Abstract
Breast cancer (BC) is the most frequently diagnosed cancer that affects women worldwide. Early detection of BC is important to improve survival rates and decrease mortality. The aim of the present study was to investigate serum biomarkers using surface-enhanced laser desorption ionization time-of-flight mass spectrometry (SELDI-TOF-MS) to distinguish patients with BC from the healthy population and patients with benign breast diseases (BBDs). A total of 62 patients with invasive ductal carcinoma, as confirmed by histopathology, and 47 non-cancerous individuals (NCIs) [16 healthy controls (HCs) and 31 patients with BBD] were enrolled in the present study. Serum protein profiles were determined by SELDI-TOF-MS using an immobilized metal affinity capture array. Serum from patients with BC were compared with that from the HC group using univariate and multivariate statistical analyses. A total of 118 clusters were generated from the individual serum. Univariate analysis revealed that 5 peaks were significantly downregulated (m/z 1,452, 2,670, 3,972, 5,354 and 5,523; P<0.001) and 4 were upregulated (m/z 6,850, 7,926, 8,115 and 8,143; P<0.001) in patients with BC compared with the HC group. A comparison of patients with BC and patients with BBD revealed an additional 9 protein peaks. Among these, 3 peaks (m/z 3,972, 5,336 and 11,185) were significantly downregulated and 6 peaks (m/z 4,062, 4,071, 4,609, 6,850, 8,115 and 8,133) were significantly upregulated. A total of 3 peaks [mass-to-change ratio (m/z) 3,972, 6,850 and 8,115 (BC2)] were common in both sets. The results of the present study suggest that a 4 protein peak set [m/z 3,972, 6,850 and 8,115 (BC2) and 8,949 (BC3)] could be used to distinguish patients with BC from NCI.
Keywords: breast cancer, early diagnosis, biomarker, proteomics, surface-enhanced laser desorption ionization time-of-flight mass spectrometry
Introduction
Breast cancer (BC) is among the most frequent cancers in women in worldwide, and is the second leading cause of cancer-related mortality in women (1). Breast cancer may generally spread to distant locations, which affects curability of the cancer (2). Therefore, accurate and early detection of BC, particularly when pre-symptomatic, is a crucial factor in attaining a higher survival rate and improved prognosis for patients (3). Mammography is a generally preferred method in the detection of BC; however, in young women and women with dense breast tissue, mammographic screening may be less sensitive (4). In these cases, magnetic resonance imaging, an alternative screening approach, may be more sensitive than mammography (5). Blood-based screening tests would be cost-effective and efficient as an application for large-scale screenings. Studies on the specific molecular targets (oncogenes, tumor suppressor genes, growth factors, tumor antigens or other gene products) in BC make possible the application of blood-based screening approaches for BC. Further improvements in protein expression analysis and proteomics methods have led to the development of serum based diagnostics and prognostics for many types of cancer (6). One of these methods, surface-enhanced laser desorption ionization (SELDI), enables the identification of biomarkers in physiological fluid samples to distinguish patients with patients from healthy individuals (7). Although certain biomarkers may be used to identify BC, few of them have been validated for clinical use. Previous studies have reported a number of potential serum biomarkers that may be used to diagnose BC; BC1 (4.3 kDa), BC2 (8.1 kDa) and BC3 (8.9 kDa) (8,9). The structural definitions of these biomarkers are as follows: BC1, inter-α-trypsin inhibitor heavy chain H4; BC2, C3a des-arginine-C terminal truncated peptide; and BC3, C3a des-arginine (10). A number of other potential biomarkers have been identified and demonstrated to have high diagnostic accuracy (7,11,12). However, validating these proteins is difficult due to different protocols for sample handling, patient populations and tumor characteristics.
The aim of the present study was to investigate and develop an efficient biomarker panel to distinguish patients with BC from healthy individuals and patients with benign breast disease (BBD) using SELDI time-of-flight mass spectrometry (SELDI-TOF-MS).
