Leong et al. identified ARID5B as a collaborating oncogenic factor involved in the transcriptional program in T-ALL. ARID5B positively regulates the expression of TAL1 and its regulatory partners and also activates the expression of the oncogene MYC.
Keywords: T-cell acute lymphoblastic leukemia, TAL1, ARID5B, MYC, core regulatory circuit
Abstract
The oncogenic transcription factor TAL1/SCL induces an aberrant transcriptional program in T-cell acute lymphoblastic leukemia (T-ALL) cells. However, the critical factors that are directly activated by TAL1 and contribute to T-ALL pathogenesis are largely unknown. Here, we identified AT-rich interactive domain 5B (ARID5B) as a collaborating oncogenic factor involved in the transcriptional program in T-ALL. ARID5B expression is down-regulated at the double-negative 2–4 stages in normal thymocytes, while it is induced by the TAL1 complex in human T-ALL cells. The enhancer located 135 kb upstream of the ARID5B gene locus is activated under a superenhancer in T-ALL cells but not in normal T cells. Notably, ARID5B-bound regions are associated predominantly with active transcription. ARID5B and TAL1 frequently co-occupy target genes and coordinately control their expression. ARID5B positively regulates the expression of TAL1 and its regulatory partners. ARID5B also activates the expression of the oncogene MYC. Importantly, ARID5B is required for the survival and growth of T-ALL cells, and forced expression of ARID5B in immature thymocytes results in thymus retention, differentiation arrest, radioresistance, and tumor formation in zebrafish. Our results indicate that ARID5B reinforces the oncogenic transcriptional program by positively regulating the TAL1-induced regulatory circuit and MYC in T-ALL, thereby contributing to T-cell leukemogenesis.
T-cell acute lymphoblastic leukemia (T-ALL) is a malignant disorder of thymic T-cell precursors (Look 1997; Armstrong and Look 2005; Aifantis et al. 2008; Belver and Ferrando 2016). T-ALL accounts for ∼15% and 25% of ALL cases in childhood and adulthood, respectively (Pui et al. 2008; Inaba et al. 2013). One of the most prevalent oncogenes in T-ALL is TAL1 (also known as SCL), a class II basic helix–loop–helix (bHLH) family transcription factor (Begley et al. 1989; Brown et al. 1990; Ferrando and Look 2000). TAL1 is normally expressed in hematopoietic stem cells (HSCs) and progenitor cells (Herblot et al. 2000; Lecuyer and Hoang 2004), whereas it is silenced during T-cell development. In contrast, TAL1 is ectopically overexpressed in 40%–60% of T-ALL cases by chromosomal translocation, intrachromosomal rearrangement, or mutations in the enhancer region (Begley et al. 1989; Chen et al. 1990; Janssen et al. 1993; Mansour et al. 2014). TAL1 forms a large transcriptional complex with E proteins (HEB and E2A), LMO family proteins (LMO1 and LMO2), LDB1, and GATA family proteins (GATA2 and GATA3) in normal HSCs and T-ALL cells (Hsu et al. 1991; Wadman et al. 1997; Rabbitts 1998). Several transcription factors, including RUNX1, MYB, and ETS family proteins, have also been implicated as regulatory partners of TAL1 (Nottingham et al. 2007; Landry et al. 2008). We reported previously that TAL1, GATA3, RUNX1, and MYB positively regulate each other in T-ALL cells, thus forming an autoregulatory loop (Sanda et al. 2012; Mansour et al. 2014). TAL1 target genes are also co-occupied by these factors, which coordinately regulate the expression of the target genes along with TAL1 (Sanda et al. 2012). These mechanisms are essentially similar to those of the transcriptional regulatory motifs previously identified in normal HSCs (Wilkinson and Gottgens 2013). However, the critical factors regulated by the TAL1 complex in T-ALL cells are largely unknown.
Recent advances in genomic research have elucidated the mutational landscape of many cancers. One family of DNA-binding factors that is frequently dysregulated across different types of cancer is the AT-rich interactive domain (ARID) family (Lin et al. 2014). The ARID of these proteins binds directly to DNA (Wilsker et al. 2002). Several ARID proteins also contain the Jumonji (jmj) domain, which has essential roles in transcription and histone modification. To date, 15 ARID family members have been identified in humans, and they are further classified into subfamilies based on the similarity of the ARID. For example, ARID1A is a subunit of the SWI/SNF chromatin remodeling complex, which is widely expressed in many human tissues (Guan et al. 2011). Accumulating evidence indicates that ARID1A is frequently mutated in a number of different cancers, including ovarian cancer, uterine cancer, gastric cancer, and hepatocellular carcinoma (Wang et al. 2004; Wu and Roberts 2013). Functional analyses have indicated that ARID1A acts as a tumor suppressor that is essential for regulating cell cycle progression (Nagl et al. 2005; Guan et al. 2012; Wu et al. 2014). In contrast, Jumonji ARID1A (JARID1A; also known as KDM5A) has been implicated as a potential oncogene. JARID1A regulates gene transcription by demethylating histone H3 at Lys4 (Christensen et al. 2007). A fusion protein that consists of JARID1A and nucleoporin-98 (NUP98) has been reported in patients with hematopoietic malignancies (van Zutven et al. 2006). This potent oncogenic protein induces deregulation of specific histone modifications, thus leading to hematopoietic differentiation arrest and acute myeloid leukemia (AML) in mouse models (Wang et al. 2009).
Another example is ARID5B, an ARID family member that recognizes the core DNA sequence motif AAT(C/T) (Whitson et al. 1999). This protein has been known to play a role in the cell growth and differentiation of B-lymphocyte progenitors (Lahoud et al. 2001). Several groups have shown that ARID5B interacts with the histone demethylase PHD finger protein 2 (PHF2) and histone deacetylases (HDACs) (Baba et al. 2011; Hata et al. 2013; Joshi et al. 2013). Recent genome-wide association studies have revealed that several single-nucleotide polymorphisms (SNPs) within the ARID5B gene are significantly associated with risk for B-cell ALL (B-ALL) (Papaemmanuil et al. 2009; Trevino et al. 2009). However, the detailed molecular functions of ARID5B and its roles in normal T-cell development and leukemogenesis have not yet been elucidated.
Here, we report that ARID5B is a critical transcriptional target of the TAL1 complex in T-ALL and plays important roles in the transcriptional regulatory program and T-cell leukemogenesis. The ARID5B gene is directly regulated by TAL1 under a superenhancer (SE), and its expression is associated with TAL1. ARID5B frequently co-occupies its target genes with the TAL1 complex, which positively regulates target gene expression. Additionally, ARID5B promotes the expression of the oncogene MYC. Forced expression of ARID5B in immature thymocytes results in thymus retention, differentiation arrest, radioresistance, and tumor formation in zebrafish.
