Abstract
The aim of the present study was to measure the expression of microRNA (miRNA)-506-3p in the peripheral blood of patients with hypertension and to determine the biological functions and mechanisms of action of miR-506-3p. A total of 61 patients with primary hypertension were included in the present study. Peripheral blood was collected from all patients, as well as 31 healthy subjects who were included in a control group. The expression of miR-506-3p in peripheral blood was determined by reverse transcription-quantitative polymerase chain reaction. Human umbilical vein endothelial cells (HUVECs) were transfected with miR-506-3p mimics or miR-506-3p inhibitor. The proliferation and migration of HUVECs were determined using cell-counting kit 8 and Transwell assays, respectively. The cell cycle and apoptosis of HUVECs were detected by flow cytometry. The expression of Beclin1 (BECN1) protein, a potential target of miR-506-3p, was measured using western blotting. A dual-luciferase reporter assay was performed to determine the interaction between BECN1 and miR-506-3p. It was demonstrated that miR-506-3p expression in the peripheral blood of patients with patients was upregulated and dependent on the severity of hypertension. miR-506-3p overexpression inhibited the proliferation and migration of HUVECs. In addition, miR-506-3p inhibited the transition from the G1 phase to the S-phase in HUVECs. Overexpression of miR-506-3p promoted the apoptosis of HUVECs. Notably, miR-506-3p downregulated the expression of BECN1 by directly binding to its 3′-untranslated region. The present study demonstrated that miR-506-3p expression is elevated in the peripheral blood of patients with hypertension and is associated with the severity of hypertension. By downregulating BECN1 expression, miR-506-3p aggravates injury in vascular endothelial cells by inhibiting the proliferation and migration of HUVECs, as well as promoting their apoptosis.
Keywords: microRNA-506-3p, hypertension, Beclin1
Introduction
Hypertension is a common chronic cardiovascular disease clinically characterized by an elevated blood pressure and is an independent risk factor for cardiac and cerebral vascular diseases (1–3). The incidence of hypertension increases each year, particularly among young people (4); however, its molecular mechanism of action remains unclear. As an important component of the blood circulation and immune system, vascular endothelial cells are widely involved in biological processes, including blood pressure regulation, blood vessel formation, anticoagulation, fibrinolysis and inflammation (5–7). Previous studies have demonstrated that vascular endothelial cell dysfunction serves an important role in the onset and development of hypertension (8,9). Endothelial cells not only act as a physical barrier but also secrete a variety of substances that affect the function of relaxation and contraction of blood vessels. During sustained hypertension, endothelial cells are damaged and the expression of inflammatory factors is increased (10,11). Therefore, investigating the molecular mechanism of vascular endothelial cells in the onset and development of hypertension may help to improve the diagnosis and treatment of hypertension.
MicroRNA (miRNA) are a class of non-coding small RNA molecules consisting of 18–23 nucleotides. miRNA regulates the expression of target mRNA at the post-transcriptional level by binding to the 3′-untranslated region (UTR) of target gene mRNA (12,13). miRNA participates in a wide variety of biological processes, including cell proliferation, differentiation, apoptosis, energy metabolism, hormone secretion and development (14,15). miRNA serves an important biological role in cardiovascular diseases and affects the progression of hypertension by regulating a variety of tissues and organs, including myocardial tissue, vascular wall tissue, fundus tissue and kidney (16). Furthermore, miRNA serves important regulatory roles in cardiac hypertrophy and heart failure (17,18) and its expression is closely associated with arrhythmia and atherosclerotic plaque formation. As miRNA expression is stable in peripheral blood, it may act as a potential diagnostic marker for various diseases, including myocardial infarction and cancer (19). The upregulation of miR-1 levels in the peripheral blood is closely associated with myocardial injury (20). However, the mechanism by which miRNA induces endothelial injury in hypertension remains unclear.
It has been demonstrated that the recently identified miRNA molecule miR-506 is closely associated with proliferation, drug resistance, invasion and metastasis in tumors. The upregulation of miR-506 increases drug resistance in colon cancer cells (21) and in neuroglioma, miR-506 inhibits the proliferation and metastasis of tumor cells by targeting signal transducer and activator of transcription 3 (22). In addition, miR-506 participates in angiogenesis, suggesting that it may serve a role in cardiovascular disease (23). Beclin1 (BECN1) is a key gene in the regulation of autophagy activity and participates in the onset and development of different tumors by directly regulating autophagy activity (24). In most tumors, BECN1 expression is downregulated and BECN1 overexpression inhibits tumor growth (25,26). It remains unclear whether miR-506-3p is associated with BECN1 expression. Therefore, the aim of the present study was to investigate the effect of miR-506-3p in the onset and development of hypertension at the clinical and cellular levels.
Materials and methods
Patients
A total of 61 patients with primary hypertension admitted to Zaozhuang Municipal Hospital (Zaozhuang, China) between December 2014 and January 2016 were included in the present study. Peripheral blood was collected from all patients, as well as 31 healthy subjects who were included in a control group. The 61 patients included 34 males and 27 females, with a mean age of 62.5 years (median, 61 years; range, 46–81 years). Diagnosis of hypertension was determined when systolic blood pressure ≥160 mmHg and/or diastolic blood pressure ≥100 mmHg without the use of antihypertensive drugs, in accordance with the 2010 Chinese Guidelines for the Management of Hypertension (27). The 61 patients were divided into grades I, II and III according to the severity of hypertension (27). Patients with grade I had mild hypertension (systolic blood pressure of 140–159 mmHg and/or diastolic blood pressure of 90–99 mmHg), patients with grade II had moderate hypertension (systolic blood pressure of 160–179 mmHg and/or diastolic blood pressure of 100–109 mmHg) and patients with grade III had severe hypertension (systolic blood pressure of ≥180 mmHg and/or diastolic blood pressure of ≥110 mmHg). There were 23, 27 and 11 patients with grades I, II and III, respectively. The duration of hypertension for each patient was >5 years. All procedures were approved by the Ethics Committee of Zaozhuang Municipal Hospital (Zaozhuang, China) and written informed consent was obtained from all patients or their families.
