
Figure 4.
Activation of RhoA restores cell shape and migration in Girdin knockdown cells. Confocal immunofluorescence (top) and reflectance (bottom) images showing actin filament (red) organization of (a) control; (b) Girdin knockdown, and (c) RhoA Activator treated Girdin knockdown MDA-MB-231 cells during 3D microtrack migration, Inset magnification 2X, White arrows indicate presence of actin stress fibers (red) in control and RhoA activator treated Girdin knockdown cells; (d) Average speed of control siRNA, RhoA activated control siRNA, Girdin siRNA and RhoA activated Girdin siRNA-treated cells during migration through microtrack; (e) average aspect ratio and (f) average circularity of control and RhoA treated Girdin knockdown cells; # of cells per treatment = 44–45, *p value < 0.05, ‘n.s.’ **p value > 0.05, Scale bars = 25 µm.
Discussion and Conclusions
In this study, we demonstrated that Girdin, a protein known to be involved in numerous cellular processes including cancer metastasis, regulates unidirectional MDA-MB-231 cell migration in 3D in vitro collagen microtracks. Our results indicate that Girdin-depleted MDA-MB-231 cells migrate with significantly slower speed and less directional persistence in 3D collagen microtracks compared to control cells. In addition, in the absence of Girdin, cells fail to achieve the elongated migratory morphology that is generally observed when control cells migrate through the 3D collagen microtracks. Cells exhibit lower aspect ratio and higher circularity compared to highly migratory control cells. Similarly, Girdin-deficient cells are unable to polarize and orient their cell body in the direction of migration based on our analysis of the location of the Golgi apparatus. Furthermore, Girdin knockdown cells have impaired actin filament organization that can be partially restored through pharmacological activation of RhoA, which also partially restores migration ability in Girdin-depleted cells.
Cell migration is a complex process that involves organization and activation of various internal molecular machineries within the cell. Cell body polarization and actin cytoskeleton remodeling are two essential processes that enable a cell to move forward during cell migration.17,19,27,39,48 During directional migration, cells must become polarized by establishing a front-rear orientation, and continuously reorganize the actin cytoskeleton to move the cell body forward and coordinate directed migration.31,38,39,46,48 It is well-known that Partitioning defective (Par) proteins constitute a signaling pathway that helps cells to polarize and orient themselves during migration.17 Interestingly, Girdin is a protein that not only physically interacts with Par3 to play a role in establishing cell polarity, but it is also an actin binding protein.18,21,35 In the absence of Girdin, we observed impaired cell migration in 3D collagen microtracks suggesting that knockdown of Girdin impedes migration by disrupting cell polarization and modifying actin organization.
Previous migration studies on Girdin indicate that a reduction in Girdin expression impairs cell migration ability in both 2D and 3D migration platforms.18,20 When we compared migration within 3D collagen microtracks to migration within 3D isotropic collagen matrices, we found a similar result. Knockdown of Girdin with siRNA resulted in reduced migration speed and persistence in both 3D collagen isotropic matrices and 3D collagen microtracks. However, Girdin-depleted cells migrated with a relatively increased average migration speed in 3D collagen microtracks compared to the average speed of both the Girdin-depleted cells and the control siRNA-treated cells in 3D collagen matrices. This increased speed and persistence is likely because cells encounter minimal physical barriers, less resistance and are only allowed to move in one direction while migrating in preformed collagen microtracks. However, there was no visible formation of actin stress fibers or cortical actin in the absence of Girdin, and cells exhibited rugged boundaries at the cell edges and a more circular and non-migratory morphology. Girdin-depleted cells failed to reorganize actin cytoskeleton and elongate the cell body to form lamellipodia at the leading edge which are critical for directional cell migration.24 Previously, we observed that when actomyosin contractility regulators are inhibited, cell speed decreases in microtracks but migration is not completely stalled.2 Of the inhibitors we previously tested, cell motility only stops completely when actin polymerization is inhibited.2 Altogether, these data suggest that both the architecture of the matrix and the molecular machinery in the cell determine the migratory phenotype of cells.
