Reginato et al. report that the nonhomologous end joining factor Ku limits the exonuclease activity of Mre11 and promotes its endonuclease to cleave 5′-terminated DNA strands at break sites. Following initial endonucleolytic cleavage past the obstacle, Exo1 specifically extends the resection track, leading to the generation of long 3′ overhangs that are required for homologous recombination.
Keywords: homologous recombination, DNA nuclease, yMre11, Sae2, Ku70–80, DNA end resection
Abstract
DNA double-strand break repair by homologous recombination is initiated by DNA end resection, which is commenced by the Mre11–Rad50–Xrs2 complex and Sae2 in yeast. Here we report that the nonhomologous end joining factor Ku limits the exonuclease activity of Mre11 and promotes its endonuclease to cleave 5′-terminated DNA strands at break sites. Following initial endonucleolytic cleavage past the obstacle, Exo1 specifically extends the resection track, leading to the generation of long 3′ overhangs that are required for homologous recombination. These experiments provide mechanistic insights into how short-range and long-range DNA end resection enzymes overcome obstacles near broken DNA ends to initiate recombination.
DNA double-strand breaks (DSBs) arise accidentally or in a programmed manner (Jackson and Bartek 2009; Lam and Keeney 2014). Depending on the mechanism of DSB formation, DNA breaks can either be chemically clean or contain various secondary DNA structures, chemical adducts, or protein blocks. DSBs can be repaired by either template-independent nonhomologous end joining (NHEJ), which is often mutagenic (Chang et al. 2017), or template-dependent homologous recombination (HR), which is largely accurate (Kowalczykowski 2015). How these pathways deal with obstacles at the break ends remains poorly defined.
The DSB repair by HR is initiated by DNA end resection, which specifically degrades 5′-terminated DNA at DSB sites to produce 3′ ssDNA overhangs, which are then protected by the ssDNA-binding protein replication protein A (RPA) (White and Haber 1990; Cejka 2015). Following the exchange of RPA for the strand exchange protein RAD51, the RAD51 nucleoprotein filament invades template dsDNA to mediate the homology-directed repair (Kowalczykowski 2015).
DNA end resection in yeast Saccharomyces cerevisiae is initiated by the Mre11–Rad50–Xrs2 (MRX) complex containing the Mre11 nuclease that functions in conjunction with Sae2 (Moreau et al. 2001; Mimitou and Symington 2008; Zhu et al. 2008). It has been proposed that Mre11 first cleaves the 5′-terminated DNA strand at the broken end endonucleolytically, which is followed by the 3′ → 5′ exonuclease of Mre11 back toward the DNA end (Neale et al. 2005; Garcia et al. 2011; Shibata et al. 2014). In addition to the nucleolytic activity of MRX–Sae2 in the vicinity of the broken ends, the complex has an additional structural role to recruit components of two long-range resection pathways that function downstream (Cejka et al. 2010; Nicolette et al. 2010; Niu et al. 2010; Shim et al. 2010; Cannavo et al. 2013). These are dependent on Exo1 or Dna2 nuclease, which function in a redundant manner and are capable of resecting DNA thousands of nucleotides in length in the 5′ → 3′ direction. Genetic experiments showed that the short-range resection by MRX–Sae2 and in particular the nuclease activity of Mre11 can be bypassed during the processing of clean endonuclease-induced DNA breaks (Llorente and Symington 2004). Instead, the Mre11 nuclease is important for the processing of DNA ends with covalent protein blocks, such as stalled topoisomerases or Spo11 in meiosis (Neale et al. 2005; Aparicio et al. 2016; Hoa et al. 2016).
The NHEJ factor Ku is recruited rapidly and independently of MRX to DSBs (Lisby et al. 2004; Wu et al. 2008). Ku was shown to limit DNA end resection, particularly in the G1 phase (Clerici et al. 2008). Deletion of genes coding for the Ku subunits Yku70 or Yku80 partially suppressed the ionizing radiation sensitivity of sae2 or mre11 mutants (Clerici et al. 2008; Bonetti et al. 2010; Mimitou and Symington 2010; Foster et al. 2011; Langerak et al. 2011). These studies suggested that MRX–Sae2 directly counteract the Ku heterodimer at the DNA ends, particularly in the G2 phase. The MRX–Sae2 complex thus helps to initiate resection also in the case of noncovalently bound obstacles as well as when the broken ends contain secondary DNA structures or lesions induced by ionizing radiation (Lobachev et al. 2002; Mimitou and Symington 2010). As MRX also directly promotes NHEJ (Chen et al. 2001), Sae2 is likely the factor that channels MRX function toward the recombination pathway. However, the mechanism of interplay of MRX and Sae2 with Ku at DNA ends remains undefined (Lisby and Rothstein 2009).