Materials and methods
Patient characteristics
This prospective study included 62 patients with histopathologically confirmed BC with invasive ductal carcinoma and 47 non-cancerous individuals (NCIs). The study was approved by the Ataturk Training and Research Hospital Ethics Committee of Katip Celebi University (Izmir, Turkey) and performed in accordance with the Declaration of Helsinki. Among the patients with BC, 27 were in the early stages of disease (stages I and II) and 35 patients were in the advanced stages (stages III and IV). All patients were staged using the American Joint Committee on Cancer (AJCC) 6th edition staging manual (13). In the NCI group, 16 subjects were healthy controls (HCs) and 31 were patients with BBD. Patients with carcinoma in situ were not enrolled in the present study. Informed consent was obtained from all patients and control subjects. Demographic and clinical characteristics of the participants are presented in Table I. Patients did not receive radiotherapy or chemotherapy prior to surgery. Patients with BC had no evidence of chronic obstructive pulmonary disease, diabetes mellitus or malignancy at other sites. For women in the HC group who had breast examinations, the mammography and ultrasound results were normal.
Table I.
Demographics of patients and control subjects enrolled in the present study.
| Age (years) | ||||
|---|---|---|---|---|
| Groups | n | Median | Range | Carbohydrate antigen 15-3 level (U/ml) |
| Breast cancer | 24.59±15.79 | |||
| Stages I and II | 27 | 53.7 | 37–73 | 16.77±2.50 |
| Stages III and IV | 35 | 53.5 | 41–61 | 30.62±18.89 |
| Non-cancerous individuals | ||||
| Healthy controls | 16 | 38.7 | 21–52 | 15.92±4.8 |
| Benign breast disease | 31 | 41.3 | 23–71 | 16.01±5.27 |
Laboratory analysis
A standardized protocol was used for sample collection and processing. Blood samples were collected in a BD Vacutainer P100 v1.1 Tube (8.5 ml; BD Biosciences, Franklin Lakes, NJ, USA) and centrifuged at 3,000 × g and 4°C for 15 min. Samples were subsequently aliquoted and stored at −80°C for further analysis. An automatic electrochemistry luminescence immunoassay system (ECLIA, Elecsys 2010 system; Roche Diagnostics GmbH, Basel, Switzerland) was used to determine the serum carbohydrate antigen 15-3 (CA15-3 level), according to the manufacturer's instructions. The cut-off value of CA15-3 was 25 U/ml, with values >25 U/ml considered to be positive.
Serum protein profiling
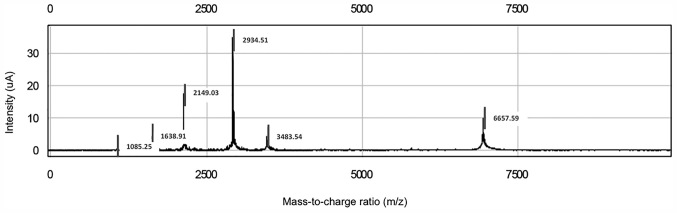
Serum protein profiles were determined using immobilized metal affinity capture arrays (IMAC30; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Samples were incubated with 50 µl of 100 mM CuSO4 solution for 5 min, rinsed with distilled water and washed three times for 10 min with binding solution (200 µl; comprising 500 mM NaCl and 100 mM NaH2PO4/NaOH; pH 7.0). Samples were subsequently diluted with dilution solution (9 M urea, 50 mM Tris/HCl, pH 9.0, 2% w/v CHAPS (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at a ratio of 5:1 and vortexed at medium mixing mode for 30 min at 4°C. A total of 100 µl binding buffer was added into 10 µl of this mixture. From this, 100 µl was applied to each spot on IMAC30 chips. The chips were incubated at room temperature for 60 min on the horizontal shaker (900 rpm) and subsequently washed 4 times with 200 µl binding solution for 10 min on the horizontal shaker. Chips were washed with 1 mM HEPES, pH 7.0 and stored at room temperature until dry. Matrix solution (1 µl) including 50% saturated solution of sinapinic acid (Fluka Chemie AG, Buchs, Switzerland) in 50% acetonitrile and 0.5% trifluoroacetic acid (both from Sigma-Aldrich; Merck KGaA), was added to each well and stored at room temperature until dry. Chips were analyzed by using the Protein Biological System IIc SELDI-TOF (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The generation of TOF spectra was achieved by 192 laser shots with laser intensity of 220 eV and a detector sensitivity of 9. Peaks between 3,000 and 50,000 Da were recorded, with a maximum peak of 200,000 Da. The external calibration of the instrument was achieved using the all-in-one peptide molecular mass standard (Bio-Rad Laboratories, Inc.) as presented in Fig. 1. The mass accuracy was 0.08% with this system. All mass spectra were calibrated internally. Peak intensities were normalized to total ion flow.
