This Outlook by Gnügge and Symington discusses the studies by Wang et al. and Reginato et al., which report that a variety of physiological protein blocks, including Ku, RPA, and nucleosomes, stimulate MRX–Sae2 endonuclease cleavage in vitro. These studies have important implications for how cells deal with a range of barriers to end resection and highlight the importance of Sae2 in activating MRX cleavage at the correct cell cycle stage.
Keywords: nuclease, MRX–Sae2, Ku70–Ku80, RPA, nucleosome, homologous recombination, DNA end resection
Abstract
The yeast Mre11–Rad50–Xrs2 (MRX) complex and Sae2 function together to initiate DNA end resection, an essential early step in homology-dependent repair of DNA double-strand breaks (DSBs). In this issue of Genes & Development, Wang and colleagues (pp. 2331–2336) and Reginato and colleagues (pp. 2325–2330) report that a variety of physiological protein blocks, including Ku, RPA, and nucleosomes, stimulate MRX–Sae2 endonuclease cleavage in vitro. These studies have important implications for how cells deal with a range of barriers to end resection and highlight the crucial role of Sae2 in activating MRX cleavage at the correct cell cycle stage.
Chromosomal double-strand breaks (DSBs) are toxic DNA lesions that must be accurately repaired to maintain genome integrity; failure to properly mend DSBs can result in loss of genetic information, chromosome rearrangements, or even cell death. Typically, cells repair DSBs by nonhomologous end joining (NHEJ) or homologous recombination (HR). NHEJ directly ligates DSB ends, whereas HR uses extensive homology and templated DNA synthesis to restore the broken chromosome. HR is activated during the S and G2 phases of the cell cycle to ensure error-free repair from a sister chromatid template. HR initiates by nucleolytic degradation of the 5′ terminated strands in a process termed end resection (for review, see Symington 2016). End resection generates 3′ ssDNA tails, substrates for Rad51 to catalyze homologous pairing and exchange of DNA strands and for activation of the DNA damage checkpoint.
Genetic studies in yeast implicate the Mre11–Rad50–Xrs2 (MRX) complex and Sae2 in early steps of resection (Xrs2 and Sae2 are replaced by NBS1 and CtIP, respectively, in mammalian cells). In vitro, Mre11 exhibits 3′–5′ dsDNA-specific exonuclease and ssDNA-specific endonuclease activities (Paull and Gellert 1998). The current model for resection initiation is by MRX-catalyzed incision of the 5′-terminated strands internal to the ends in a reaction stimulated by cyclin-dependent kinase (CDK) phosphorylated Sae2 (Huertas et al. 2008; Cannavo and Cejka 2014). The resulting nicks are entry sites for the Mre11 3′ exonuclease to degrade back to the DSB and for more extensive processing of 5′ strands by either Exo1, a 5′–3′ exonuclease, or the combined activities of the Sgs1 helicase and the Dna2 endonuclease (Fig 1; Garcia et al. 2011).
Figure 1.
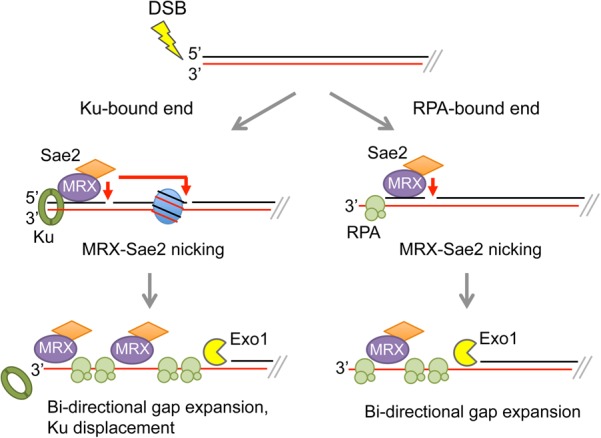
The MRX–Sae2 nuclease nicks 5′-terminated strands of DSBs at diverse protein barriers. In S–G2-phase cells, Sae2 is phosphorylated by CDK and activates the MRX endonuclease to incise 5′-terminated strands at Ku- or RPA-bound ends or adjacent to nucleosomes. The MRX 3′–5′ exonuclease and the Exo1 5′–3′ exonuclease expand the resulting nicks to create long tracts of ssDNA for HR. MRX 3′–5′ degradation is anticipated to displace Ku from ends. Note that only one side of a DSB is shown.