We previously demonstrated that phosphorylated Sae2 stimulates the endonuclease activity of Mre11 within the MRX complex on synthetic substrates with streptavidin-blocked DNA ends (Cannavo and Cejka 2014). Here, we show that the highly abundant Ku heterodimer protects free DNA ends from the 3′ → 5′ exonuclease of Mre11 and instead promotes the 5′ endonucleolytic cleavage. This is followed by the 3′ → 5′ exonuclease of Mre11 back toward the DNA end. We also show that the substrate cleaved by MRX–Sae2 is suitable for subsequent processing by Exo1 but not by an unrelated exonuclease, directly demonstrating bidirectional resection. Our experiments reveal the interplay of the MRX–Sae2 nuclease complex with physiological obstacles such as Ku and RPA at the break ends and reconstitute a DNA end resection pathway where both short-range and long-range nucleases are activated sequentially.
Results and Discussion
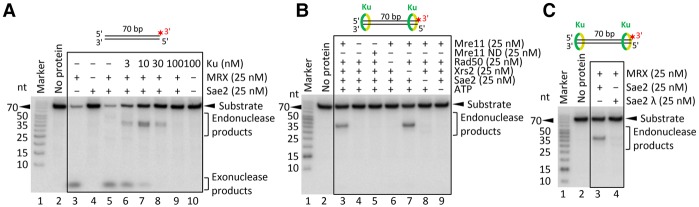
Both Ku and MRX are recruited quickly and independently to DNA ends in both G1 and S/G2 phases of the cell cycle (Lisby et al. 2004; Wu et al. 2008). Previously, we showed that recombinant MRX and Sae2 preferentially cleave the 5′-terminated DNA strand in the vicinity of streptavidin-blocked DNA ends (Cannavo and Cejka 2014). To investigate the interplay of MRX–Sae2 and Ku, we expressed and purified the recombinant S. cerevisiae Yku70–80 heterodimer (Ku) (Supplemental Fig. S1A). Ku binds dsDNA (Supplemental Fig. S1B), as established previously (Blier et al. 1993; Gottlieb and Jackson 1993). The recombinant MRX complex degraded a 70-base-pair (bp)-long dsDNA substrate primarily using the 3′ → 5′ exonuclease activity of Mre11, releasing the radioactive label from the 3′ end (Fig. 1A, lane 3) independently of Sae2 (Fig. 1A, lane 5). Strikingly, when we titrated Ku into the reaction, we observed an inhibition of the Mre11 exonuclease and the appearance of novel endonuclease products (Fig. 1A, lanes 6–8). Very high Ku concentrations fully protected the substrate (Fig. 1A, lane 9), as multiple Ku molecules likely occlude the dsDNA substrate and thus prevent the access of MRX (Fig. 1A, lane 9). Similar behavior was observed with a 100-bp-long dsDNA substrate (Supplemental Fig. S1C). These results indicate that in the presence of Ku, DNA ends are likely to be efficiently processed not by the 3′ → 5′ exonuclease of Mre11 but rather by its endonuclease activity. The endonucleolytic clipping depends on the nuclease of Mre11, as the nuclease-dead Mre11 125–126 HD/LV (Mre11 ND) variant did not support this cleavage (Fig. 1B, lane 5). Furthermore, Sae2, Rad50, and ATP were required, while Xrs2 was dispensable under our experimental conditions, in agreement with previous data (Fig. 1B; Supplemental Fig. S1D; Cannavo and Cejka 2014; Oh et al. 2016).
Figure 1.