Results
The TAL1 complex positively regulates the ARID5B gene in T-ALL cells
In our previous study, we identified the genome-wide occupancies of TAL1 and its regulatory partners (E2A, HEB, LMO1, GATA3, RUNX1, and MYB) in T-ALL cells by chromatin immunoprecipitation (ChIP) combined with sequencing (ChIP-seq) and microarray analyses (Sanda et al. 2012). In the present study, we sought to identify critical downstream targets that are abnormally activated by all of these factors in T-ALL cells. For this purpose, we newly performed an RNA sequencing (RNA-seq) analysis to more comprehensively analyze gene expression profiles.
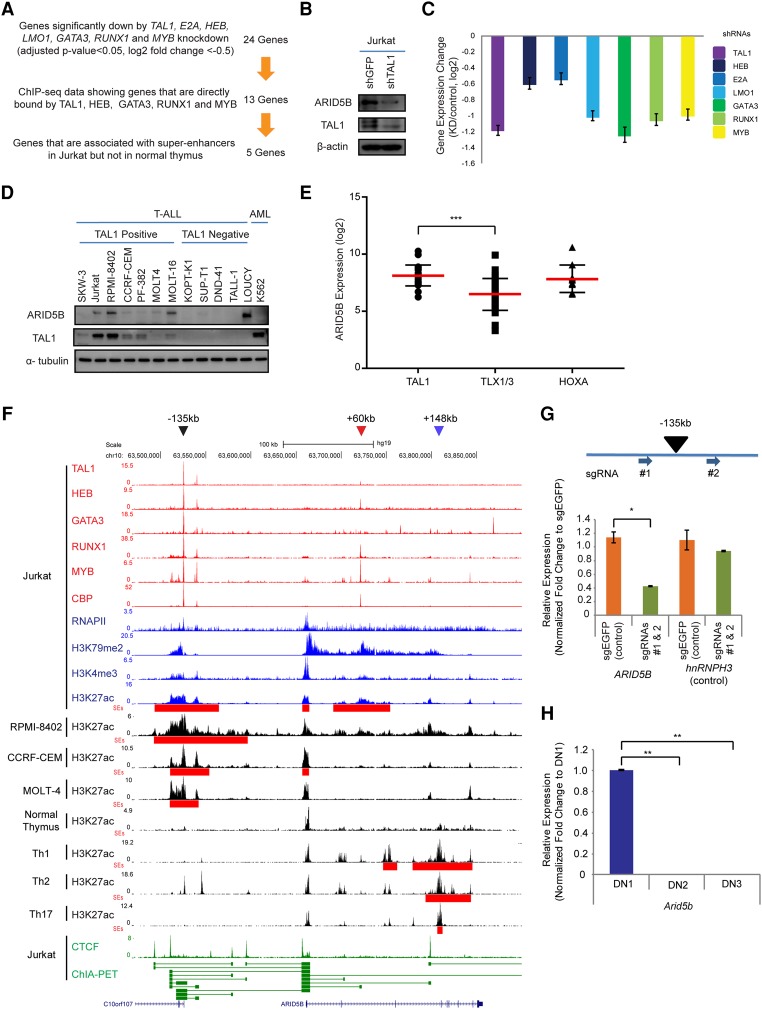
First, we genetically knocked down TAL1 and each of its regulatory partners by the lentivirus-mediated delivery of shRNA in a T-ALL cell line (Jurkat). We selected genes that were significantly down-regulated after the knockdown of each of the seven factors (TAL1, E2A, HEB, LMO1, GATA3, RUNX1, and MYB) based on the criteria of an adjusted P-value of <0.05 and a log2 fold change of less than −0.5 between two control samples and two knockdown samples for each factor. A total of 24 genes was commonly regulated by all seven factors (Fig. 1A). We then filtered for genes that were identified to be directly bound by TAL1, HEB, GATA3, RUNX1, and MYB in our ChIP-seq analysis. The ChIP-seq data for LMO1 and E2A were not considered owing to the poor quality of the antibodies. Through these criteria, 13 genes were selected, including ALDH1A2 and NKX3-1, which have been reported previously as TAL1 targets in T-ALL cells (Supplemental Fig. S1A; Ono et al. 1998; Kusy et al. 2010). Among these genes, five were associated with SEs—which are clusters of enhancers determined by ChIP-seq analysis for histone H3 Lys27 acetylation (H3K27ac) (Hnisz et al. 2013; Loven et al. 2013; Whyte et al. 2013)—in Jurkat cells but not in the normal human thymus. This selection identified candidate genes that may be aberrantly activated by the TAL1 complex in T-ALL cells.
Figure 1.
The TAL1 complex positively regulates the ARID5B gene in T-ALL cells. (A) Filtering criteria for identifying genes directly activated by the TAL1 transcriptional complex and associated with the SE in the T-ALL cell line Jurkat. We first selected 24 genes that are commonly regulated by all seven transcription factors involved in the TAL1 complex. (B) Western blot analysis for protein expression of ARID5B, TAL1, and β-actin (loading control) in Jurkat cells on day 3 after shRNA-expressing lentivirus infection. (C) mRNA expression level of ARID5B after knockdown of TAL1, HEB, E2A, LMO1, GATA3, RUNX1, and MYB in Jurkat cells analyzed by RNA-seq. (D) Western blot analysis for protein expression of ARID5B, TAL1, and α-tubulin (loading control) in T-ALL and AML cell lines. (E) The mRNA expression level of ARID5B in primary T-ALL cases analyzed by microarray analysis using a publicly available data set (Homminga et al. 2011). T-ALL cases were classified into subgroups based on the expression of transcription factors (TAL1, TLX1/3, and HOXA). (***) P < 0.001 by two-sample two-tailed t-test. (F) ChIP-seq gene tracks showing the binding locations of the TAL1 complex (TAL1, HEB, GATA3, RUNX1, and MYB), the coactivator protein CBP, and RNA polymerase II (RNAPII) at the genomic region around the ARID5B gene in a T-ALL cell line (Jurkat). ChIP-seq data for H3K79me2 and H3K4me3 were used to represent transcription initiation and elongation, respectively. H3K27ac and SEs (red bars) for T-ALL cell lines (Jurkat, RPMI-8402, CCRF-CEM, and MOLT-4) and normal T cells (thymus; Th1, Th2, and Th17) are shown. The ChIP-seq profiles of CTCF and cohesin in Jurkat were analyzed using a chromatin–chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) interaction data set reported by Hnisz et al. (2016). The horizontal green lines linking two bars illustrate a chromatin–chromatin interaction. Black, red, and blue arrowheads indicate SEs around the −135-kb, +60-kb, and +148-kb regions, respectively. (G) Single-guide RNAs (sgRNAs: #1 and #2) were designed to knock out the −135-kb element and deliver it by lentiviral infection into Jurkat cells. The mRNA expression of ARID5B and its neighboring gene, hnRNPH3 (control), in knockout cells was measured on day 6 after lentivirus infection by quantitative RT–PCR (qRT–PCR) analysis. The relative gene expression was normalized to GAPDH expression. (*) P < 0.05 by two-sample two-tailed t-test. (H) Expression of the mouse Arid5b gene in normal thymocytes at different stages: double-negative 1 (DN1; CD44+CD25−), DN2 (CD44+CD25+), and DN3 (CD44−CD25+). The mRNA expression of Arid5B was analyzed by qRT–PCR and normalized to β-actin expression. Fold change values compared with the expression in DN1 cells are shown as the mean ± standard deviation (SD) of duplicate samples. (**) P < 0.01 by two-sample two-tailed t-test.