The Golgi apparatus, the location of which is known to be an indicative of cell polarization,19 was not positioned away from the nucleus in the absence of Girdin during microtrack migration. This finding supports results from a previous study demonstrating that the actin cytoskeleton promotes the symmetry-breaking process, which permits the establishment of a polarized distribution of regulatory molecules.27 In this study, we observed impairment of the actin cytoskeletal organization and Golgi positioning in Girdin-deficient cells. The cytoskeleton structure is involved in localization of the Golgi within the cell body, and the Golgi orientation leads to directional cell migration.5,34 As such, these data suggest that Girdin contributes to directional migration in the 3D collagen microtracks by promoting actin cytoskeleton organization and orienting the Golgi within the cell body.
Disruption of actin polymerization results in changes to cytoskeletal dynamics and morphological phase transition.32 RhoA activation is known to directly induce actin polymerization by structurally changing the actin monomers to filaments and facilitate cell migration.16,42 Our results show that activation of RhoA can partially overcome Girdin-mediated disruption of actin stress fibers and cortical actin. Interestingly, with the activation of RhoA, actin cytoskeletal structural integrity was restored and cells exhibited shapes with higher aspect ratio and less circularity, more closely resembling control cells. However, despite these cytoskeletal and morphological changes, cells were only partially able to increase average migration speed with RhoA activation. Additionally, we observed that activation of RhoA impeded migration speed of control siRNA treated cells. This result is consistent with previous studies, where authors demonstrated that excessive Rho activity can inhibit cell migration by increasing stress fiber formation, adhesion and preventing focal adhesion turnover.1 In the absence of Girdin, when actin stress fiber organization is compromised and cell migration is impaired, activation of RhoA helped to counteract migration inability by forming adequate stress fibers within the cell to partially restore cell migration. But in control cells, excess stress fiber formation by RhoA activation had the opposite effect on speed, cell migration speed was significantly reduced.
Our results indicate that RhoA is only one of several important downstream effectors of Girdin that help in promoting cell migration. Certainly, there are other regulators that are affected by Girdin deficiency that could also potentially be responsible for the impaired migration in the presence of RhoA. For example, Signal transducer and activator of transcription-3 (STAT3) is known to promote directional cell migration and is a central regulator of cancer metastasis.37,45 Interestingly, STAT3 signaling is activated by RhoA pathway, and also is enhanced by Girdin’s function as a guanine exchange factor, in a RhoA independent pathway.4,14 Moreover, Girdin depletion has showed to reduce active RhoA.26 In our study, activation of RhoA partially restored migration in the absence of Girdin, but upregulation of STAT3 can further promote migration in the microtracks. In addition, it would be interesting to examine microtubule dynamics in Girdin-deficient cells. Microtubules are important for directed cell migration as they are involved in maintaining cell polarization and orientation.7 On the other hand, Girdin contains a hook domain that interacts with microtubules.22 Our study indicates that Girdin depleted cells fail to orient Golgi Apparatus in the direction of migration. Further investigation of the inter-relationship between microtubules and Girdin during directional migration may help further elucidate the mechanism by which girdin mediates migration.
In summary, our data suggest that Girdin helps to promote actin cytoskeleton organization, maintain morphological cell polarity, and enable cell migration in 3D collagen microtracks. In addition, RhoA activation restores cell shape and migration in the absence of Girdin. Our findings demonstrate a way to disrupt the directional migration in interstitial spaces or microtracks within the ECM by targeting Girdin.
Acknowledgment
The authors would like to acknowledge the use of equipment and resources at the Cornell NanoScale Science and Technology Facility (CNF).