Ku bound to DNA ends limits the exonuclease and stimulates the endonuclease activity of MRX–Sae2. (A) A representative experiment showing the effect of Ku on the nuclease activities of MRX–Sae2. The recombinant proteins, as indicated, were incubated with 3′-labeled oligonucleotide-based dsDNA of 70 bp in length. The reaction products were separated by denaturing polyacrylamide gel (15%) electrophoresis. The red asterisk indicates the location of the 32P label. The positions of exonuclease and endonuclease products are indicated at the right. (B) The endonucleolytic cleavage near Ku (10 nM)-bound DNA ends requires the nuclease of Mre11, Rad50, Sae2, and ATP. A representative experiment is shown. (C) Sae2 needs to be phosphorylated in order to promote MRX cleavage near Ku (10 nM)-bound DNA ends. The reaction in lane 3 included phosphorylated Sae2. The reaction in lane 4 contained λ phosphatase-treated Sae2. A representative experiment is shown.
Genetic experiments suggested that Ku inhibits DNA end resection, particularly in the G1 phase of the cell cycle. In G2, Ku is inhibitory when overexpressed but not when present at physiological levels (Clerici et al. 2008). Efficient resection in G2 cells is dependent on the nuclease of Mre11 and also on Exo1 (Bonetti et al. 2010; Mimitou and Symington 2010; Foster et al. 2011). DNA end resection is under cyclin-dependent kinase (CDK) control, with the main target being S267 of Sae2 at S/G2 (Huertas et al. 2008). In accord with this, we observed the endonucleolytic cleavage near Ku-bound ends when using recombinant phosphorylated Sae2 but not when Sae2 was treated with λ phosphatase during purification (Fig. 1C, all experiments in this work use phosphorylated Sae2 unless indicated otherwise). These experiments demonstrate that phosphorylated Sae2 stimulates MRX to cleave near Ku-bound DNA ends, which likely recapitulates events that occur during the G2 phase in vivo.
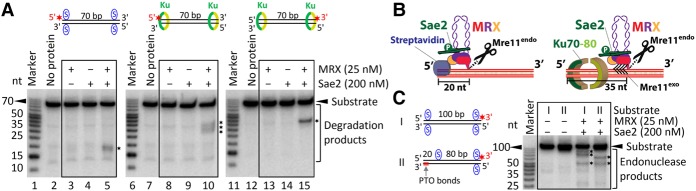
Previously, we observed that MRX cleaved the streptavidin-bound DNA ∼20 nucleotides (nt) away from the 5′ DNA end (Fig. 2A, lane 5; Cannavo and Cejka 2014). Therefore, unlike the 3′ → 5′ exonuclease of Mre11, the Mre11 endonuclease degrades DNA with the right polarity for recombination (White and Haber 1990; Cannavo and Cejka 2014; Cejka 2015). In contrast to streptavidin-blocked ends, Ku-bound DNA was cleaved further away, ∼35 nt from the 5′ DNA end (Fig. 2A, lane 10, Fig. 2B). This is in agreement with the Ku DNA-binding site size of ∼30–35 bp (Blier et al. 1993), which likely occludes a longer DNA stretch than terminally bound streptavidin (Fig. 2B). The cleavage position did not change when using a longer, 100-bp-long DNA substrate (up to ∼35 nt from the 5′ end) (Supplemental Fig. S1E), demonstrating the preferential cleavage of the 5′-terminated strand. Instead, the site of DNA cleavage changed when we shifted the position of biotin–streptavidin away from the DNA end (Fig. 2C), suggesting that MRX–Sae2 cuts DNA near obstacles located also internal to DNA ends.
Figure 2.
Obstacles located at or internal to DNA ends determine the position of the endonucleolytic cleavage by MRX–Sae2. (A) A representative nuclease assay showing the processing of the 5′-labeled DNA substrate of 70 bp in length blocked with streptavidin (designated by the blue S) at DNA ends or bound by 30 nM Ku by MRX–Sae2. The endonucleolytic DNA cleavage products are indicated at the right with black asterisks. See also Supplemental Figure S1F. (B) A schematic representation of MRX–Sae2-dependent cleavage of streptavidin-bound versus Ku-bound DNA ends. The black arrows in the case of Ku-bound ends indicate exonucleolytic degradation following endonucleolytic clipping. (C) MRX–Sae2-dependent cleavage of DNA substrates of 100 bp in length with end-bound (substrate I) or internally bound (substrate II) streptavidin; see the cartoons at the left. The endonucleolytic DNA cleavage products are indicated by black asterisks. The phosphorothioate (PTO) bonds at the 3′ end of the bottom oligonucleotide were included to prevent the degradation by the Mre11 exonuclease. A representative experiment is shown.