Interestingly, the shortlisted genes included ARID5B, which has not been implicated previously in T-ALL. ARID5B is positively regulated by the TAL1 complex because knockdown of TAL1 down-regulated ARID5B protein expression in T-ALL cells (Jurkat) (Fig. 1B). Knockdown of the TAL1 regulatory partners also resulted in a significant down-regulation of ARID5B expression at the mRNA level (Fig. 1C). The protein expression of ARID5B was associated with TAL1 expression in T-ALL cell lines (Fig. 1D). We next analyzed ARID5B expression among different T-ALL subgroups using a data set for primary T-ALL samples (Homminga et al. 2011). This result demonstrated that the mRNA expression of ARID5B was significantly higher in TAL1-positive T-ALL cases than in TLX1/3-positive cases (Fig. 1E), which compose another major subgroup of T-ALL (Belver and Ferrando 2016). ARID5B was also highly expressed in HOXA-positive cases. Indeed, one cell line (LOUCY), which has been known to express HOXA genes (Van Vlierberghe et al. 2008), expressed a high level of ARID5B but not TAL1 (Fig. 1D). These results indicated that ARID5B is positively regulated by TAL1 in T-ALL cells and can also be activated independently of TAL1 in HOXA-positive cases.
The enhancer located 135 kb upstream of the ARID5B gene is activated in T-ALL
Importantly, the ARID5B gene locus was highly activated in Jurkat cells, as shown by active histone marks (H3K79me2, H3K4me3, and H3K27ac) and the DNA binding of a coactivator protein, CBP, as well as RNA polymerase II (RNAPII) (Fig. 1F). An element located ∼135 kb upstream of the ARID5B gene (“−135 kb”) was found to be bound by TAL1 and its regulatory partners (Fig. 1F, black arrowhead). This locus was associated with a SE in multiple TAL1-positive T-ALL cell lines (Jurkat, RPMI-8402, CCRF-CEM, and MOLT-4) but not in the normal human thymus or CD4+ single-positive (SP) T cells (Th1, Th2, and Th17) (Fig. 1F, red bars). An additional SE was located within the ARID5B gene around the +60-kb region in Jurkat (Fig. 1F, red arrowhead), but this locus was very weakly bound by TAL1 or GATA3. There was another SE within the ARID5B gene around the +148-kb region that was observed specifically in CD4+ SP T cells but not in T-ALL cells. Importantly, a chromatin–chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) for the cohesin protein demonstrated a cis-regulatory interaction between the −135-kb element and the ARID5B transcriptional start site (TSS) (Fig. 1F, bottom, green line). In fact, genetic knockout of the −135-kb element by the CRISPR/Cas9 method resulted in a marked down-regulation of ARID5B gene expression (Fig. 1G; Supplemental Fig. S1B–E). Thus, these results indicate that the TAL1-bound region at −135 kb is the enhancer controlling ARID5B expression in T-ALL cells.
In agreement with this result, an analysis using the Gene Expression Commons database (Seita et al. 2012) demonstrated that mouse Arid5b expression was down-regulated in the CD4−CD8− double-negative 2–4 (DN2–4) stages of immature T cells, whereas it was expressed in progenitor cells, DN1, CD4+CD8+ double-positive (DP), and SP mature T cells (Supplemental Fig. S1F). We independently validated this result and confirmed that Arid5b was down-regulated during the transition from DN1 to DN2 in mice (Fig. 1H). Additionally, we analyzed an RNA-seq data set for different stages of human hematopoietic cells (Casero et al. 2015) and found that mRNA expression of human ARID5B was also down-regulated at DN stage compared with DP and SP stages (Supplemental Fig. S1G). Together, these results indicated that ARID5B is normally down-regulated at DN2–4 stages of thymocytes, while it is aberrantly activated by the TAL1 complex in T-ALL.
ARID5B overexpression supports the survival of TAL1-positive T-ALL cells
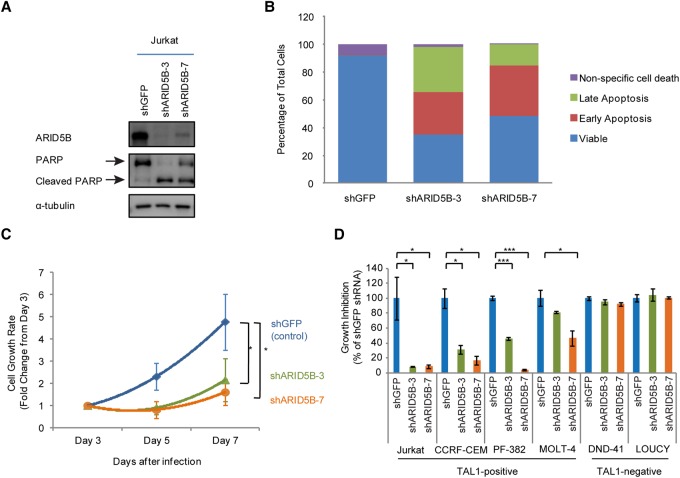
We next examined whether ARID5B has an oncogenic function that is required for cell growth, survival, or proliferation in T-ALL cells. To analyze phenotypes after the loss of the ARID5B protein, we selected two different shRNAs (#3 and #7) specifically targeting the ARID5B gene and determined the ARID5B protein level after lentiviral transduction (Fig. 2A). Both shRNAs decreased the expression of the ARID5B protein compared with the control shRNA (shGFP). We next measured apoptosis and cell viability upon gene knockdown. A Western blot analysis showed an increase in the cleaved form of PARP (Fig. 2A), which is a marker of apoptosis. In agreement with this finding, we also detected an increase in the cell population undergoing early and late apoptosis based on annexin V staining (Fig. 2B). Analysis of the cell viability over 7 d after shRNA-expressing lentivirus transduction showed that ARID5B knockdown inhibited the growth rate of Jurkat cells (Fig. 2C). Overexpression of the anti-apoptotic protein BCL2 restored cell viability (Supplemental Fig. S2A) and completely rescued apoptosis (Supplemental Fig. S2B) in Jurkat cells after ARID5B knockdown, thus confirming that the growth-inhibiting effect after ARID5B knockdown was due primarily to apoptotic cell death. A cell cycle analysis revealed an increase in arrested cells at the sub-G1 phase with a decrease in the number of cells in the G1, S, and G2/M phases after ARID5B knockdown (Supplemental Fig. S2C).
Figure 2.