Cells
Human umbilical vein endothelial cells (HUVECs; Cell Bank; Chinese Academy of Sciences, Shanghai, China) and HEK293T cells (Cell Bank; Chinese Academy of Sciences) were cultured in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C and 5% CO2. Upon reaching 70–90% confluence, cells were seeded into 24-well plates at 1×105 cells/well. HUVECs were divided into three groups: A negative control (NC) group, a miR-506-3p mimics group (transfection with miR-506-3p mimics) and a miR-506-3p inhibitor group (co-transfected with miR-506-3p mimics and inhibitor).
Transfection of cells
Prior to transfection, HUVECs were cultured in serum-free DMEM until they reached 90% confluence. In the first vial, 2.5 µl miR-506-3p mimics (25 pmol/µl; miR-506-3p mimics group; HanBio, Shanghai, China; sequences not supplied), or 2.5 µl miR-506-3p mimics (25 pmol/µl; HanBio; sequences not supplied) and 2.5 µl miR-506-3p inhibitor (25 pmol/µl; miR-506-3p inhibitor group; HanBio; sequences not supplied) were mixed with 50 µl Opti Mem I medium (Thermo Fisher Scientific, Inc.). In the second vial, 1 µl Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) was mixed with 50 µl Opti Mem I medium. Vials were left to stand for 5 min, the two vials were then combined and left to stand at room temperature for a further 20 min. Subsequently, mixtures were added to cells in each respective group. After 6 h, the medium was replaced with fresh DMEM containing 10% fetal bovine serum. Following cultivation for 48 h, cells were collected for further assays.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Cells (1×105) were trypsinized and mixed with 0.5 ml TRIzol (Thermo Fisher Scientific, Inc.) for lysis. Total RNA was then extracted using the phenol chloroform method (28). The purity of RNA was determined by A260/A280 using ultraviolet spectrophotometry (Nanodrop ND2000, Thermo Fisher Scientific, Inc.). Subsequently, cDNA was obtained by reverse transcription using the Reverse Transcription System (Takara Biotechnology, Co., Ltd., Dalian, China) using 0.5 µg RNA and stored at −20°C. The expression of miR-506-3p was determined using a SYBR PrimeScript miRNA RT-PCR kit (Takara Biotechnology, Co., Ltd.) and U6 (forward, 5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′) was used as an internal reference. The reaction system (20 µl) contained 10 µl qRT-PCR-Mix, 0.5 µl upstream primer (5′-GGCACCCTTCTGAGTAGA−3′), 0.5 µl downstream universal primer (provided by the kit), 2 µl cDNA and 7 µl ddH2O. The reaction protocol was: Initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation 95°C for 1 min and annealing at 60°C for 30 sec. The reaction was performed on ABI StepOne Plus Real-Time PCR system (Thermo Fisher Scientific, Inc.). The 2−ΔΔCq method (29) was used to calculate the relative expression of miR-506-3p against U6 and each sample was measured in triplicate.
Cell counting kit-8 (CCK-8) assay
At 24 h after transfection, HUVECs were trypsinized and seeded into 96-well plates at a density of 1×103/well. At 24, 48 and 72 h, the medium was discarded and the cells were washed with sterile phosphate-buffered saline twice. Subsequently, DMEM and 10% CCK-8 reaction reagent (Beyotime Institute of Biotechnology, Haimen, China) were added. Following incubation at 37°C for 1 h, the absorbance of each well was measured at 490 nm and cell proliferation curves were plotted.
Flow cytometry
At 48 h after transfection, HUVECs were trypsinized, collected and washed twice with precooled phosphate-buffered saline. Cellular apoptosis was measured by flow cytometry using a FITC Annexin V apoptosis detection kit I (BD Biosciences, Franklin Lakes, NJ, USA), following the manufacturer's instructions. Early apoptotic cells were identified by single-positive staining with Annexin V, necrotic cells were identified by single-positive staining with propidium iodide and late apoptotic cells were identified by double-positive staining with propidium iodide and Annexin V. To determine the cell cycle, a cell cycle assay kit (BD Biosciences) was employed. Cells were incubated with 200 µl liquid A at room temperature for 10 min, followed by 150 µl liquid B at room temperature for 10 min and finally 120 µl liquid C at room temperature in the dark for 10 min. A flow cytometer was used for detection and the results were analyzed using ModFit v3.2 software (http://www.vsh.com).
Transwell assay
Transwell chambers (8 µm; 24-wells; Corning Inc., Corning, NY, USA) were used to evaluate the migration ability of HUVECs. Cells were collected by trypsin digestion and resuspended to a density of 5×105 cells/ml using DMEM containing 0.1% bovine serum albumin (Beyotime Institute of Biotechnology). The cell suspension (200 µl) was added to the upper chamber. In the lower chamber, 500 µl DMEM supplemented with 10% fetal bovine serum was added. Following 24 h cultivation, cells in the upper chamber were wiped using a cotton swab. Cells that moved to the other side of the chamber were fixed with 4% formaldehyde at room temperature for 10 min. Following staining with Giemsa at room temperature for 1 min, the number of cells in five randomly selected in fields were counted under a light microscope (magnification, ×200) and averaged.