Careful examination of assays with Ku-blocked DNA ends revealed differences in product lengths when comparing reactions carried out with 5′- or 3′-labeled DNA substrates. In case of the 5′-labeled strand, MRX–Sae2 consistently gave rise to “fuzzy” signals, indicating end products of various lengths (Fig. 2A, lane 10). In contrast, MRX–Sae2 nuclease led to a “sharp” band when using a substrate with the same strand but 3′-labeled (Fig. 2A, lane 15; see also Supplemental Fig. S1F). This likely indicates that MRX–Sae2 initially cleave endonucleolytically at a relatively precise position away from the 5′ end determined by the end-bound Ku and then continue degrading DNA back toward the 5′ end, likely using the 3′ → 5′ exonuclease of Mre11, which terminates at various positions (Fig. 2B). We believe that the exonucleolytic degradation following endonucleolytic cleavage is more likely to occur with Ku that has the capacity to slide on DNA rather than with tightly bound streptavidin.
Stretches of ssDNA are bound by the RPA in vivo. To investigate whether RPA can direct DNA cleavage by MRX–Sae2, we compared the processing of blunt-ended DNA and DNA with an overhang by MRX–Sae2 in the absence or presence of RPA (Fig. 3A,B; Supplemental Fig. S2A). Without RPA, the substrates were preferentially degraded by the 3′ → 5′ exonuclease activity of Mre11 independently of Sae2 (Fig. 3A,B). Strikingly, when ssDNA overhangs were present, the inclusion of RPA led to the stimulation of endonucleolytic DNA cleavage in conjunction with Sae2 (Fig. 3A,B; Supplemental Fig. S2A). In particular, we observed that RPA-coated 3′ overhangs stimulated MRX–Sae2 cleavage of the opposite 5′-terminated DNA strand (Fig. 3A, lane 5). At the same time, RPA inhibited the 3′ → 5′ exonucleolytic processing of DNA with overhangs (Fig. 3B, cf. lanes 3,5 and 6,8), which can be also seen with a ssDNA oligonucleotide (Supplemental Fig. 2B,C). In contrast, we did not see stimulation of the endonuclease activity of MRX–Sae2 by RPA in the case of blunt-ended DNA (Fig. 3A,B). RPA bound to ssDNA overhangs thus also directs preferential 5′ DNA cleavage by MRX–Sae2 while protecting 3′ overhangs from degradation.
Figure 3.
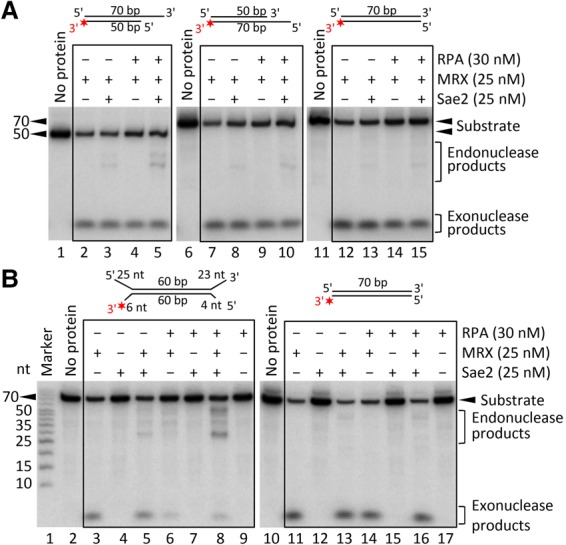
RPA bound to ssDNA directs the nuclease cleavage of adjacent dsDNA by the MRX–Sae2 complex. (A) Representative nuclease assays showing the degradation of 3′ tailed (PC216 and PC216_C_50 nt), 5′ tailed (oligonucleotides PC216_50 nt and PC217), and blunt-ended DNA (PC216 and PC217) by MRX–Sae2 without or with RPA. The red asterisk indicates the location of the 32P label. (B) Representative nuclease assays as in A showing the degradation of tailed (oligonucleotides PC204 and PC217) and blunt-ended (PC216 and PC217) DNA by MRX–Sae2 with or without RPA.