ARID5B expression supports the survival of TAL1-positive T-ALL cells. (A) Western blot analysis for protein expression of ARID5B, PARP, and α-tubulin (loading control) in Jurkat cells on day 3 after the transduction of lentivirus expressing shGFP (control), shARID5B-3, or shARID5B-7. (B) Detection of apoptosis based on annexin V by flow cytometry analysis in Jurkat cells on day 3 after transduction with lentivirus expressing shRNA. (Early apoptosis) Annexin V-positive and propidium iodide (PI)-negative; (late apoptosis) annexin V-positive and PI-positive; (nonspecific cell death) annexin V-negative and PI-positive. The data represent the mean of duplicate samples. (C) Cell viability of Jurkat cells was measured by CellTiter Glo assay at days 3, 5, and 7 after lentivirus infection. The cell growth rate (fold change) compared with day 3 is shown as the mean ± SD of triplicate samples. (*) P < 0.05 by two-sample two-tailed t-test. (D) Growth inhibition of Jurkat, CCRF-CEM, PF-382, MOLT-4, DND-41, and LOUCY cells on day 7 after lentivirus infection. Growth inhibition was calculated based on cell viability on day 7 compared with day 3. The data are shown as the mean ± SD of triplicate samples. (*) P < 0.05; (***) P < 0.001 by two-sample two-tailed t-test.
To determine whether these phenotypes are shared among other T-ALL cell lines, we analyzed cell viability after shRNA knockdown in multiple T-ALL cell lines (Jurkat, CCRF-CEM, PF-382, MOLT-4, DND-41, and LOUCY). Importantly, significant growth inhibition was observed after ARID5B knockdown in four cell lines (Jurkat, CCRF-CEM, PF-382, and MOLT-4) (Fig. 2D) expressing both the ARID5B and TAL1 proteins (Fig. 1D). This phenotype was not observed in DND-41 cells, which do not express either ARID5B or TAL1. It is also noteworthy that LOUCY cells, which express ARID5B and HOXA but not TAL1 (Fig. 1D), were insensitive to ARID5B knockdown. In agreement with the results described above, we observed apoptotic cell death in T-ALL cell lines that were inhibited after ARID5B knockdown (Supplemental Fig. S2D).
ARID5B bindings are associated predominantly with active transcription
We next examined the mode of action by which activation of ARID5B affects transcription in T-ALL cells. Another group had reported previously that ARID5B interacts with HDAC1 and HDAC2 proteins, thus epigenetically silencing gene expression in other cell types (Joshi et al. 2013). In fact, we were able to detect the binding of ARID5B to HDAC1 and HDAC2 but not to HDAC3 or HDAC4 in Jurkat cells (Supplemental Fig. S3A). Several genes that have been known to be epigenetically silenced by HDACs, including the tumor suppressors EGR1, EGR2, and CDKN1A, were up-regulated after ARID5B knockdown (Supplemental Fig. S3B). This up-regulation was also observed after treatment with a small-molecule HDAC inhibitor (SAHA) that inhibits both HDAC1 and HDAC2 (Supplemental Fig. S3C). Thus, this result suggested that ARID5B is possibly involved in gene silencing in T-ALL cells.
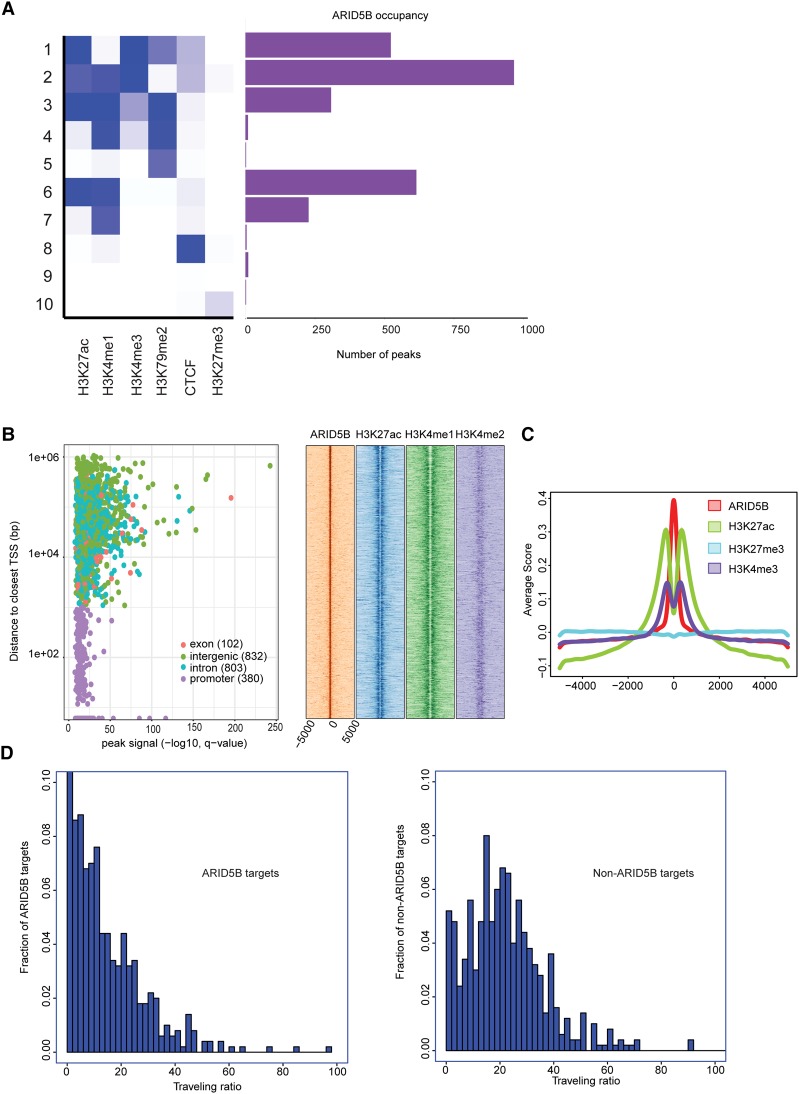
To comprehensively analyze the association between DNA binding of ARID5B and transcriptional status, we performed a ChIP-seq analysis of the ARID5B protein in Jurkat cells. Surprisingly, we found that DNA binding of ARID5B often coexisted with active histone marks. Classification of chromatin state, defined by the combination of various histone marks, indicated that active histone marks such as H3K27ac, H3K4me1, and H3K4me3 were found at many ARID5B-bound regions (Fig. 3A). Approximately 18% (380 out of 2117 peaks) of ARID5B bindings were found in promoter regions, while higher ARID5B peak signals were found in nonpromoter regions, including exons, introns, and intergenic regions (Fig. 3B, left). The density plot analysis for nonpromoter regions revealed that ARID5B-bound regions were frequently present with H3K27ac and H3K4me1 signals (Fig. 3B, right). The metagene analysis also demonstrated the enrichment of those active histone marks, but not of a repressive mark (H3K27me3), at the ARID5B-bound loci in T-ALL cells (Fig. 3C).
Figure 3.