Conflicts of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This work was supported by the National Science Foundation (NSF)—National Institute of Health Physical and Engineering Sciences in Oncology (PESO) award (Award Number 1233827) and by National Institutes of Health (Award Number HL127499) to Cynthia Reinhart-King. In addition, this was work also supported by National Science Foundation Graduate Fellowship to Aniqua Rahman-Zaman.
Abbreviations
- ECM
Extracellular matrix
- siRNA
Short interfering RNA
- Par-3
Partitioning defective-3
- STAT3
Signal transducer and activator of transcription-3
References
- 1.Besson A, Gurian-West M, Schmidt A, Hall A, Roberts JM. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 2004;18(8):862–876. doi: 10.1101/gad.1185504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carey SP, Rahman A, Kraning-Rush CM, Romero B, Somasegar S, Torre OM, Williams RM, Reinhart-King CA. Comparative mechanisms of cancer cell migration through 3D matrix and physiological microtracks. Am. J. Physiol -Cell Physiol. 2015;308(6):C436–C444. doi: 10.1152/ajpcell.00225.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey SP, Kraning-Rush CM, Williams RM, Reinhart-King CA. Biophysical control of invasive tumor cell behavior by extracellular matrix microarchitecture. Biomaterials. 2012;33(16):4157–4165. doi: 10.1016/j.biomaterials.2012.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debidda M, Wang L, Zang H, Poli V, Zheng Y. A role of STAT3 in Rho GTPase-regulated cell migration and proliferation. J. Biol. Chem. 2005;280(17):17275–17285. doi: 10.1074/jbc.M413187200. [DOI] [PubMed] [Google Scholar]
- 5.Egea G, Serra-Peinado C, Gavilán MP, Ríos RM. Cytoskeleton and Golgi-apparatus interactions: a two-way road of function and structure. Cell Health Cytoskelet. 2015;7:37–54. doi: 10.2147/CHC.S57108. [DOI] [Google Scholar]
- 6.Enomoto A, Ping J, Takahashi M. Girdin, a novel actin-binding protein, and its family of proteins possess versatile functions in the Akt and Wnt signaling pathways. Ann. N. Y. Acad. Sci. 2006;1086(1):169–184. doi: 10.1196/annals.1377.016. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Manneville S. Microtubules in cell migration. Annu. Rev. Cell Dev. Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- 8.Fraley SI, Feng Y, Krishnamurthy R, Kim D-H, Celedon A, Longmore GD, Wirtz D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 2010;12(6):598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedl, P., and K. Wolf. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. jcb-200909003, 2009. [DOI] [PMC free article] [PubMed]
- 10.Friedl P, Wolf K. Tube travel: the role of proteases in individual and collective cancer cell invasion. Cancer Res. 2008;68(18):7247–7249. doi: 10.1158/0008-5472.CAN-08-0784. [DOI] [PubMed] [Google Scholar]
- 11.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer. 2003;3(5):362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 12.Friedl Peter, Sahai Erik, Weiss Stephen, Yamada Kenneth M. New dimensions in cell migration. Nat. Rev. Mol. Cell Biol. 2012;13(11):743–747. doi: 10.1038/nrm3459. [DOI] [PubMed] [Google Scholar]
- 13.Fukata M, Nakagawa M, Kaibuchi K. Roles of Rho-family GTPases in cell polarisation and directional migration. Curr. Opin. Cell Biol. 2003;15:590–597. doi: 10.1016/S0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J. Biol. Chem. 2015;290(11):6697–6704. doi: 10.1074/jbc.R114.613414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Gαi3 triggers cell migration via regulation of GIV. J. Cell Biol. 2008;182(2):381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giehl K, Keller C, Muehlich S, Goppelt-Struebe M. Actin-mediated gene expression depends on RhoA and Rac1 signaling in proximal tubular epithelial cells. PLoS ONE. 2015;10(3):e0121589. doi: 10.1371/journal.pone.0121589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu F, Wang L, He J, Liu X, Zhang H, Li W, Fu L, Ma Y. Girdin an actin-binding protein, is critical for migration, adhesion, and invasion of human glioblastoma cells. J. Neurochem. 2014;131(4):457–469. doi: 10.1111/jnc.12831. [DOI] [PubMed] [Google Scholar]