Figure 1.
All-in-one peptide molecular mass standard for six peptides, including 1084.247-[arginine 8]-vasopressin, 1637.903-somatostatin, 2147.500-dynorphin A, 2933.500-adrenocorticotropic hormone [1–24] human, 3495.941-insulin B-chain (bovine) and 7033.614-hirudin.
Bioinformatics analysis
The Epi Info-7 program was used to calculate sample size Spectral data were collected using Protein Chip Data Manager 3.0 software (Bio-Rad Laboratories, Inc.), which was also used for data processing and univariate statistical analysis. Internal mass calibration and normalization of peak intensities were performed based on total ion flow. Automatic baseline correction was used. For peak selection, the very low mass region (0–1,500 Da) overlapping with single-photon absorptiometry peaks was excluded. The default average filter with a setting of ×0.2 expected peak width was used for data filtering. Eligible mass peaks were with signal/noise >5. Mass window for peak clustering was 0.3% of the peak mass. Inter-group comparison of each peak cluster was performed using the one-way Mann-Whitney U test. Areas under the receiver operating characteristic (ROC) curve (AUC) were calculated for each peak cluster. A total of 118 clusters were obtained; the cluster mass was 0.5% and the signal-to-noise ratio for the second pass was 1 for the settings used for cluster completion.
Statistical analysis
All continuous variables are presented as the mean ± standard deviation. Comparisons between groups were achieved by two-sample t-tests and Pearson's χ2 tests. Relative peak intensity levels were compared using the Student's t-test. Univariate analysis using the Mann-Whitney U test was used to compare sera from patients with BC with the NCI group. ROC analysis was achieved by the calculation of AUC. SPSS 21.0 (IBM Corp., Armonk, NY, USA) was used for all statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Reproducibility
Each serum sample was spotted on eight locations on one chip, to attain a reliable analysis technique. The coefficient variations of m/z values were 0.03–0.07 and the protein intensity was 0.34–0.79 for randomly selected peaks. The reproducibility of SELDI ProteinChip assays was assessed by running 16 pooled normal human serum samples. The mean variance coefficient based on pooled sera was lower <12%. Variation in day-to-day sampling and instrumentation or chip variations were negligible.
Serum protein profiling of the BC group compared with the HC group
For the study group, 118 peaks per spectrum were obtained, with masses ranging from 2 to 20 kDa. Of these, no single peak could be used to distinguish BC from HC serum. From the 118 peaks, 9 protein peaks at m/z 1,452, 2,670, 3,972, 5,354, 5,523, 6,850, 7,926, 8,143 and 8,115 (BC2) were automatically selected as splitters. A total of 5 peaks were decreased (m/z 1,452, 2,670, 3,972, 5,354 and 5,523; P<0.001; Table II) and 4 were significantly increased (m/z 6,850, 7,926, 8,115 and 8,143; P<0.001; Table II) in patients with BC compared with the HC group.
Table II.
Surface-enhanced laser desorption ionization time-of-flight mass spectrometry results for BC and HC groups.
| Peak intensity | |||||
|---|---|---|---|---|---|
| Protein peaks mass-to-change ratio | Trend | BC (n=62) | HC (n=16) | Area under curve | P-value |
| 1452 | Decreased | 154.74±88.02 | 262.18±124.99 | 0.775 | <0.001 |
| 2670 | Decreased | 19.88±17.01 | 42.80±20.85 | 0.796 | <0.001 |
| 3972 | Decreased | 62.29±36.27 | 104.06±45.17 | 0.781 | <0.001 |
| 5354 | Decreased | 20.69±18.48 | 50±28.64 | 0.805 | <0.001 |
| 5523 | Decreased | 8.94±0.68 | 17±0.76 | 0.806 | <0.001 |
| 6850 | Increased | 44.55±2.99 | 21.95±1.6 | 0.781 | <0.001 |
| 7926 | Increased | 70.86±36.19 | 37.56±17.54 | 0.796 | <0.001 |
| 8115 (BC2) | Increased | 97.65±10.15 | 22.76± 13.85 | 0.751 | <0.001 |
| 8143 | Increased | 138.37±80.33 | 62.19±35.95 | 0.781 | <0.001 |
BC, breast cancer; HC, healthy control.