Western blot analysis
HUVECs were trypsinized and collected prior to mixing with 100 µl precooled radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) containing 1% phenylmethylsulfonyl fluoride for 15 min lysis at 4°C. Subsequently, cells were centrifuged at 12,000 × g and 4°C for 10 min. The supernatant was used to determine protein concentration using a bicinchoninic acid protein concentration determination kit [RTP7102, Real-Times (Beijing) Biotechnology Co., Ltd., Beijing, China]. Subsequently, samples were mixed with 5X loading buffer prior to denaturation in a boiling water bath for 10 min. Protein samples (20 µg) were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis at 100 V. The resolved proteins were transferred to polyvinylidene difluoride membranes on ice (300 mA, 1 h) and blocked with 5% skimmed milk at room temperature for 1 h. Then, membranes were incubated with rabbit anti-human polyclonal primary antibodies for BECN1 (1:1,000; cat. no. ab62557) and GAPDH (1:1,000; cat. no. ab8245; both Abcam, Cambridge, UK) at 4°C overnight. Following six washes with phosphate-buffered saline and Tween-20 (5 min each), membranes were incubated with polyclonal goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:4,000; cat. no. ab6721; Abcam) for 1 h at room temperature prior to six washes with phosphate-buffered saline and Tween-20 (5 min each). Subsequently, membranes were developed using an enhanced chemiluminescence detection kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Image lab v3.0 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to acquire and analyze imaging signals. Relative protein content was expressed against GAPDH.
Bioinformatics prediction
Bioinformatics prediction is a powerful tool for the study of the functions of miRNA. To determine the regulatory mechanism of BECN1, miRanda (http://www.microrna.org/microrna/home.do), TargetScan (http://www.targetscan.org), PiTa (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) and PICTA (http://pictar.mdc-berlin.de/) were used to predict the miRNA molecules that may regulate BECN1. The results indicated that miR-506-3p may potentially regulate BECN1 (Fig. 1).
Figure 1.
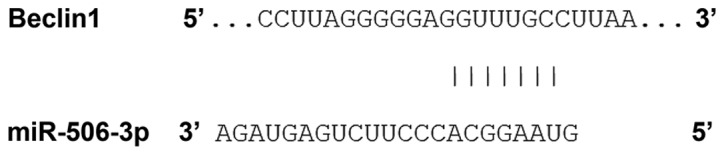
Bioinformatics prediction of direct interactions between miR-506-3p and BECN1. Bioinformatics prediction is a powerful tool for the study of the functions of miRNAs. To understand the regulatory mechanism of BECN1 in hypertension, miRanda (http://www.microma.org/rnicroma/home.do), TargetScan (http://www.targetscan.org), PiTa (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) and PICTA (http://pictar.mdc-berlin.de/) were used to predict the miRNA molecules that may regulate BECN1. It was demonstrated that miR-506-3p was able to regulate BECN1. miRNA/miR, microRNA; BECN1, Beclin1.
Dual-luciferase reporter assay
Using the results from bioinformatics, wild-type (WT) and mutant seed regions of miR-506-3p in the 3′-UTR of BECN1 gene were chemically synthesized in vitro. SpeI and HindIII restriction sites were added and subsequently cloned into pMIR-REPORT luciferase reporter plasmids (Thermo Fisher Scientific, Inc.). Plasmids (1 µg) with WT or mutant 3′-UTR DNA sequences were co-transfected with miR-506-3p mimics (100 nM; Sangon Biotech, Shanghai, China) into HEK293T cells. Following cultivation for 24 h, cells were lysed using a dual-luciferase reporter assay kit (Promega Corporation, Madison, WI, USA) according to the manufacturer's manual and fluorescence intensity was measured with a GloMax 20/20 luminometer (Promega Corporation). Using Renilla luciferase activity as an internal reference, the fluorescence values of each group of cells were measured.
Statistical analysis
Statistical analysis was performed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). Measurement data were expressed as the mean ± standard deviation. Differences between two groups were compared using Student's t-test. For comparison of multiple groups, ANOVA was used, followed by a post hoc comparisons test using Tukey's/Dunnett's method. Each test was performed in triplicate. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-506-3p in the peripheral blood of hypertension patients is upregulated and dependent on hypertension staging
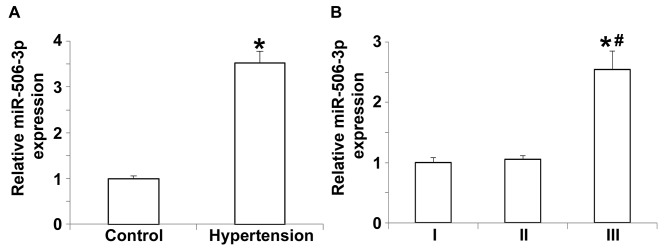
To measure the expression of miR-506-3p in the peripheral blood, RT-qPCR was performed. The data indicated that miR-506-3p expression in patients with hypertension was significantly higher compared with the control group (P<0.05; Fig. 2A). Further analysis indicated that the expression of miR-506-3p in patients with grade III hypertension was significantly higher than in patients with grade I or II hypertension (P<0.05; Fig. 2B), whereas miR-506-3p expression did not differ between patients with grade I and II hypertension (P>0.05; Fig. 2B). These results demonstrate that miR-506-3p expression in the peripheral blood of patients with hypertension is upregulated and that this upregulation is dependent on hypertension staging.
Figure 2.
Relative expression of miR-506-3p in peripheral blood as determined by reverse transcription-quantitative polymerase chain reaction. (A) Relative expression of miR-506-3p in healthy subjects and patients with hypertension. *P<0.05 vs. control. (B) Relative expression of miR-506-3p in patients with hypertension at grades I, II and III. *P<0.05 vs. grade I and #P<0.05 vs. grade II. miR, microRNA.
Overexpression of miR-506-3p inhibits the proliferation of HUVECs
To determine the effect of miR-506-3p on the proliferation of HUVECs, a CCK-8 assay was performed. The data indicated that the absorbance of HUVECs transfected with miR-506-3p inhibitor was significantly higher than that of HUVECs in the NC group 24 and 48 h after transfection (P<0.05; Fig. 3). By contrast, the absorbance of HUVECs transfected with miR-506-3p mimics did not differ from that of HUVECs in the NC group at 24 and 48 h (P>0.05; Fig. 3). These results indicate that the overexpression of miR-506-3p inhibits the proliferation of HUVECs.
Figure 3.