- 19.Hehnly H, Xu W, Chen JL, Stamnes M. Cdc42 regulates microtubule-dependent golgi positioning. Traffic. 2010;11(8):1067–1078. doi: 10.1111/j.1600-0854.2010.01082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huynh J, Bordeleau F, Kraning-Rush CM, Reinhart-King CA. Substrate stiffness regulates PDGF-induced circular dorsal ruffle formation through MLCK. Cell. Mol. Bioeng. 2013;6(2):138–147. doi: 10.1007/s12195-013-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang P, Enomoto A, Jijiwa M, Kato T, Hasegawa T, Ishida M, Sato T, Asai N, Murakumo Y, Takahashi M. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer Res. 2008;68(5):1310–1318. doi: 10.1158/0008-5472.CAN-07-5111. [DOI] [PubMed] [Google Scholar]
- 22.Kenakin T. Oligomerization and allosteric modulation in G-protein coupled receptors. Cambridge: Academic Press; 2012. [Google Scholar]
- 23.Kraning-Rush CM, Carey SP, Lampi MC, Reinhart-King CA. Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D. Integr. Biol. 2013;5(3):606–616. doi: 10.1039/c3ib20196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 2014;15(9):577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 25.Leyme A, Marivin A, Garcia-Marcos M. GIV/Girdin (Gα-interacting, vesicle-associated protein/Girdin) creates a positive feedback loop that potentiates outside-in integrin signaling in cancer cells. J. Biol. Chem. 2016;291(15):8269–8282. doi: 10.1074/jbc.M115.691550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leyme, A., A. Marivin, L. Perez-Gutierrez, L. T. Nguyen, and M. Garcia-Marcos. Integrins activate trimeric G proteins via the nonreceptor protein GIV/Girdin. J. Cell Biol. jcb-201506041, 2015. [DOI] [PMC free article] [PubMed]
- 27.Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat. Rev Mol. Cell Biol. 2008;9(11):860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Zhang Y, Xu H, Zhang R, Li H, Lu P, Jin F. Girdin protein: a new potential distant metastasis predictor of breast cancer. Med. Oncol. 2012;29(3):1554–1560. doi: 10.1007/s12032-011-0087-6. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Sanchez I, Kalogriopoulos N, Lo I-C, Kabir F, Midde KK, Wang H, Ghosh P. Focal adhesions are foci for tyrosinebased signal transduction via GIV/Girdin and G proteins. Mol. Biol. Cell. 2015;26(24):4313–4324. doi: 10.1091/mbc.E15-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macara IG. Parsing the polarity code. Nat. Rev. Mol. Cell Biol. 2004;5:220–231. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- 31.Mak M, Spill F, Kamm RD, Zaman MH. Single-cell migration in complex microenvironments: mechanics and signaling dynamics. J. Biomech. Eng. 2015;138:1–8. doi: 10.1115/1.4032188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mak, M., M. H. Zaman, R. D. Kamm, and T. Kim. Interplay of active processes modulates tension and drives phase transition in self-renewing, motor-driven cytoskeletal networks. Nat. Commun. 7 2016. [DOI] [PMC free article] [PubMed]
- 33.Meyer, A. S., S. K. Hughes-Alford, J. E. Kay, A. Castillo, A. Wells, F. B. Gertler, and D. A. Lauffenburger. 2D protrusion but not motility predicts growth factor–induced cancer cell migration in 3D collagen. J. Cell Biol. jcb-201201003, 2012. [DOI] [PMC free article] [PubMed]