Serum protein profiling of the BC group compared with the BBD group
The SELDI-TOF-MS results for the BC and BBD groups revealed that 3 protein peaks (m/z 3,972, 5,336, and 11,185) were significantly decreased and 6 (m/z 4,062, 4,071, 4,609, 6,850, 8,115, and 8,133) were significantly increased in the BC group compared with the BBD group (P<0.01; Table III). Of these, the protein peak at 8,115 Da (BC2) was one of the previously defined biomarkers of BC (8,9).
Table III.
Surface-enhanced laser desorption ionization time-of-flight mass spectrometry results for BC and BBD groups.
| Peak intensity | |||||
|---|---|---|---|---|---|
| Protein peaks mass-to-change ratio | Trend | BC (n=62) | BBD (n=31) | Area under curve | P-value |
| 3972 | Decreased | 62.29±36.27 | 123.57±73.94 | 0.783 | <0.001 |
| 4062 | Increased | 127.77±115.72 | 36.76±22.98 | 0.721 | <0.001 |
| 4071 | Increased | 118.95±79.82 | 59.03±23.58 | 0.740 | <0.001 |
| 4609 | Increased | 92.38±40.01 | 67.23±16.39 | 0.702 | 0.001 |
| 5336 | Decreased | 45.74±39.68 | 93.61±57.77 | 0.783 | <0.001 |
| 6850 | Increased | 44.55±2.99 | 25.75±11.68 | 0.721 | <0.001 |
| 8115 (BC2) | Increased | 97.65±10.15 | 31.53±23.77 | 0.698 | 0.002 |
| 8133 | Increased | 131.77±114.87 | 48.46±23.45 | 0.698 | 0.001 |
| 11185 | Decreased | 6.02±8.75 | 16.04±15.94 | 0.769 | <0.001 |
BC, breast cancer; BBD, benign breast disease.
Serum protein profiling of the BC group compared with the NCI group
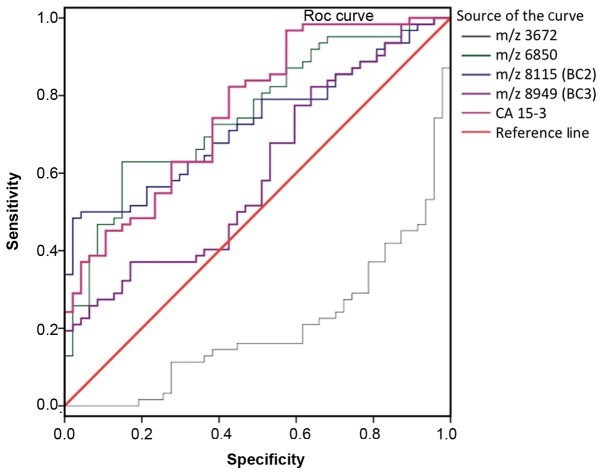
SELDI-TOF-MS results revealed peaks at m/z 3,972, 6,850, 8,115 (BC2) and 8,949 Da (BC3) for patients with BC compared with NCI (Table IV). ROC analysis of each candidate tumor marker as a biomarker to distinguish between the BC and NCI groups is presented in Fig. 2. The AUC values for peaks are as follows: 0.799 for m/z 3,972, 0.752 for m/z 6,850, 0.732 for m/z 8,115 (BC2), 0.603 for m/z 8,949 (BC3), and 0.762 for CA15-3. The ROC data revealed that the protein peaks at 3,972 and 6,850 in patients with BC were novel biomarkers suitable for diagnosis. The sensitivity rates of 3,972, 6,850, 8,115 (BC2) and 8,949 Da (BC3) peaks were 63.0, 83.87, 66.13 and 77.0%, respectively (P<0.05). Sensitivity and specificity values of CA15-3 were determined as 25.81 and 97.87%, respectively, whereas the sensitivity and specificity of the serum protein profiling test (4 peak pattern tests and CA15-3) were 98.39 and 100%, respectively (data not shown).