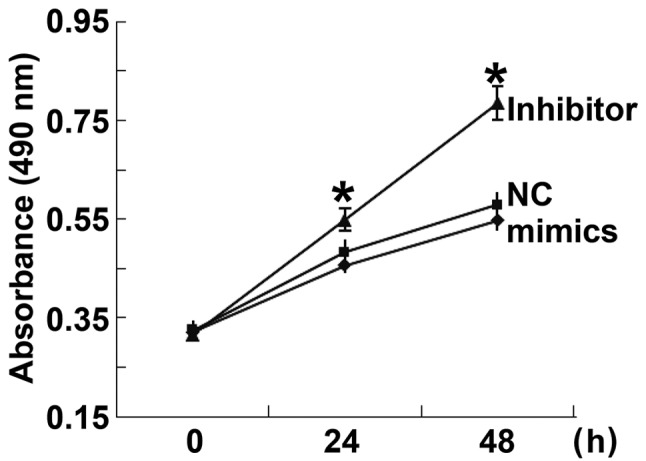
Proliferation of HUVECs at 24 and 48 h after transfection. A cell counting kit-8 assay was used to determine the proliferation of HUVECs transfected with miR-506-3p mimics or miR-506-3p inhibitor. The absorbance of each well was measured at 490 nm with a microplate reader and cell proliferation curves were plotted. *P<0.05 vs. NC group. miR, microRNA; HUVECs, human umbilical vein endothelial cells; NC, negative control.
Overexpression of miR-506-3p inhibits the migration ability of HUVECs
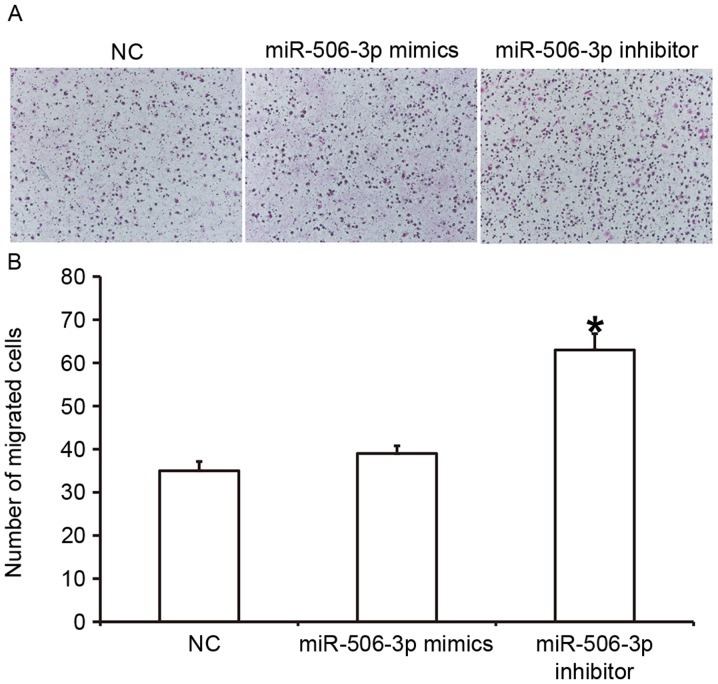
To determine the effect of miR-506-3p on the migration ability of HUVECs, a Transwell assay was conducted (Fig. 4). The data indicated that the number of HUVECs that passed through the chamber membrane in the miR-506-3p inhibitor group was significantly higher than in the NC group (P<0.05; Fig. 4B). However, the number of HUVECs did not differ significantly between the miR-506-3p mimics group and the NC group (P>0.05; Fig. 4B). These results suggest that the overexpression of miR-506-3p inhibits the migration ability of HUVECs.
Figure 4.
Effect of miR-506-3p on the migration ability of HUVECs. (A) Images of migrated HUVECs (magnification, ×200). (B) The number of migrated HUVECs. A Transwell assay was conducted to determine the migration ability of HUVECs transfected with miR-506-3p mimics or miR-506-3p inhibitor. *P<0.05 vs. NC group. HUVECs, human umbilical vein endothelial cells; NC, negative control; miR, microRNA.
miR-506-3p regulates the G1/S phases of HUVECs
To determine how miR-506-3p regulates the cell cycle of HUVECs, flow cytometry was used. The data indicated that the proportion of cells in G1 in the miR-506-3p inhibitor group (49.5±2.1%) was significantly lower than in the NC group (63.7±3.8%; P<0.05), whereas the proportion of cells in the S-phase in miR-506-3p inhibitor group (38.2±1.8%) was significantly higher than in the NC group (21.5±3.9%; P<0.05; Fig. 5). There were no differences in the proportion of cells in each phase of the cell cycle between the miR-506-3p mimics group and the NC group. These results indicate that miR-506-3p may suppresses the proliferation of HUVECs by regulating the G1/S phase transition.
Figure 5.
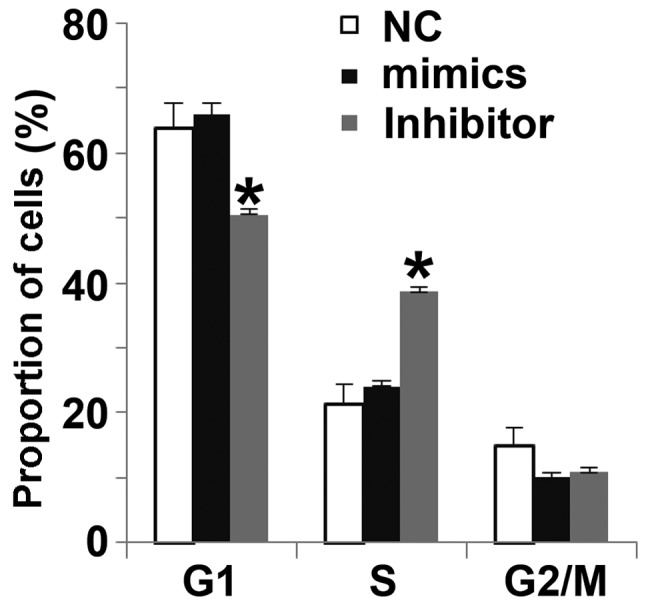
Effect of miR-506-3p on the cell cycle of HUVECs. Flow cytometry was used to detect the proportion of cells in each phase. HUVECs were transfected with miR-506-3p mimics or miR-506-3p inhibitor. *P<0.05 vs. NC group. HUVECs, human umbilical vein endothelial cells; NC, negative control; miR, microRNA.