Previous biochemical studies by Cannavo and Cejka (2014) demonstrated that biotin–streptavidin linkages at the ends of linear duplex substrates induced MRX endonucleolytic cleavage of the 5′ strands in a reaction stimulated by Sae2. This observation raised the question of whether physiological protein blocks would also stimulate MRX–Sae2-catalyzed incision. The Ku complex, an essential component of the NHEJ pathway, is rapidly recruited to DSBs in cells and protects ends from degradation by Exo1 (Symington 2016). In the accompanying studies (Reginato et al. 2017; Wang et al. 2017), the investivators show that Ku is as effective as biotin–streptavidin in stimulating endonucleolytic cleavage by MRX–Sae2 (Fig 1). Furthermore, Ku inhibited Mre11-catalyzed 3′–5′ degradation at DNA ends. In agreement with previous work, the clipping reaction depended on ATP hydrolysis by Rad50 and phosphorylated Sae2. These findings are consistent with cooperation between Ku and MRX to promote NHEJ in G1-phase cells, whereas, in S–G2-phase cells, when a sister chromatid is available and Sae2 is activated by CDK, Ku stimulates endonucleolytic clipping by MRX–Sae2, thereby committing cells to HR.
Binding of RPA to ssDNA overhangs had an effect similar to that of Ku binding; exonucleolytic degradation was impeded, while endonucleolytic scission was stimulated. Additionally, Wang et al. (2017) used substrates with terminal hairpin structures and found that RPA binding enhanced MRX–Sae2 clipping next to the structures. These data support a model in which the MRX complex can reinitiate resection in case the long-range resection machinery becomes disengaged or encounters an obstacle and have important implications for removal of fold-back structures that are precursors to palindromic duplications and other complex rearrangements.
DSB processing in cells impaired for long-range resection as well as recent genome-wide resection analysis in meiotic cells suggested that MRX–Sae2 nicking occurs preferentially between nucleosomes (Mimitou and Symington 2008; Zhu et al. 2008; Mimitou et al. 2017). To test this hypothesis using their in vitro system, Wang et al. (2017) positioned a single nucleosome on a 232-base-pair substrate. Indeed, they identified novel scission sites corresponding to MRX–Sae2 cleavage adjacent to the nucleosome (Fig 1). These data nicely explain the 100- to 200-nucleotide incremental cleavages detected at endonuclease-induced DSBs in exo1Δ sgs1Δ cells.
The bidirectional resection model posits that the Mre11 3′–5′ exonuclease chews back from the MRX–Sae2-generated nick and that Exo1 degrades the 5′-terminated strand (Garcia et al. 2011; Symington 2016). Importantly, Reginato et al. (2017) show that Exo1 efficiently degrades from the nick generated by MRX–Sae2 in a coupled reaction. The bacteriophage T7 exonuclease was ineffective in chewing from the MRX–Sae2-catalyzed nick even though it showed activity equivalent to that of Exo1 at DNA ends and nicks. By devising a substrate with an internal nick, Wang et al. (2017) were able to demonstrate a stimulatory role for Sae2 in Mre11-catalyzed degradation from the nick. In contrast to MRX incision, this activity was not dependent on phosphorylated Sae2 or Rad50 ATPase activity. Together, these findings provide strong biochemical support for the bidirectional resection model.
The studies by Wang et al. (2017) and Reginato et al. (2017) demonstrate how MRX–Sae2 interacts with natural proteins bound to DSB ends and with the long-range machinery and confirm the importance of Sae2 to restrict resection initiation to the appropriate cell cycle stage. The findings raise several interesting questions: How is the endonuclease activity of MRX controlled not only temporally but also locally to the vicinity of DSB ends? Given that MRX–Sae2 is able to incise the 5′ DNA strand next to a multitude of stably bound protein obstacles, such as nucleosomes and RPA, how is spurious nicking prevented? It also remains to be determined how Sae2 and Rad50 coordinate to target Mre11 nicking to the 5′-terminated strands at DSBs.
Acknowledgments
Studies in the Symington laboratory are supported by grants from the National Institutes of Health (GM041784 and CA174653).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.310771.117.
References
- Cannavo E, Cejka P. 2014. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature 514: 122–125. [DOI] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ. 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Yamada S, Keeney S. 2017. A global view of meiotic double-strand break end resection. Science 355: 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979. [DOI] [PubMed] [Google Scholar]
- Reginato G, Cannavo E, Cejka P. 2017. Physiological protein blocks direct the Mre11–Rad50–Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev (this issue) 10.1101/gad.308254.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. 2016. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 51: 195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Daley JM, Kwon Y, Krasner DS, Sung P. 2017. Plasticity of the Mre11–Rad50–Xrs2–Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev (this issue) 10.1101/gad.307900.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]