Table IV.
Biomarkers used to determine patients with BC compared with NCI.
| Protein peak | |||||
|---|---|---|---|---|---|
| Peak intensity mass-to-change ratio | Trend | BC (n=62) | NCI (n=47) | Area under curve | P-value |
| 3972 | Decreased | 62.29±36.27 | 116.93±65.71 | 0.799 | <0.001 |
| 6850 | Increased | 44.55±2.99 | 24.45±13.52 | 0.752 | <0.001 |
| 8115 (BC2) | Increased | 97.65±10.15 | 28.54±21.19 | 0.732 | <0.001 |
| 8949 (BC3) | Increased | 185.28±95.47 | 148.26±62.16 | 0.603 | 0.016 |
| Carbohydrate antigen 15-3 | Increased | 24.59 ±15.79 | 15.72±4.72 | 0.762 | <0.001 |
BC, breast cancer; NCIs, non-cancerous individuals.
Figure 2.
ROC curve for each protein peak identified using SELDI-TOF-MS in patients with breast cancer compared with non-cancerous individuals. Area under curve values were as follows: m/z 3,972, 0.700; m/z 6,850, 0.752; m/z 8,115, 0.732; m/z 8,949, 0.603; CA15-3, 0.762. ROC, receiver operating characteristic; SELDI-TOF-MS, surface-enhanced laser desorption ionization time-of-flight mass spectrometry; CA15-3, carbohydrate antigen 15-3.
Bioinformatics analysis
A logistic regression curve was calculated using the equation [y = 2.438 + 0.020(m/z 397) + 0.033(m/z 6850) + 0.011(m/z 8115) + 0.154(CA15-3)]. As presented in Table V, the minimum increase was 0.98-fold [odds ratio (OR) 0.982] at 3,972 Da, and the maximum increase was 1.17-fold (OR 1.167) at CA15-3. Considering the results of multivariable logistic regression, a set of 4 protein peaks was selected to diagnose BC. The 4 peaks were 3,972, 6,850, 8,115 (BC2) and 8,949 (BC3), as well as CA15-3. Of these, 8,115 (BC2), 8,949 (BC3) and CA15-3 have been previously reported as biomarkers (8,9). The peaks at m/z 3,972 and 6,850 may therefore be novel candidate markers for distinguishing patients with BC from NCI.
Table V.
Results of multivariable logistic regression of risk factors for breast cancer.
| Covariate mass-to-change ratio | Odds ratio | 95% Confidence limit | P-value | Diagnostic cut-off |
|---|---|---|---|---|
| 3972 | 0.982 | 0.968–0.995 | 0.009 | 63.54 |
| 6850 | 1.051 | 1.008–1.096 | 0.019 | 59.27 |
| 8115 (BC2) | 1.021 | 1.000–1.042 | 0.047 | 25.33 |
| 8949 (BC3) | 0.991 | 0.981–1.002 | 0.115 | 140.94 |
| Carbohydrate antigen 15-3 | 1.167 | 1.036–1.314 | 0.011 | 19.21 |
Discussion
Proteomics are widely used in BC studies to identify diagnostic markers; however, the numbers of validated clinical markers for BC is limited (13). Several genes have previously been reported to be upregulated in BC compared with control tissues (14,15). In addition, proteomic methods are easy to apply in the clinic to diagnose patients (16,17). Serum SELDI-TOF-MS protein profiling is a powerful method used for biomarker discovery and can be used to distinguish patients with BC from NCI with high sensitivity and specificity (18). It has previously been reported that monitoring protein-based markers in blood and urine samples may be used for cancer staging (19). In the past decade, a number of studies have utilized SELDI-TOF to identify proteomic patterns in biological fluids, including serum, urine and nipple aspirates (20–22).