Overexpression of miR-506-3p promotes the apoptosis of HUVECs
To determine the effect of miR-506-3p on the apoptosis of HUVECs, flow cytometry was used (Fig. 6). The results demonstrated that the apoptotic rate of HUVECs transfected with miR-506-3p inhibitor was significantly lower than that of the NC group (P<0.05; Fig. 6B), whereas the apoptotic rate of HUVECs transfected with miR-506-3p mimics did not differ from that in the NC group (P>0.05; Fig. 6B). These results suggest that the overexpression of miR-506-3p promotes the apoptosis of HUVECs.
Figure 6.
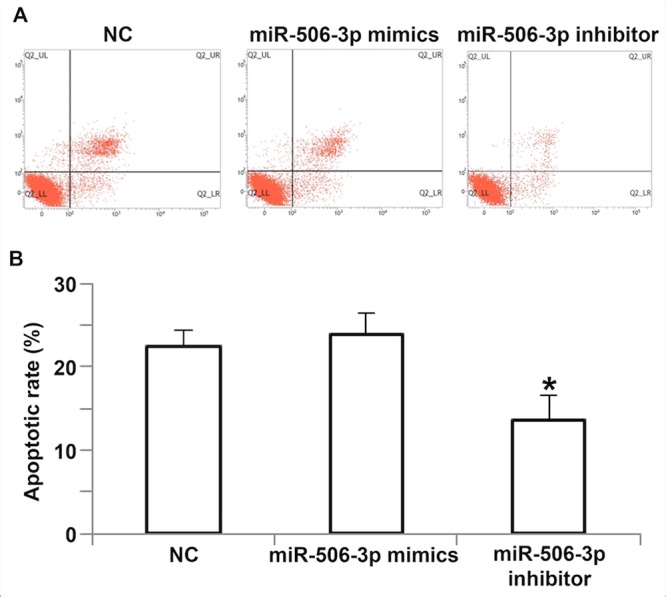
Effect of miR-506-3p on the apoptosis of HUVECs. (A) Flow cytometry plots of HUVECs transfected with miR-506-3p mimics or miR-506-3p inhibitor. (B) The apoptotic rate of HUVECs transfected with miR-506-3p mimics or miR-506-3p inhibitor. *P<0.05 vs. NC group. HUVECs, human umbilical vein endothelial cells; NC, negative control; miR, microRNA.
miR-506-3p exerts its biological role by regulating the expression of BECN1 protein
To determine the expression of BECN1, which is a target protein of miR-506-3p, western blotting was performed. The results demonstrated that levels of BECN1 in HUVECs transfected with miR-506-3p inhibitor were significantly higher than in the NC group (P<0.05), however the expression of BECN1 in HUVECs transfected with miR-506-3p mimics did not differ from that in the NC group (P>0.05; Fig. 7). The result indicates that miR-506-3p may exert its biological role by regulating the expression of BECN1.
Figure 7.
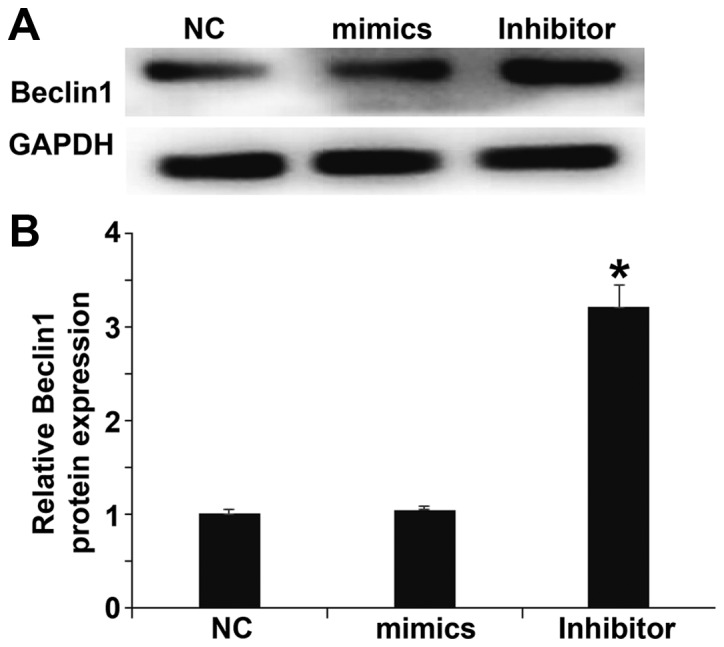
Effect of miR-506-3p on the expression of BECN1. (A) Western blots of BECN1 protein. (B) Relative expression of BECN1 protein in HUVECs transfected with miR-506-3p mimics or miR-506-3p inhibitor. *P<0.05 vs. NC group. HUVECs, human umbilical vein endothelial cells; NC, negative control; miR, microRNA; BECN1, Beclin1.
miR-506-3p regulates the expression of BECN1 by binding to the 3′-UTR of BECN1
To identify the interaction between miR-506-3p and BECN1, a dual-luciferase reporter assay was performed. The data demonstrated that transfection with miR-506-3p mimics and pMIR-REPORT in the WT group led to significantly reduced fluorescence intensity compared with the NC (P<0.05). By contrast, fluorescence intensity in the mutant group did not differ significantly from that in the NC (P>0.05; Fig. 8). These results indicate that miR-506-3p regulates the expression of BECN1 by binding to the 3′-UTR of BECN1.
Figure 8.