According to the DNA end resection model, endonucleolytic cleavage by MRX–Sae2 creates entry sites for the long-range DNA end resection enzymes such as Exo1 (Cejka 2015). Previously, it had been demonstrated that MRX has a structural (i.e., nuclease independent) role to recruit Exo1 to DNA ends and stimulate its exonuclease activity (Nicolette et al. 2010; Cannavo et al. 2013). This function is likely relevant for the processing of “clean” DNA ends. In contrast, genetic experiments also suggested that the endonucleolytic cleavage by Mre11 precedes resection by Exo1 in the case of blocked DNA ends, such as Spo11-bound ends in meiosis or Ku-bound ends in mitotic cells in G2 (Keeney and Kleckner 1995; Neale et al. 2005; Bonetti et al. 2010; Mimitou and Symington 2010; Foster et al. 2011). Streptavidin bound at DNA ends inhibited resection by Exo1, as expected (Supplemental Fig. S3A). The exonuclease activity was intrinsic to Exo1 because nuclease-dead Exo1 D173A possessed no activity (Supplemental Fig. S3B; Tran et al. 2002). To investigate the interplay of the MRX–Sae2 nuclease complex with Exo1, we carried out reactions with streptavidin-blocked dsDNA and MRX–Sae2 with or without Exo1. While Exo1 did not affect the efficiency of the endonucleolytic cleavage by MRX–Sae2, it was clearly capable of extending the degradation of products resulting from clipping by MRX–Sae2 (Fig. 4A). We observed that the same concentration of Exo1 (25 nM) was required to efficiently degrade DNA downstream from the MRX–Sae2 cleavage as well as free DNA (cf. lane 10 in Fig. 4B and lane 5 in Supplemental Fig. S3A), showing that the DNA degradation by Exo1 following MRX–Sae2 clipping is very efficient.
Figure 4.
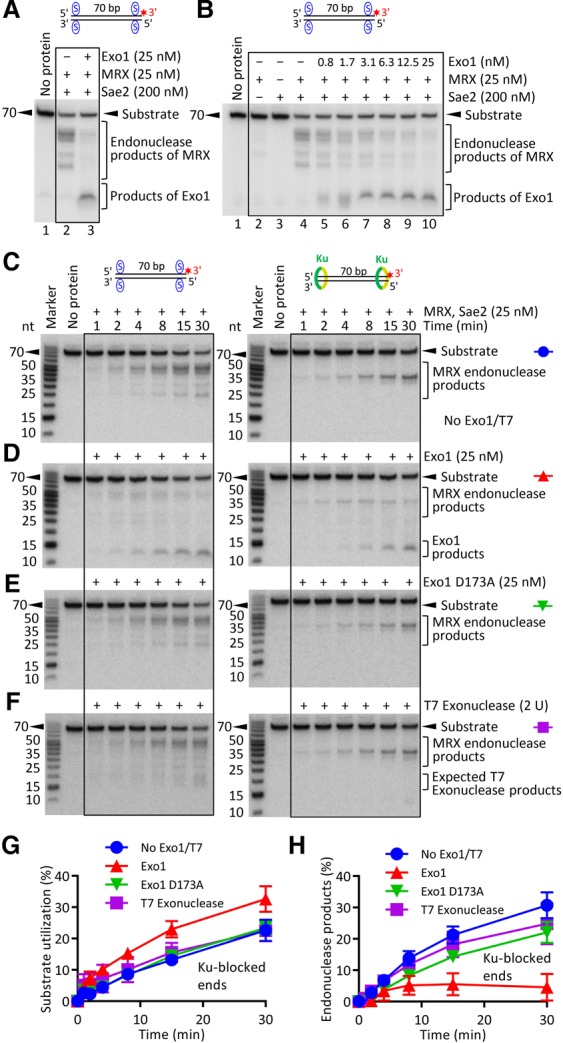
Exo1 efficiently and specifically functions downstream from the MRX–Sae2 nuclease in the processing of dsDNA with blocked ends. (A) A representative experiment showing the processing of streptavidin-blocked dsDNA by MRX–Sae2 in the absence (lane 2) or presence (lane 3) of Exo1. (B) Representative DNA clipping reactions with MRX–Sae2 with various concentrations of Exo1, as indicated. (C–F) Exo1 degrades the MRX–Sae2 nuclease products in a specific manner. Representative kinetic nuclease assays with MRX–Sae2 and either no additional exonuclease (C), Exo1 (D), Exo1 D173A (nuclease-dead; E), or T7 exonuclease (F). The DNA was either blocked by streptavidin (left) or bound by 30 nM Ku (right). (G) Quantitation of experiments such as those from C–F showing overall substrate utilization in reactions with Ku-blocked dsDNA. Averages are shown. n ≥ 3. Error bars indicate SEM. (H) Quantitation of experiments such as those from C–F showing the fraction of endonuclease products versus total DNA in reactions with Ku-blocked dsDNA. Averages are shown. n ≥ 3. Error bars indicate SEM.