In the present study, serum samples of 62 patients with BC and 47 NCI were analyzed using SELDI-TOF-MS. Among the 118 examined peaks, multivariate analysis revealed 9 peaks in SELDI-TOF-MS results for patients with BC compared with those with BBD, as well as 9 peaks for patients with BC compared with HC subjects. By using the common peaks in both sets, 4 protein peaks were predicted as biomarkers to distinguish patients with BC from NCI. Several mutations are known to increase susceptibility to BC; the majority of inherited cases of BC are associated with two abnormal genes, BC1 and BC2 (23). It has been identified that the protein expression of BC1/2 is associated with increased susceptibility to BC by 40–80% (24). Although several biomarker panels have been reported for various diseases using SELDI and ProteinChip arrays (25), there are few validated markers. The first validation study by Li et al (8) proposed the use of BC2 and BC3 as biomarkers for BC. Decreases in BC1 and increased expression of BC2 and BC3 in patients with BC have been reported in several studies (22,26,27). BC2 and BC3 have also been reported to possess independent diagnostic merit to distinguish between malignant disease, benign disease and control groups (9). BC3 was therefore included in the protein panel for analysis.
CA15-3 is the most widely used serum marker in patients with BC (22). Serum CA15-3 has been extensively studied, mainly to monitor the response of BC to treatment or to detect early relapse in BC follow-up (28). A study by Gautam et al (29) reported that elevated concentrations of CA15-3 may be a useful and reliable diagnostic and prognostic indicator, as well as an indicator of treatment efficacy (29). The number of metastatic sites has been demonstrated to affect levels of CA15-3 (30). As CA15-3 is the most widely used serum tumor marker, it was included in the diagnostic panel in the present study.
Two peaks, at m/z values 3,972 and 6,850 Da, were identified for the first time as potential biomarkers of BC using SELDI-TOF-MS. These peaks were demonstrated to be able to distinguish patients with BC from HC subjects and patients with BBD. The peak at 3,972 Da was significantly decreased and the peak at 6,850 Da was significantly increased. Previous studies have reported 4.3, 8.1 and 8.9 kDa proteins as biomarkers of BC; however, these results differ to those of the present study (8,11,22,26). A 3.8 kDa protein, close to m/z 3,972, was reported to be highly sensitive for the diagnosis of BC by Chung et al (12). A number of serum-based candidate biomarkers have been identified in different studies (8,10,22,31). Studies may have some differences in terms of inclusion/exclusion criteria, biologic samples, preparation protocols, arrays and analytical settings, which will affect the reproducibility and robustness of results. SELDI-TOF-MS proteomic profiling has been proposed as a promising high-throughput technology with potential applications in BC screening, detection and prognostication. In some studies, detected ion peaks were identified as different due to variations in population demographics, stage of disease, collection and storage conditions of samples, and analytical procedures (11,32,33). Aside from the most common proteins (BC1, BC2, BC3 and CA15-3), many of the studied peaks had poor sensitivities and specificities and alone were insufficient to diagnose BC. Furthermore, discrepancies in the peaks have been reported for several studies. For example, peaks at m/z 4,276 and 4,292 were reported by Winden et al (11) to be decreased in patients with BC, which was consistent with other studies; however, the peak at m/z 8,941 was reported to be decreased, whereas it was increased in previous studies (8,22,26). A review by Muthu et al (34), which summarized the features of SELDI-TOF-MS and its application in cancer biomarker discovery, reported that the technique has not yet been sufficiently investigated in different types of cancer and other diseases (34).
In present study, the resultant 4 peaks panel together with CA15-3 was demonstrated to have good sensitivity and specificity for the diagnosis of BC. These results indicate that protein peaks at 3,972 and 6,850 Da may be the novel candidate biomarkers for the diagnosis of BC. In conclusion, 4 peak clusters which are significantly different in patients with BC compared with NCI were identified using SELDI-TOF-MS, The identified peaks: 3,972, 6,850, 8,115 (BC2) and 8,949 Da (BC3) combined with CA15-3 expression may be used as a protein-profiling test to diagnose BC. However, further investigation using a larger sample size should be performed to verify these results.