ARID5B-bound regions are associated predominantly with active transcription. (A) Ten different chromatin states were first defined using published ChIP-seq data sets for various histone marks and CTCF in Jurkat cells. The number of peaks of ARID5B for each state is shown. (B) Each region bound by ARID5B was first mapped to the closest gene and classified into one of four categories: promoter, exon, intron, or intergenic. (Left) A scatter plot shows all ARID5B-bound regions plotted by distance from the closest TSS (Y-axis) and ARID5B signals (X-axis). (Right) The density plots show distributions (base pairs) of H3K27ac, H3K4me1, and H3K4me2 at nonpromoter regions bound by ARID5B in Jurkat cells. (C) Metagene analysis showing distribution (base pairs) of H3K27ac, H3K27me3, and H3K4me3 at the ARID5B-bound nonpromoter regions in Jurkat cells. (D) Histograms were made for the fraction of ARID5B targets/nontargets with the given traveling ratio values for the 500 active genes with the highest ARID5B signals (left) and the 500 active genes with the lowest ARID5B signals (right).
Additionally, we analyzed the association of ARID5B binding with the transcriptional state based on the occupancy of RNAPII. We measured the “traveling ratio” (TR), which compares RNAPII density at the promoter region with that at the gene body (Rahl et al. 2010). In principle, a lower TR value represents a higher level of RNAPII binding along the gene body, thus indicating that the gene undergoes transcription elongation. We first selected “active” genes that are bound by RNAPII with H3K4me3 and H3K79me2 marks and then classified them into two groups: one with the highest ARID5B signals (ARID5B targets) and the other with the lowest ARID5B signals (non-ARID5B targets) (Supplemental Fig. S3D). Interestingly, ARID5B targets showed lower TR values than non-ARID5B targets (P-value < 2.2 × 10−16 by the Kolmogorov-Smirnov test) (Fig. 3D). This result further suggested that ARID5B binding is associated with active transcription in T-ALL cells.
Of note, knockdown of ARID5B did not significantly change the total level of H3K27ac (Supplemental Fig. S3E). This suggested that ARID5B may affect transcriptional status at specific target sites. ARID5B has been also reported to interact with the histone demethylase PHF2, which removes a repressive histone mark (Baba et al. 2011). However, we could not detect binding between ARID5B and PHF2 proteins in an endogenous setting in Jurkat cells (Supplemental Fig. S3F) or in 293T cells after cotransfection (Supplemental Fig. S3G). This result suggested that the binding partners of ARID5B could be different depending on the cell type.
ARID5B transcriptionally activates the oncogene MYC in T-ALL cells
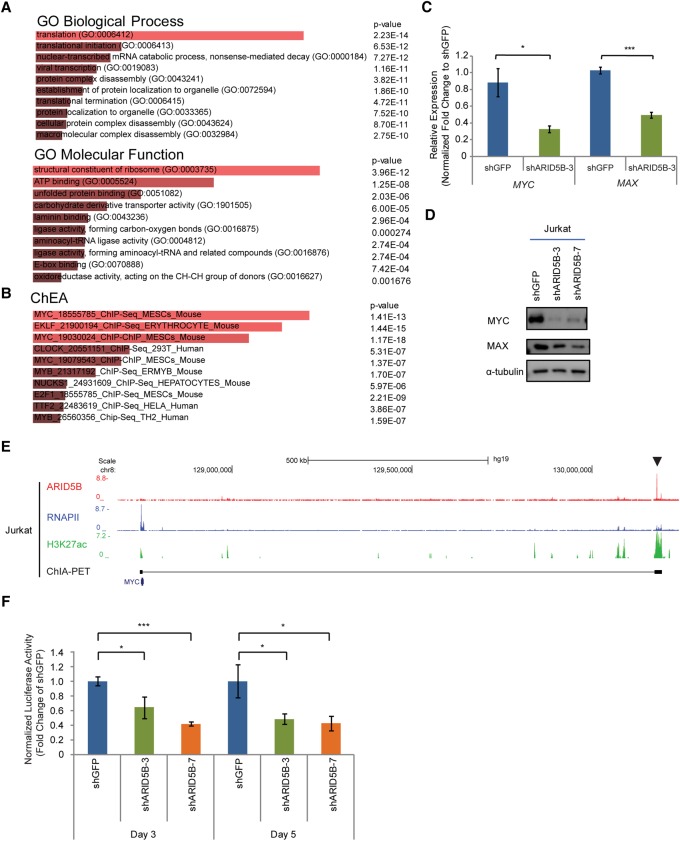
Our results suggested that ARID5B serves as a transcriptional activator in T-ALL cells. Hence, we next aimed to identify target genes positively regulated by ARID5B. We performed an RNA-seq analysis in Jurkat cells transduced with control or ARID5B shRNAs by lentiviral infection. We harvested RNAs at an early time point (day 3 after virus infection) to reduce indirect effects caused by changes in downstream factors or due to cell death. We first selected all genes that were significantly down-regulated by ARID5B knockdown (those with an adjusted P-value of <0.05 and a log2 fold change of less than −0.5 between two control and two knockdown samples) (see Supplemental Table S1 for down-regulated and up-regulated genes). A gene ontology (GO) analysis with the Enrichr tool (Chen et al. 2013) revealed that genes down-regulated by ARID5B knockdown were significantly enriched in the categories of protein translation, synthesis, and ribosomal genes (Fig. 4A; Supplemental Table S2). We next compared the same set of genes with targets of various transcription factors determined by a ChIP enrichment analysis (ChEA) (Chen et al. 2013). Interestingly, genes that were down-regulated by ARID5B knockdown were enriched in MYC target genes (Fig. 4B). Notably, these GO and ChEA categories were not identified in a similar analysis for genes that were positively regulated by TAL1 or its regulatory partners (GATA3, RUNX1, and MYB) at the same time point that we analyzed (Supplemental Fig. S4A; Supplemental Table S2). Hence, our results suggested that these signatures could be unique to ARID5B knockdown.
Figure 4.
ARID5B transcriptionally activates the oncogene MYC in T-ALL cells. (A,B) GO analysis (A) and ChEA (B) were performed in the Enrichr program using 1202 genes that were significantly down-regulated after ARID5B knockdown (with an adjusted P-value of <0.05 and a log2 fold change of less than −0.5 between two control and two knockdown samples). The top 10 terms ranked by combined score are shown. (C) mRNA expression of MYC and MAX in Jurkat cells on day 3 after ARID5B knockdown was measured by qRT–PCR analysis. The relative gene expression was normalized to the ERCC spike-in exogenous control (E130). The data represent the mean ± SD of duplicate samples. (*) P < 0.05; (***) P < 0.001 by two-sample two-tailed t-test. (D) Western blot analysis for protein expression of MYC, MAX, and α-tubulin (loading control) in Jurkat cells on day 3 after lentivirus transduction. (E) The ChIP-seq gene tracks represent the binding of ARID5B and RNAPII, the presence of H3K27ac, and chromatin interactions determined by ChIA-PET (cohesin) analysis at the NOTCH1-driven MYC enhancer (N-Me) region (arrowhead) in Jurkat cells. See the legend for Figure 1F for details. (F) Luciferase activity driven by the N-Me was measured after ARID5B knockdown in Jurkat cells. The luciferase activity on days 3 and 5 was normalized to shGFP (control). The data represent the mean ± SD of duplicate samples. (*) P < 0.05; (***) P < 0.001 by two-sample two-tailed t-test.