To determine the specificity of DNA degradation by Exo1 downstream from MRX–Sae2, we performed kinetic experiments with unblocked, streptavidin-blocked, or Ku-blocked dsDNA (Fig. 4C–H; Supplemental Figure S3D–J). In the absence of Exo1, the endonucleolytic products of MRX–Sae2 appeared in a time-dependent manner (Fig. 4C,H; Supplemental Fig. S3J). Inclusion of Exo1, but not nuclease-dead Exo1 D173A, resulted in further degradation of most of these cleavage products (Fig. 4D,E,H; Supplemental Fig. S3J). This showed that Exo1 can function directly downstream from MRX–Sae2 clipping also during the processing of Ku-bound DNA ends. We next used the T7 exonuclease, which is a 5′ → 3′ DNA exonuclease acting on dsDNA from ends or nicks as Exo1. We selected T7 exonuclease concentration with higher specific activity than 25 nM Exo1 on unprotected DNA (2 U of T7 exonuclease) (Supplemental Fig. S3, cf. A and C). This amount of T7 exonuclease led to degradation of DNA with unblocked ends similar to that by Exo1 in conjunction with MRX–Sae2 (Supplemental Fig. S3E,G,H). Strikingly, the inclusion of 2 U of T7 exonuclease into the MRX–Sae2 reactions resulted in only a minimal degradation of the endonucleolytic cleavage products downstream from MRX–Sae2 in the case of both streptavidin-blocked and Ku-blocked DNA (Fig. 4, cf. C and F; Supplemental Fig. S3J). In contrast, both 25 nM Exo1 and 2 U of T7 exonuclease were sufficient to degrade ∼90% of nicked circular DNA (Supplemental Fig. S3K). This suggested that Exo1 functions downstream from MRX–Sae2 clipping in a highly specific manner and that the endonucleolytic cleavage sites are likely protected from unscheduled degradation (Fig. 4H; Supplemental Fig. S3J). In all cases, the efficiency of DNA cleavage by MRX–Sae2 was not notably affected by the addition of the exonucleases, as seen by overall substrate utilization (Fig. 4G; Supplemental Fig. S3I).
Together, our results implicate that MRX in conjunction with phosphorylated Sae2 is a versatile complex that is likely to cleave 5′-terminated DNA strands at DSBs past various covalently or noncovalently bound polypeptides. We show that MRX–Sae2 efficiently cleaves near Ku-blocked DNA ends, which shows how recombination competes with NHEJ in the S/G2 phase when Sae2 is phosphorylated and thus capable of promoting endonucleolytic DNA cleavage by MRX. As MRX also promotes NHEJ, our results support a model in which DNA ends that are initially bound by Ku and MRX first channel DSB repair to NHEJ (Chen et al. 2001). In cases when this does not occur in a timely fashion, the subsequent recruitment of phosphorylated Sae2 and the endonucleolytic cleavage of DNA by MRX then direct the repair toward HR (Shibata et al. 2014).