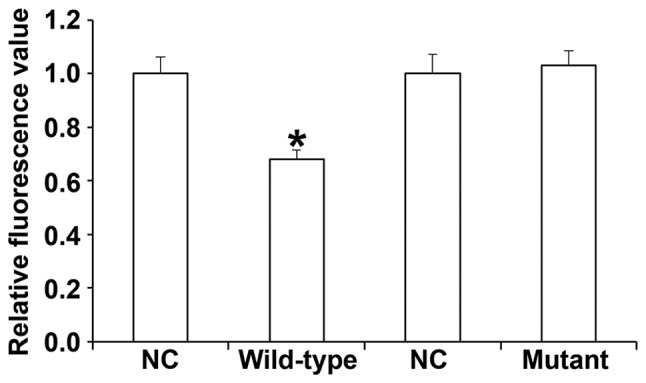
Fluorescence values of HUVECs transfected with WT or mutant 3′-UTR DNA sequences of BECN1 as well as miR-506-3p. A dual-luciferase reporter assay was used to evaluate the interaction between miR-506-3p and BECN1. *P<0.05 vs. NC group. HUVECs, human umbilical vein endothelial cells; NC, negative control; miR, microRNA; WT, wild-type; 3′-UTR, 3′-untranslated region; BECN1, Beclin1.
Discussion
Hypertension is a complex disease caused by the interaction between environmental and genetic factors. In recent years, studies have demonstrated that miRNAs are closely associated with the onset and development of hypertension (30,31). The expression of some miRNA molecules in vascular tissues is increased markedly. For example, miR-204 is highly expressed in vascular tissues and helps to determine blood pressure by regulating the proliferation and apoptosis of vascular smooth muscle cells (32,33). In addition, miR-155 regulates the expression of angiotensin II type 1 receptor, affects the relaxation and contraction of vascular endothelial cells and promotes the onset and development of hypertension (34). It has been demonstrated that miR-122 influences the expression of the amino acid transporter gene SLC7A by changing the small nucleotide polymorphism site, disrupting nitric oxide metabolism, regulating vascular endothelial cell function and promoting the onset of primary hypertension (35). The results of the current study demonstrate that miR-506-3p expression is upregulated in the peripheral blood of patients with hypertension and is dependent on the stage of hypertension.
It has been demonstrated that patients with hypertension develop vascular injury in the early course of the disease. Furthermore, vascular endothelial cell injury and dysfunction are important signs of hypertension and may even occur prior to the onset of hypertension (36). miRNAs serve important roles in endothelial cell injury and dysfunction. miR-34a promotes pulmonary arterial hypertension by regulating the proliferation and apoptosis of vascular endothelial cells (37). The in vitro experiments performed in the current study demonstrate that miR-506-3p inhibits the proliferation, G1/S phase transition and migration of HUVECs, and promotes the apoptosis of HUVECs.
BECN1 has three structural domains, Bcl-2-homology-3, the central coiled-coil domain and the evolutionarily conserved domain, by which it interacts with other molecules (38). BECN1 is essential for the formation of autophagosomes and regulates the formation and maturation of intracellular autophagosomes by mediating the localization of other autophagy proteins in phagophores (39). Elevated autophagy activity is associated with the upregulation of BECN1 expression and downregulating BECN1 expression inhibits autophagy activity (40). Additionally, it has been demonstrated that BECN1 is closely associated with tumorigenesis and organ damage (41,42).
In the current study, bioinformatics analysis indicated that BECN1 is a potential target gene of miR-506-3p. BECN1 is a homologous gene of autophagy-related 6 in yeast and regulates autophagy by forming a complex with phosphoinositide 3-kinase (38). Previous studies have demonstrated that autophagy is closely associated with vascular endothelial injury (43,44). The activation of autophagy contributes to the survival and proliferation of vascular endothelial cells under stress. In the present study, the results of the dual-luciferase reporter assay indicated that miR-506-3p directly regulates the expression of BECN1 and that BECN1 is a target gene of miR-506-3p.
Acknowledgements
The authors would like to thank Professor Chunting Wang from the Department of Intensive Care Unit, Shandong Provincial Hospital Affiliated to Shandong University for kindly providing helpful advice in the conception, design, statistical analysis and preparation of the manuscript.