- 34.Millarte, V., and H. Farhan. The Golgi in cell migration: regulation by signal transduction and its implications for cancer cell metastasis. Scientific World J. 2012, 2012. [DOI] [PMC free article] [PubMed]
- 35.Ohara K, Enomoto A, Kato T, Hashimoto T, Isotani-Sakakibara M, Asai N, Ishida-Takagishi M, Weng L, Nakayama M, Watanabe T, Kato K. Involvement of Girdin in the determination of cell polarity during cell migration. PLoS ONE. 2012;7(5):e36681. doi: 10.1371/journal.pone.0036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 2001;13:641–648. doi: 10.1016/S0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- 37.Pan B, Shen J, Cao J, Zhou Y, Shang L, Jin S, Cao S, Che D, Liu F, Yu Y. Interleukin-17 promotes angiogenesis by stimulating VEGF production of cancer cells via the STAT3/GIV signaling pathway in non-small-cell lung cancer. Sci. Rep. 2015;5:16053. doi: 10.1038/srep16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellegrin S, Mellor H. Actin stress fibres. J. Cell Sci. 2007;120(20):3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 39.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 40.Qi M, Liu Y, Freeman MR, Solomon KR. Cholesterol-regulated stress fiber formation. J. Cell. Biochem. 2009;106:1031–1040. doi: 10.1002/jcb.22081. [DOI] [PubMed] [Google Scholar]
- 41.Rahman A, Carey SP, Kraning-Rush CM, Goldblatt ZE, Bordeleau F, Lampi MC, Lin DY, García AJ, Reinhart-King CA. Vinculin regulates directionality and cell polarity in two-and three-dimensional matrix and three-dimensional microtrack migration. Mol. Biol. Cell. 2016;27(9):1431–1441. doi: 10.1091/mbc.E15-06-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reffay M, Parrini M-C, Cochet-Escartin O, Ladoux B, Buguin A, Coscoy S, Amblard F, Camonis J, Silberzan P. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat. Cell Biol. 2014;3:217. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki K, Kakuwa T, Akimoto K, Koga H, Ohno S. Regulation of epithelial cell polarity by PAR-3 depends on Girdin transcription and Girdin–Gαi3 signaling. J. Cell Sci. 2015;128(13):2244–2258. doi: 10.1242/jcs.160879. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J. Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 45.Teng TS, Lin B, Manser E, Ng DCH, Cao X. Stat3 promotes directional cell migration by regulating Rac1 activity via its activator βPIX. J. Cell Sci. 2009;122(22):4150–4159. doi: 10.1242/jcs.057109. [DOI] [PubMed] [Google Scholar]
- 46.Tojkander S, Gateva G, Lappalainen P. Actin stress fibers–assembly, dynamics and biological roles. J. Cell Sci. 2012;125(8):1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- 47.Vallenius T. Actin stress fibre subtypes in mesenchymal-migrating cells. Open Biol. 2003;3(6):130001. doi: 10.1098/rsob.130001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J. Cell Sci. 2005;118(21):4917–4919. doi: 10.1242/jcs.02662. [DOI] [PubMed] [Google Scholar]
- 49.Weng L, Enomoto A, Ishida-Takagishi M, Asai N, Takahashi M. Girding for migratory cues: roles of the Akt substrate Girdin in cancer progression and angiogenesis. Cancer Sci. 2010;101(4):836–842. doi: 10.1111/j.1349-7006.2009.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu D, Yu D, Wang X, Yu B. F-actin rearrangement is regulated by mTORC2/Akt/Girdin in mouse fertilized eggs. Cell Prolif. 2016;49(6):740–750. doi: 10.1111/cpr.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl Acad. Sci. 2006;103(29):10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Guo W-H, Wang Y-L. Microtubules stabilize cell polarity by localizing rear signals. Proc. Natl Acad. Sci. 2014;111(46):16383–16388. doi: 10.1073/pnas.1410533111. [DOI] [PMC free article] [PubMed] [Google Scholar]