Genetic experiments showed that MRX may initially cleave further away from the ends (in some cases, up to ∼300 bp) (Garcia et al. 2011); in agreement with this, we observed that internally bound streptavidin also directed MRX–Sae2 cleavage (Fig. 2C). To this point, it is interesting that DNA end resection end points of MRX–Sae2 resection tracks in meiotic cells correlated with nucleosome positions (Mimitou et al. 2017). Our observations thus raise a possibility that nucleosomes might also direct the endonucleolytic cleavage by MRX–Sae2; however, whether this is indeed the case and whether any chromatin mark specifically regulates this cleavage remain to be determined. Here, we directly demonstrate bidirectional resection by MRX–Sae2 and Exo1: Following endonucleolytic cleavage by MRX, the complex proceeds in a 3′ → 5′ direction back toward the end (Garcia et al. 2011; Shibata et al. 2014). Concurrently, we show that cognate Exo1 is capable of continuing resecting 5′ → 3′ downstream from the MRX–Sae2 catalyzed clipping in a highly specific manner. These experiments thus reconstitute resection of blocked DNA ends where both short-range (MRX–Sae2) and long-range (Exo1) nucleases are activated sequentially.
Materials and methods
Preparation of recombinant proteins
The recombinant S. cerevisiae MRX complex, Xrs2, Exo1, and nuclease-dead Exo1 D173A were expressed in Spodoptera frugiperda 9 (Sf9) cells and purified as described previously (Cannavo et al. 2013; Cannavo and Cejka 2014; Oh et al. 2016). Mre11 and Mre11 ND were expressed in Sf9 cells using pFB-MBP-Mre11-his and pFB-MBP-Mre11 ND-his vectors, respectively, and purified similarly as described previously for Sgs1 (Cejka and Kowalczykowski 2010). Both Mre11 variants contained an MBP tag at the N terminus that was cleaved during purification as well as a 10× His-tag at the C terminus. Rad50 was expressed in Sf9 cells using a pFB-RAD50-Flag vector using anti-Flag affinity chromatography; the recombinant Rad50 construct contained a C-terminal Flag tag. Yeast RPA was expressed from p11d-scRPA vector (a kind gift from M. Wold, University of Iowa) in Escherichia coli and purified using procedures similar to those used for human RPA (Binz et al. 2006). T7 exonuclease was purchased from New England Biolabs (M0263). Yeast Sae2 was prepared as described previously (Cannavo and Cejka 2014) with the following modification: Okadaic acid (100 nM final concentration) (Calbiochem) was added to the Sf9 cells expressing Sae2 for the last 3 h before harvesting. Lysis buffer was additionally supplemented with the following phosphatase inhibitors: 25 nM okadaic acid (Calbiochem), 1 mM Na3VO4 (Sigma), 20 mM NaF (Applichem), and 15 mM Na4P2O7 (Applichem). This preserved phosphorylation of Sae2 (details provided elsewhere) (E Cannavo and P Cejka, in prep.). To prepare dephosphorylated Sae2, the Sf9 culture and lysis buffer were not supplemented with phosphatase inhibitors. Additionally, 20,000 U of λ phosphatase (New England Biolabs) for purification from 1.6 l Sf9 cells was added in the course of protein purification during the step when MBP-Sae2 was digested with Prescission protease (Cannavo and Cejka 2014); the reaction was supplemented with 1 mM MnCl2 and incubated for 30 min at room temperature and 90 min at 4°C together with Prescission protease. Mock treatment (without λ phosphatase) of phosphorylated Sae2 did not affect its capacity to promote MRX (data not shown). The preparation of the Ku complex is detailed in the Supplemental Material.
DNA substrates
The DNA substrates for the in vitro assays were used and radioactively labeled as described previously (Cannavo and Cejka 2014); the 70-bp-long dsDNA substrate with biotin labels was prepared by annealing oligonucleotides PC210 and PC211, and the 100-bp-long substrate was prepared by annealing oligonucleotides 100TOP and 100BOTTOM (Cannavo and Cejka 2014). The preparation of other substrates is detailed in the Supplemental Material.