References
- 1.Leopold JA. Catheter-based therapies for patients with medication-refractory pulmonary arterial hypertension. Circ Cardiovasc Interv. 2015;8:e003332. doi: 10.1161/CIRCINTERVENTIONS.115.003332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olatunji LA, Seok YM, Igunnu A, Kang SH, Kim IK. Combined oral contraceptive-induced hypertension is accompanied by endothelial dysfunction and upregulated intrarenal angiotensin II type 1 receptor gene expression. Naunyn Schmiedebergs Arch Pharmacol. 2016;389:1147–1157. doi: 10.1007/s00210-016-1272-0. [DOI] [PubMed] [Google Scholar]
- 3.Zheng Y, Poon CC, Yan BP, Lau JY. Pulse arrival time based cuff-less and 24-H wearable blood pressure monitoring and its diagnostic value in hypertension. J Med Syst. 2016;40:195. doi: 10.1007/s10916-016-0558-6. [DOI] [PubMed] [Google Scholar]
- 4.Gerage AM, Bededetti TR, Ritti-Dias RM, Dos Santos ACO, de Souza BCC, Almeida FA. Effectiveness of a behavior change program on physical activity and eating habits in patients with hypertension: A randomized controlled trial. J Phys Act Health. 2017:1–30. doi: 10.1123/jpah.2016-0268. [DOI] [PubMed] [Google Scholar]
- 5.Ambrozova G, Fidlerova T, Verescakova H, Koudelka A, Rudolph TK, Woodcock SR, Freeman BA, Kubala L, Pekarova M. Nitro-oleic acid inhibits vascular endothelial inflammatory responses and the endothelial-mesenchymal transition. Biochim Biophys Acta. 2016;1860:2428–2437. doi: 10.1016/j.bbagen.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souza PS, Gonçalves ED, Pedroso GS, Farias HR, Junqueira SC, Marcon R, Tuon T, Cola M, Silveira PCL, Santos AR, et al. Physical exercise attenuates experimental autoimmune encephalomyelitis by inhibiting peripheral immune response and blood-brain barrier disruption. Mol Neurobiol. 2017;54:4723–4737. doi: 10.1007/s12035-016-0014-0. [DOI] [PubMed] [Google Scholar]
- 7.Balduino Mendes AB, Giollo-Junior LT, de Andrade DO, Gregório ML, Yugar-Toledo JC, Vilela-Martin JF. How to investigate the vascular changes in resistant hypertension. Curr Hypertens Rev. 2016;12:139–147. doi: 10.2174/1573402111666150812143349. [DOI] [PubMed] [Google Scholar]
- 8.Vaillancourt M, Ruffenach G, Meloche J, Bonnet S. Adaptation and remodelling of the pulmonary circulation in pulmonary hypertension. Can J Cardiol. 2015;31:407–415. doi: 10.1016/j.cjca.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins D. Pulmonary endarterectomy: The potentially curative treatment for patients with chronic thromboembolic pulmonary hypertension. Eur Respir Rev. 2015;24:263–271. doi: 10.1183/16000617.00000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds JA, Rosenberg AZ, Smith CK, Sergeant JC, Rice GI, Briggs TA, Bruce IN, Kaplan MJ. Brief Report: Vitamin D deficiency is associated with endothelial dysfunction and increases type-1 interferon gene expression in a murine model of systemic lupus erythematosus. Arthritis Rheumatol. 2016;68:2929–2935. doi: 10.1002/art.39803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furumoto Y, Smith CK, Blanco L, Zhao W, Brooks SR, Thacker SG, Abdalrahman Z, Sciumè G, Tsai WL, Trier AM, et al. Tofacitinib ameliorates murine lupus and its associated vascular dysfunction. Arthritis Rheumatol. 2017;69:148–160. doi: 10.1002/art.39818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Shi H, Luo J, Yi Y, Yao D, Zhang X, Ma G, Loor JJ. MiR-145 regulates lipogenesis in goat mammary cells via targeting INSIG1 and epigenetic regulation of lipid-related genes. J Cell Physiol. 2017;232:1030–1040. doi: 10.1002/jcp.25499. [DOI] [PubMed] [Google Scholar]
- 13.Lu YJ, Liu RY, Hu K, Wang Y. MiR-541-3p reverses cancer progression by directly targeting TGIF2 in non-small cell lung cancer. Tumour Biol. 2016;37:12685–12695. doi: 10.1007/s13277-016-5241-5. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Guo Y, Wang C, Yu H, Yu X, Yu H. MicroRNA-142-3p inhibits chondrocyte apoptosis and inflammation in osteoarthritis by targeting HMGB1. Inflammation. 2016;39:1718–1728. doi: 10.1007/s10753-016-0406-3. [DOI] [PubMed] [Google Scholar]
- 15.Liu B, Ding JF, Luo J, Lu L, Yang F, Tan XD. Seven protective miRNA signatures for prognosis of cervical cancer. Oncotarget. 2016;7:56690–56698. doi: 10.18632/oncotarget.10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang TY, Tsai WC, Huang TS, Su SH, Chang CY, Ma HY, Wu CH, Yang CY, Lin CH, Huang PH, et al. Dysregulation of endothelial colony-forming cell function by a negative feedback loop of circulating miR-146a and −146b in cardiovascular disease patients. PLoS One. 2017;12:e0181562. doi: 10.1371/journal.pone.0181562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carr G, Barrese V, Stott JB, Povstyan OV, Jepps TA, Figueiredo HB, Zheng D, Jamshidi Y, Greenwood IA. MicroRNA-153 targeting of KCNQ4 contributes to vascular dysfunction in hypertension. Cardiovasc Res. 2016;pii:cvw177. doi: 10.1093/cvr/cvw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oury C, Servais L, Bouznad N, Hego A, Nchimi A, Lancellotti P. MicroRNAs in valvular heart diseases: Potential role as markers and actors of valvular and cardiac remodeling. Int J Mol Sci. 2016;17:pii: E1120. doi: 10.3390/ijms17071120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elia L, Condorelli G. MicroRNAs and pulmonary hypertension: A tight link. Cardiovasc Res. 2016;111:163–164. doi: 10.1093/cvr/cvw163. [DOI] [PubMed] [Google Scholar]
- 20.Yu L, Yu H, Li X, Jin C, Zhao Y, Xu S, Sheng X. P38 MAPK/miR-1 are involved in the protective effect of EGCG in high glucose-induced Cx43 downregulation in neonatal rat cardiomyocytes. Cell Biol Int. 2016;40:934–942. doi: 10.1002/cbin.10637. [DOI] [PubMed] [Google Scholar]