References
- 1.Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, Shi W, Jiang J, Yao PP, Zhu HP. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13:1387–1397. doi: 10.7150/ijbs.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahman M, Mohammed S. Breast cancer metastasis and the lymphatic system. Oncol Lett. 2015;10:1233–1239. doi: 10.3892/ol.2015.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niell BL, Freer PE, Weinfurtner RJ, Arleo EK, Drukteinis JS. Screening for breast cancer. Radiol Clin North Am. 2017;55:1145–1162. doi: 10.1016/j.rcl.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Nothacker M, Duda V, Hahn M, Warm M, Degenhardt F, Madjar H, Weinbrenner S, Albert US. Early detection of breast cancer: Benefits and risks of supplemental breast ultrasound in asymptomatic women with mammographically dense breast tissue. A systematic review. BMC Cancer. 2009;9:335. doi: 10.1186/1471-2407-9-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drukteinis JS, Mooney BP, Flowers CI, Gatenby RA. Beyond mammography: New frontiers in breast cancer screening. Am J Med. 2013;126:472–479. doi: 10.1016/j.amjmed.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baskın Y, Yiğitbaşı T. Clinical proteomics of breast cancer. Curr Genomics. 2010;11:528–536. doi: 10.2174/138920210793175930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C. The application of SELDI-TOF-MS in clinical diagnosis of cancers. J Biomed Biotechnol. 2011;2011:245821. doi: 10.1155/2011/245821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Orlandi R, White CN, Rosenzweig J, Zhao J, Seregni E, Morelli D, Yu Y, Meng XY, Zhang Z, et al. Independent validation of candidate breast cancer serum biomarkers identified by mass spectrometry. Clin Chem. 2005;51:2229–2235. doi: 10.1373/clinchem.2005.052878. [DOI] [PubMed] [Google Scholar]
- 9.Atahan K, Küpeli H, Gür S, Yiğitbaşı T, Baskın Y, Yiğit S, Deniz M, Cökmez A, Tarcan E. The value of serum biomarkers (Bc1, Bc2, Bc3) in the diagnosis of early breast cancer. Int J Med Sci. 2011;8:148–155. doi: 10.7150/ijms.8.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belluco C, Petricoin EF, Mammano E, Facchiano F, Ross-Rucker S, Nitti D, Di Maggio C, Liu C, Lise M, Liotta LA, et al. Serum proteomic analysis identifies a highly sensitive and specific discriminatory pattern in stage 1 breast cancer. Ann Surg Oncol. 2007;14:2470–2476. doi: 10.1245/s10434-007-9354-3. [DOI] [PubMed] [Google Scholar]
- 11.van Winden AW, Gast MC, Beijnen JH, Rutgers EJT, Grobbee DE, Peeters PH, van Gils CH. Validation of previously identified serum biomarkers for breast cancer with SELDI-TOF MS: A case control study. BMC Med Genomics. 2009;2:4. doi: 10.1186/1755-8794-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung L, Moore K, Phillips L, Boyle FM, Marsh DJ, Baxter RC. Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer. Breast Cancer Res. 2014;16:R63. doi: 10.1186/bcr3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singletary SE, Connolly JL. Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J Clin. 2006;56:37–47. doi: 10.3322/canjclin.56.1.37. [DOI] [PubMed] [Google Scholar]
- 14.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC, Jr, American Society of Clinical Oncology American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 15.Paweletz CP, Gillespie JW, Ornstein DK, Simone NL, Brown MR, Cole KA, Wang QH, Huang J, Hu N, Yip TT, et al. Rapid protein display profiling of cancer progression directly from human tissue using a protein biochip. Drug Dev Res. 2000;49:34–42. doi: 10.1002/(SICI)1098-2299(200001)49:1<34::AID-DDR6>3.0.CO;2-W. [DOI] [Google Scholar]
- 16.Nakanishi T, Shimizu A, Okamoto N, Ingendoh A, Kanai M. Analysis of serum protein precipitated with antiserum by matrix-assisted laser desorption ionization/time-of-flight and electrospray ionization mass spectrometry as a clinical laboratory test. J Am Soc Mass Spectrom. 1995;6:854–859. doi: 10.1016/1044-0305(95)00326-9. [DOI] [PubMed] [Google Scholar]