Indeed, we confirmed that the expression levels of MYC and its partner, MAX, were decreased at the mRNA and protein levels in T-ALL cells after ARID5B knockdown (Fig. 4C,D). A similar result was observed in BCL2-overexpressing cells (Supplemental Fig. S4B), thus indicating that the loss of MYC expression was not caused nonspecifically by cell death. Stable genetic knockout of ARID5B by the CRISPR/Cas9 method also resulted in a decrease in MYC expression to some extent (Supplemental Fig. S4C). A ChIP-seq analysis of the ARID5B protein showed that ARID5B binds directly to the NOTCH-driven MYC enhancer (N-Me), which has been reported previously to be activated by NOTCH1 in T-ALL cells (Fig. 4E, arrowhead). Validation with ChIP-PCR demonstrated that ARID5B binds at the same element in multiple T-ALL cell lines (Supplemental Fig. S4D). Knockdown of ARID5B slightly reduced the H3K27ac signal around the N-Me locus (Supplemental Fig. S4E). Analysis of transcriptional activity using a luciferase reporter assay with the N-Me sequence also demonstrated that ARID5B knockdown significantly inhibited the enhancer activity (Fig. 4F). Hence, these results indicated that ARID5B directly activates the expression of MYC in T-ALL.
Additionally, MYC overexpression partially rescued growth inhibition in Jurkat cells after ARID5B knockdown (Supplemental Fig. S4F). LOUCY cells, which are insensitive to ARID5B knockdown (Fig. 2D), do not express MYC protein (Supplemental Fig. S4G). Hence, these results suggested that up-regulation of MYC partially explained the mechanism of dependence on ARID5B for T-ALL cell growth and survival. It is noteworthy that the mouse Myc gene is normally upregulated in thymocytes after the DN3 stage (Supplemental Fig. S4H), while Arid5b is down-regulated at this stage (Fig. 1H; Supplemental Fig. S1F). Therefore, ARID5B is unlikely to be required for MYC expression at the DN3 stage during normal T-cell development but can activate MYC when overexpressed in human T-ALL cells.
ARID5B regulates the expression of TAL1 target genes in T-ALL cells
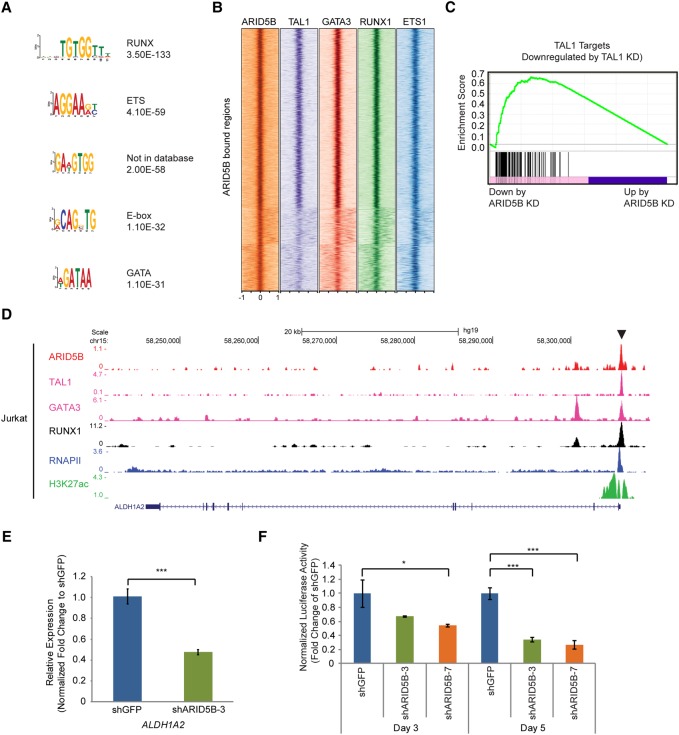
We next examined potential collaborating transcription factors sharing the same target loci. We analyzed transcription factor-binding motifs that were significantly enriched within 500 base pairs (bp) of the peak of ARID5B-binding sites in Jurkat cells. Interestingly, RUNX, ETS, E-box, and GATA motifs were identified (Fig. 5A). An analysis of these transcription factor-binding regions indicated that ARID5B target sites were frequently co-occupied by TAL1, GATA3, RUNX1, or ETS1 in T-ALL cells (Fig. 5B). A gene set enrichment analysis (GSEA) showed that many of the TAL1 target genes were also significantly down-regulated by ARID5B knockdown (Fig. 5C; Supplemental Fig. S5). For example, ARID5B binds and regulates the ALDH1A2 gene (Fig. 5D), which is a representative TAL1 target in T-ALL (Supplemental Fig. S1A). ARID5B knockdown significantly down-regulated ALDH1A2 expression (Fig. 5E). A luciferase reporter assay using the ALDH1A2 enhancer sequence demonstrated that knockdown of ARID5B led to a decrease in the activity of this enhancer over 5 d (Fig. 5F), thus indicating that ARID5B activates this TAL1-bound enhancer. These results indicated that ARID5B and the TAL1 complex coordinately regulate a very similar set of genes in T-ALL cells.
Figure 5.
ARID5B coordinately regulates the expression of TAL1 targets in T-ALL cells. (A) DNA-binding motifs significantly enriched in regions bound by ARID5B in Jurkat cells are shown with P-values. (B) Co-occupancy by TAL1, GATA3, RUNX1, and MYB at the ARID5B-bound gene loci (±1 kb from the binding sites) in Jurkat cells was analyzed by ChIP-seq. (C) A GSEA plot showing the overall pattern of gene expression changes of TAL1 target genes upon ARID5B knockdown (KD) in Jurkat cells. Each solid bar represents one gene within the gene set. (D) ChIP-seq gene tracks show the binding of ARID5B, TAL1, GATA3, RUNX1, and RNAPII and the presence of H3K27ac at the ALDH1A2 promoter in Jurkat cells. See the legend for Figure 1F for details. (E) mRNA expression of ALDH1A2 in Jurkat cells on day 3 after ARID5B knockdown was measured by qRT–PCR. The data represent the mean ± SD of duplicate samples. (***) P < 0.001 by two-sample two-tailed t-test. (F) Luciferase activity driven by the ALDH1A2 promoter was measured after ARID5B knockdown in Jurkat cells. The luciferase activity on days 3 and 5 was normalized to shGFP (control). The data represent the mean ± SD of duplicate samples. (*) P < 0.05, (***) P < 0.001 by two-sample two-tailed t-test.