Nuclease and DNA-binding assays
Unless indicated otherwise, the nuclease assays were carried out in a 15-µL volume. They were assembled on ice in reaction buffer containing 25 mM Tris-acetate (pH 7.5), 1 mM dithiothreitol, 5 mM magnesium acetate, 1 mM manganese acetate, 1 mM ATP, 80 U/mL pyruvate kinase (Sigma), 1 mM phosphoenolpyruvate, 0.25 mg/mL bovine serum albumin (New England Biolabs), and 1 nM (in molecules) DNA substrate. Where indicated, the substrate with reaction buffer was first incubated with 30 nM recombinant streptavidin (Sigma) per tetramer (in principle, 4 nM tetrameric streptavidin is sufficient to saturate 1 nM substrate with four biotin labels) or Ku (concentration as indicated) for 5 min at room temperature. The reaction was then returned on ice, where recombinant proteins were added. The reactions were incubated for 30 min at 30°C unless indicated otherwise and stopped by adding 0.5 µL of 14–22 mg/mL proteinase K (Roche), 0.5 µL of 10% (w/v) sodium dodecyl sulfate, and 0.5 µL of 0.5 M ethylenediaminetetraacetic acid (EDTA) for 30 min at 50°C. The stopped reactions were mixed with an equal volume of loading dye (95% formamide, 20 mM EDTA, 1 mg/mL bromophenol blue). The reaction products were separated by denaturing electrophoresis on 15% polyacrylamide gels (acrylamide:bisacrylamide 19:1; Bio-Rad) containing 7 M urea. Radioactively labeled low-molecular-weight marker (Affymetrix, J76410) was used where indicated. The samples were separated by electrophoresis in 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA), and the resolved gels were fixed in fixing solution (40% methanol, 10% acetic acid, 5% glycerol) for 30 min at room temperature. The gels were then dried, exposed to storage phosphor screen, and scanned by a Typhoon imager (GE Healthcare). Quantitations were carried out using ImageJ software. The Ku DNA-binding assay was carried out in the same buffer as the nuclease assays for 15 min on ice. The reactions were mixed with 4 µL of loading dye (50% glycerol, 1 mg/mL bromophenol blue) and separated by native electrophoresis in 6% polyacrylamide gels (acrylamide:bisacrylamide 19:1; Bio-Rad) in TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA). The gels were dried and analyzed as above.
Supplementary Material
Acknowledgments
We thank members of the Cejka laboratory for discussions and comments on the manuscript. This work was supported by grants from the Swiss National Science Foundation and the European Research Council to P.C.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.308254.117.
References
- Aparicio T, Baer R, Gottesman M, Gautier J. 2016. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J Cell Biol 212: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz SK, Dickson AM, Haring SJ, Wold MS. 2006. Functional assays for replication protein A (RPA). Methods Enzymol 409: 11–38. [DOI] [PubMed] [Google Scholar]
- Blier PR, Griffith AJ, Craft J, Hardin JA. 1993. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem 268: 7594–7601. [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Manfrini N, Lucchini G, Longhese MP. 2010. The MRX complex plays multiple functions in resection of Yku- and Rif2-protected DNA ends. PLoS One 5: e14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Cejka P. 2014. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature 514: 122–125. [DOI] [PubMed] [Google Scholar]
- Cannavo E, Cejka P, Kowalczykowski SC. 2013. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11–Rad50–Xrs2 to DNA end resection. Proc Natl Acad Sci 110: E1661–E1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P. 2015. DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J Biol Chem 290: 22931–22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Kowalczykowski SC. 2010. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J Biol Chem 285: 8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. 2010. DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2. Nature 467: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. 2008. The Yku70–Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 9: 810–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SS, Balestrini A, Petrini JH. 2011. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol Cell Biol 31: 4379–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ. 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. 1993. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72: 131–142. [DOI] [PubMed] [Google Scholar]
- Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsusui K, et al. 2016. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol Cell 64: 580–592. [DOI] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Kleckner N. 1995. Covalent protein–DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc Natl Acad Sci 92: 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC. 2015. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol 7: a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I, Keeney S. 2014. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 7: a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P. 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet 7: e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R. 2009. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 8: 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS. 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29: 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Yamada S, Keeney S. 2017. A global view of meiotic double-strand break end resection. Science 355: 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Morgan EA, Symington LS. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159: 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. 2010. Mre11–Rad50–Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol 17: 1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Al-Zain A, Cannavo E, Cejka P, Symington LS. 2016. Xrs2 dependent and independent functions of the Mre11-Rad50 complex. Mol Cell 64: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, et al. 2014. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell 53: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 29: 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Erdeniz N, Dudley S, Liskay RM. 2002. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst) 1: 895–912. [DOI] [PubMed] [Google Scholar]
- White CI, Haber JE. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J 9: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Topper LM, Wilson TE. 2008. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.