- 21.Tong JL, Zhang CP, Nie F, Xu XT, Zhu MM, Xiao SD, Ran ZH. MicroRNA 506 regulates expression of PPAR alpha in hydroxycamptothecin-resistant human colon cancer cells. FEBS Lett. 2011;585:3560–3568. doi: 10.1016/j.febslet.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Peng T, Zhou L, Zuo L, Luan Y. MiR-506 functions as a tumor suppressor in glioma by targeting STAT3. Oncol Rep. 2016;35:1057–1064. doi: 10.3892/or.2015.4406. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Liu Z, Dong S, Zhang J, Tan J, Wang Y, Ge C, Li R, Xue Y, Li M, et al. miR-506 inhibits epithelial-to-mesenchymal transition and angiogenesis in gastric cancer. Am J Pathol. 2015;185:2412–2420. doi: 10.1016/j.ajpath.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Yang S, Li H, Yin Z, Fan J, Zhao Y, Gong W, Yan M, Wang DW. Mir30c is involved in diabetic cardiomyopathy through regulation of cardiac autophagy via BECN1. Mol Ther Nucleic Acids. 2017;7:127–139. doi: 10.1016/j.omtn.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Shenawy HA. Expression of Beclin-1, an autophagy-related marker, in chronic hepatitis and hepatocellular carcinoma and its relation with apoptotic markers. APMIS. 2016;124:229–237. doi: 10.1111/apm.12498. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Wang Y, Qian L, Wang X, Gu H, Dong X, Huang S, Jin M, Ge H, Xu C, Zhang Y. RNF216 contributes to proliferation and migration of colorectal cancer via suppressing BECN1-dependent autophagy. Oncotarget. 2016;7:51174–51183. doi: 10.18632/oncotarget.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu LS. Writing Group of 2010 Chinese Guidelines for the Management of Hypertension: 2010 Chinese guidelines for the management of hypertension. Zhonghua Xin Xue Guan Bing Za Zhi. 2011;39:579–615. (In Chinese) [PubMed] [Google Scholar]
- 28.Nwokeoji AO, Kilby PM, Portwood DE, Dickman MJ. RNASwift: A rapid, versatile RNA extraction method free from phenol and chloroform. Anal Biochem. 2016;512:36–46. doi: 10.1016/j.ab.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Vashukova ES, Glotov AS, Fedotov PV, Efimova OA, Pakin VS, Mozgovaya EV, Pendina AA, Tikhonov AV, Koltsova AS, Baranov VS. Placental microRNA expression in pregnancies complicated by superimposed pre-eclampsia on chronic hypertension. Mol Med Rep. 2016;14:22–32. doi: 10.3892/mmr.2016.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klimczak D, Jazdzewski K, Kuch M. Regulatory mechanisms in arterial hypertension: Role of microRNA in pathophysiology and therapy. Blood Press. 2017;26:2–8. doi: 10.3109/08037051.2016.1167355. [DOI] [PubMed] [Google Scholar]
- 32.Potus F, Graydon C, Provencher S, Bonnet S. Vascular remodeling process in pulmonary arterial hypertension, with focus on miR-204 and miR-126 (2013 Grover Conference series) Pulm Circ. 2014;4:175–184. doi: 10.1086/675980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruffenach G, Chabot S, Tanguay VF, Courboulin A, Boucherat O, Potus F, Meloche J, Pflieger A, Breuils-Bonnet S, Nadeau V, et al. Role for runt-related transcription factor 2 in proliferative and calcified vascular lesions in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2016;194:1273–1285. doi: 10.1164/rccm.201512-2380OC. [DOI] [PubMed] [Google Scholar]
- 34.Yang LX, Liu G, Zhu GF, Liu H, Guo RW, Qi F, Zou JH. MicroRNA-155 inhibits angiotensin II-induced vascular smooth muscle cell proliferation. J Renin Angiotensin Aldosterone Syst. 2014;15:109–116. doi: 10.1177/1470320313503693. [DOI] [PubMed] [Google Scholar]
- 35.Kamo Y, Ichikawa T, Miyaaki H, Uchida S, Yamaguchi T, Shibata H, Honda T, Taura N, Isomoto H, Takeshima F, Nakao K. Significance of miRNA-122 in chronic hepatitis C patients with serotype 1 on interferon therapy. Hepatol Res. 2015;45:88–96. doi: 10.1111/hepr.12317. [DOI] [PubMed] [Google Scholar]
- 36.Yoruk A, Bisognano JD, Gassler JP. Baroreceptor stimulation for resistant hypertension. Am J Hypertens. 2016;29:1319–1324. doi: 10.1093/ajh/hpw074. [DOI] [PubMed] [Google Scholar]
- 37.Wang MM, Fang MX, Chen LG, Wang HQ, Liu HJ, Tang HL. Differential expression of microRNA in endothelial cells incubated with serum of hypertension patients with blood-stasis syndrome. Chin J Integr Med. 2015;21:817–822. doi: 10.1007/s11655-015-2053-1. [DOI] [PubMed] [Google Scholar]
- 38.Kumar A, Singh UK, Chaudhary A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7:1535–1542. doi: 10.4155/fmc.15.88. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Tao Y, Huang Y, Zhan K, Xue M, Wang Y, Ruan D, Liang Y, Huang X, Lin J, et al. Catalase ameliorates diabetes-induced cardiac injury through reduced p65/RelA-mediated transcription of BECN1. J Cell Mol Med. 2017 Jun 23; doi: 10.1111/jcmm.13252. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuno S, Bogaard HJ, Gomez-Arroyo J, Alhussaini A, Kraskauskas D, Cool CD, Voelkel NF. MicroRNA-199a-5p is associated with hypoxia-inducible factor-1a expression in lungs from patients with COPD. Chest. 2012;142:663–672. doi: 10.1378/chest.11-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goyal A, Gubbiotti MA, Chery DR, Han L, Iozzo RV. Endorepellin-evoked autophagy contributes to angiostasis. J Biol Chem. 2016;291:19245–19256. doi: 10.1074/jbc.M116.740266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi JJ, Han XF, Cai XL, Li XZ, Zhang LQ, Feng X, Liu DY. Expression and clinical significance of autophagy-related gene Beclin1 and P62 in nasal polyps. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2016;51:428–432. doi: 10.3760/cma.j.issn.1673-0860.2016.06.006. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 43.Baek JH, Jung J, Seo J, Kim JH, Kim J. Bacterial overexpression and denaturing purification of VPS34-binding domain of Beclin 1. J Microbiol Biotechnol. 2016;26:1808–1816. doi: 10.4014/jmb.1604.04085. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Jiang W, Hu Y, Da Z, Zeng C, Tu M, Deng Z, Xiao W. MicroRNA-199a-5p inhibits cisplatin-induced drug resistance via inhibition of autophagy in osteosarcoma cells. Oncol Lett. 2016;12:4203–4208. doi: 10.3892/ol.2016.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]