ARID5B regulates the expression of the TAL1 complex in T-ALL cells
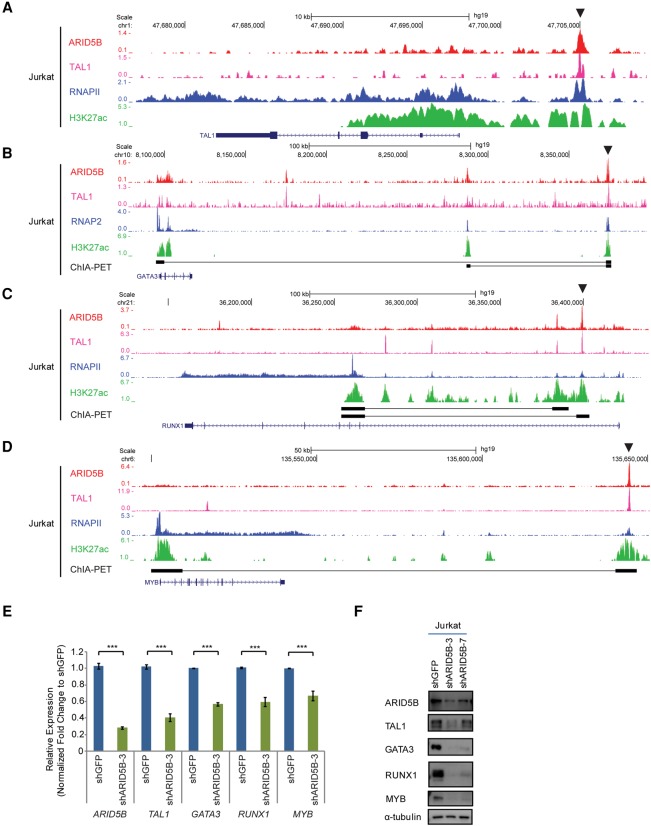
Interestingly, we also observed that ARID5B regulates the expression of four major components of the TAL1 complex (namely, TAL1, GATA3, RUNX1, and MYB) in Jurkat cells. ARID5B and TAL1 co-occupy the regulatory elements of these genes in Jurkat cells, as determined by ChIP-seq analysis (Fig. 6A–D). We validated this result in other cell lines by ChIP-PCR (Supplemental Fig. S6A–D). Knockdown of ARID5B resulted in reductions of the H3K27ac signals at those enhancer loci (Supplemental Fig. S6E–H) and down-regulation of all four factors at the mRNA (Fig. 6E) and protein levels (Fig. 6F). Genetic knockout of ARID5B by the CRISPR/Cas9 method also produced a similar result (Supplemental Fig. S6I). ARID5B knockdown in BCL2-overexpressing cells also showed down-regulation of TAL1 (Supplemental Fig. S6J), thus indicating that this was not caused nonspecifically by cell death. These results indicate that ARID5B can positively regulate the expression of the TAL1 complex and downstream target genes.
Figure 6.
ARID5B positively regulates the expression of the TAL1 complex in T-ALL cells. (A–D) ChIP-seq gene tracks showing the binding of ARID5B, TAL1, and RNAPII as well as the H3K27ac signal at the enhancer/promoter regions of TAL1 (A), GATA3 (B), RUNX1 (C), and MYB (D) in Jurkat cells. See the legend for Figure 1F for details. (E) mRNA expression of ARID5B, TAL1, GATA3, RUNX1, and MYB in Jurkat cells on day 3 after ARID5B knockdown was measured by qRT–PCR. The relative gene expression was normalized to the ERCC spike-in exogenous control (E130). The data represent the mean ± SD of duplicate samples. (***) P < 0.001 by two-sample two-tailed t-test. (F) Western blot analysis for protein expression of ARID5B, TAL1, GATA3, RUNX1, MYB, and α-tubulin (loading control) in Jurkat cells on day 3 after infection with lentivirus expressing shRNA.
Overexpression of ARID5B leads to thymus retention and the development of T-cell lymphoma in zebrafish
Our results in T-ALL cell lines strongly suggest that ARID5B plays an oncogenic role in T-ALL cell growth and survival by regulating the gene expression program. This result prompted us to analyze the tumorigenic ability of ARID5B in vivo using animal models. The ARID5B gene sequence is fairly well conserved between humans and zebrafish (Supplemental Fig. S7A). Therefore, we transduced the human ARID5B gene with a fluorescent marker (mCherry) specifically into the zebrafish thymus under the rag2 promoter (Supplemental Fig. S7B). After successfully generating founder lines in which the human ARID5B gene was transferred into germline tissues (Supplemental Fig. S7C), we analyzed thymus size and tumor development in the offspring.
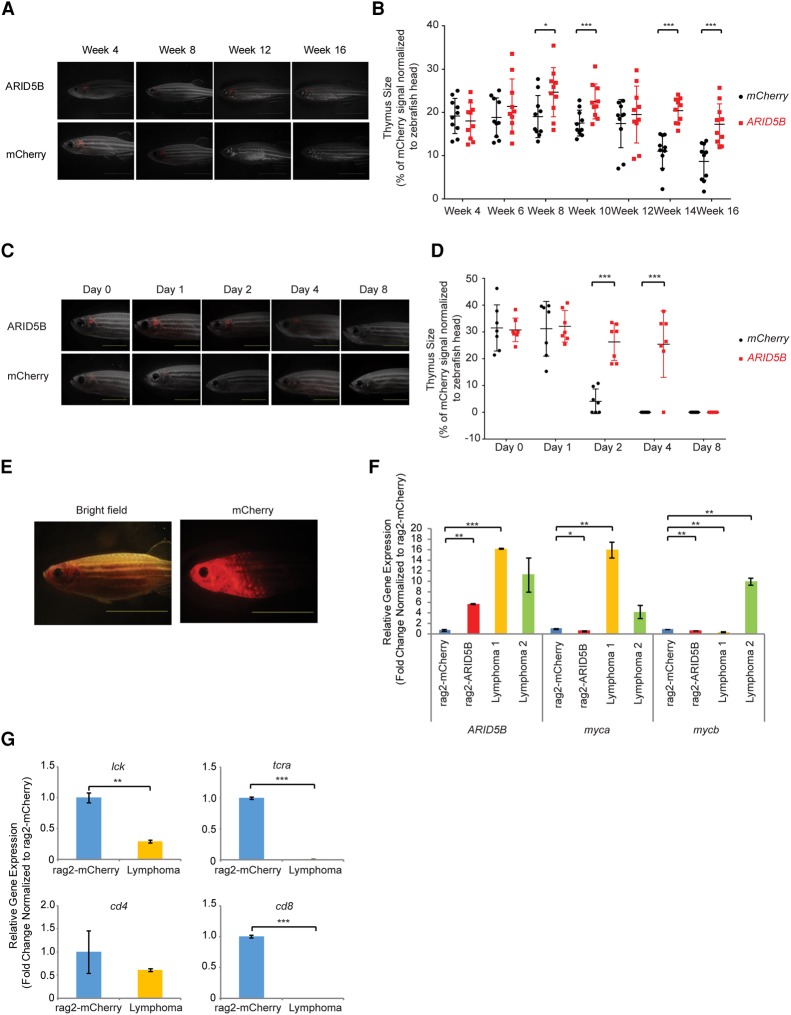
Interestingly, we found that the thymus was retained in ARID5B transgenic lines. In general, zebrafish undergo age-related thymic involution, a process in which a progressive regression of thymic cells is observed after 3 mo (Fig. 7A), similar to the processes in other vertebrates. In contrast, the ARID5B transgenic zebrafish exhibited a relatively larger thymus size at 2 mo than control fish did (mCherry only), and this effect persisted at 4 mo (Fig. 7A,B). Moreover, the mCherry signal in the thymus was observed up to 4 d after γ irradiation in the ARID5B transgenic line, whereas the signal in control fish immediately disappeared (Fig. 7C,D). These results indicated that overexpression of ARID5B promotes the survival of thymic T cells in zebrafish.