- 17.Lacey JM, Bergen HR, Magera MJ, Naylor S, O'Brien JF. Rapid determination of transferrin isoforms by immunoaffinity liquid chromatography and electrospray mass spectrometry. Clin Chem. 2001;47:513–518. [PubMed] [Google Scholar]
- 18.Vlahou A, Laronga C, Wilson L, Gregory B, Fournier K, McGaughey D, Perry RR, Wright GL, Jr, Semmes OJ. A novel approach toward development of a rapid blood test for breast cancer. Clin Breast Cancer. 2003;4:203–209. doi: 10.1016/S1526-8209(11)70627-9. [DOI] [PubMed] [Google Scholar]
- 19.Yao F, Zhang C, Du W, Liu C, Xu Y. Identification of gene-expression signatures and protein markers for breast cancer grading and staging. PLoS One. 2015;10:e0138213. doi: 10.1371/journal.pone.0138213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diamandis EP. Point: Proteomic patterns in biological fluids: Do they represent the future of cancer diagnostics? Clin Chem. 2003;49:1272–1275. doi: 10.1373/49.8.1272. [DOI] [PubMed] [Google Scholar]
- 21.Higgins SA, Matloff ET, Rimm DL, Dziura J, Haffty BG, King BL. Patterns of reduced nipple aspirate fluid production and ductal lavage cellularity in women at high risk for breast cancer. Breast Cancer Res. 2005;7:R1017–R1022. doi: 10.1186/bcr1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 23.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, et al. The Breast Cancer Linkage Consortium Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vos S, Moelans CB, van Diest PJ. BRCA promoter methylation in sporadic versus BRCA germline mutation-related breast cancers. Breast Cancer Res. 2017;19:64. doi: 10.1186/s13058-017-0856-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neagu M, Constantin C, Tanase C, Boda D. Patented biomarker panels in early detection of cancer. Recent Pat Biomark. 2011;1:10–24. doi: 10.2174/2210310411101010010. [DOI] [Google Scholar]
- 26.Mathelin C, Cromer A, Wendling C, Tomasetto C, Rio MC. Serum biomarkers for detection of breast cancers: A prospective study. Breast Cancer Res Treat. 2006;96:83–90. doi: 10.1007/s10549-005-9046-2. [DOI] [PubMed] [Google Scholar]
- 27.Atalay C, Kubilay D. The results of screening with SELDI-TOF-MS method in breast cancer diagnosis. J Breast Health. 2011;7:106–108. [Google Scholar]
- 28.Stearns V, Yamauchi H, Hayes DF. Circulating tumor markers in breast cancer: Accepted utilities and novel prospects. Breast Cancer Res Treat. 1998;52:239–259. doi: 10.1023/A:1006137619153. [DOI] [PubMed] [Google Scholar]
- 29.Gautam A, Verma S, Pantola C, Verma S. Utility of CA 15-3 as diagnostic and prognostic marker in breast cancer. IOSR-JDMS. 2015;14:17–20. [Google Scholar]
- 30.Geng B, Liang MM, Ye XB, Zhao WY. Association of CA 15-3 and CEA with clinicopathological parameters in patients with metastatic breast cancer. Mol Clin Oncol. 2015;3:232–236. doi: 10.3892/mco.2014.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeidan BA, Cutress RI, Hastie C, Mirnezami AH, Packham G, Townsend PA. SELDI-TOF-MS proteomics in breast cancer. Clin Proteomics. 2009;5:133–147. doi: 10.1007/s12014-009-9032-5. [DOI] [Google Scholar]
- 32.Yin P, Lehmann R, Xu G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal Bioanal Chem. 2015;407:4879–4892. doi: 10.1007/s00216-015-8565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandramouli K, Qian PY. Proteomics: Challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics. 2009;2009:239204. doi: 10.4061/2009/239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muthu M, Vimala A, Mendoza OH, Gopal J. Tracing the voyage of SELDI-TOF-MS in cancer biomarker discovery and its current depreciation trend-need for resurrection? Trends Analyt Chem. 2016;76:95–101. doi: 10.1016/j.trac.2015.10.004. [DOI] [Google Scholar]