Figure 7.
Overexpression of ARID5B leads to thymus retention, radioresistance, and the development of T-cell lymphoma in zebrafish. (A) Representative microscopy images of rag2-mCherry and rag2-ARID5B zebrafish at weeks 4, 8, 12, and 16. The merged fluorescent and bright-field images are shown. Bar, 5 mm. (B) Quantification of rag2-mCherry and rag2-ARID5B transgenic zebrafish thymus size at weeks 4, 6, 8, 10, 12, 14, and 16. The thymic area (indicated by the mCherry signal) was normalized to the size of the zebrafish head. Thymic involution was observed in the zebrafish at 3 mo after fertilization. (*) P < 0.05; (***) P < 0.001 by two-sample two-tailed t-test. (C) Representative microscopy images of rag2-mCherry (control) and rag2-ARID5B transgenic zebrafish after γ irradiation. The mCherry fluorescent signal was captured on day 0 (before γ irradiation) and days 1, 2, 4, and 8 (after γ irradiation). The merged fluorescent and bright-field images are shown. Bar, 5 mm. (D) Quantification of rag2-mCherry and rag2-ARID5B transgenic zebrafish thymus size after the fish were subjected to γ irradiation. The thymic area was normalized to the size of the zebrafish head. (***) P < 0.001 by two-sample two-tailed t-test. (E) Representative image of rag2-ARID5B transgenic zebrafish that developed a lymphoma phenotype. Bar, 10 mm. (F) mRNA expression of ARID5B, myca, and mycb in the thymic cells of rag2-mCherry (control) zebrafish, normal rag2-ARID5B zebrafish, and two rag2-ARID5B zebrafish with lymphoma was analyzed by qRT–PCR. The relative gene expression was normalized to that of ef1a (internal control). The data represent the mean ± SD of duplicate samples. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 by two-sample two-tailed t-test. (G) mRNA expression levels of lck, t-cell receptor α (tcra), cd4, and cd8 in the thymic cells of rag2-mCherry (control) zebrafish and rag2-ARID5B zebrafish with lymphoma were analyzed by qRT–PCR. The relative gene expression was normalized to that of ef1a (internal control). The data represent the mean ± SD of duplicate samples. (**) P < 0.01; (***) P < 0.001 by two-sample two-tailed t-test.
Strikingly, two out of 38 (∼5%) transgenic zebrafish from one founder developed lymphoma after 6 mo (Fig. 7E). A quantitative RT–PCR (qRT–PCR) analysis confirmed the expression of the human ARID5B gene in these zebrafish (Fig. 7F). Importantly, the zebrafish myca and mycb genes, which are orthologs of the human MYC gene, were found to be highly expressed in the lymphoma tissue obtained from each diseased zebrafish compared with control thymus tissue (bulk) (Fig. 7F). The expression of myca or mycb was not elevated in the nontransformed thymuses of ARID5B transgenic fish. This result suggested that ARID5B overexpression alone might not be sufficient to induce myc expression. We also found that expression of zebrafish lck, t-cell receptor α (tcra), cd4, and cd8 genes was down-regulated in tumor samples compared with the control thymus (Fig. 7G), indicating that tumor cells exhibited differentiation arrest. Together, our results indicated that ARID5B promotes cell survival and contributes to T-cell leukemogenesis in zebrafish, similar to the results observed in human T-ALL cells.
Discussion
Deregulation of transcription factor genes plays a central role in the molecular pathogenesis of T-ALL (Look 1997; Armstrong and Look 2005; Aifantis et al. 2008). A number of genetic lesions involving transcription factors such as TAL1 and NOTCH1 have been implicated in T-ALL (Look 1997; Armstrong and Look 2005; Tosello and Ferrando 2013). Subsequent activation of the oncogenic transcriptional program initiates and promotes T-cell leukemogenesis. Therefore, the identification and characterization of the oncogenic transcriptional machinery are crucial for understanding the molecular pathogenesis of T-ALL and determining potential therapeutic targets.
In this study, we identified ARID5B as a critical target directly regulated by the TAL1 complex in T-ALL cells. ARID5B binds primarily to promoters and enhancers with active histone marks. ARID5B has been reported previously to interact with the histone demethylase PHF2, which removes a repressive histone mark (Baba et al. 2011; Hata et al. 2013). However, we did not observe binding between ARID5B and PHF2 proteins in T-ALL cells. We assume that interaction of ARID5B with histone-modifying proteins may be a tissue-specific or context-dependent occurrence. Importantly, ARID5B positively regulates the expression of the TAL1 complex in T-ALL. ARID5B also frequently binds to regulatory elements in TAL1 target genes, such as ALDH1A2, and coordinately regulates their expression. ARID5B knockdown reduced H3K27ac levels at the target gene loci but did not affect global levels of H3K27ac. Therefore, we assume that ARID5B may affect transcriptional or epigenetic status at specific target loci that have been activated already by preceding transcription factors such as TAL1. Our result implicates ARID5B as a second-tier factor that likely reinforces or stabilizes the gene expression program in malignant T cells. We also found that ARID5B inhibits gene expression. ARID5B inhibition restored the expression of several tumor suppressor genes, thus providing additional evidence that ARID5B acts as an oncogenic factor in T-ALL. Further investigation is necessary to identify interacting partners of ARID5B in T-ALL cells. Of note, we also observed that ∼18% of ARID5B bindings were present in promoter regions. Although we focused mainly on the function of ARID5B at enhancers of critical gene loci in this study, it would be of interest to analyze the potential function of ARID5B at promoter regions.
We also found that ARID5B positively regulates the oncogene MYC in human T-ALL cells. MYC is one of the most prevalent oncogenes in various cancers, including T-ALL, regulating cell growth, proliferation, protein synthesis, and metabolism (Dang 2013; Gabay et al. 2014). In human T-ALL, MYC is induced predominantly through activating mutations of NOTCH1 (Weng et al. 2006; Herranz et al. 2014). Inactivating mutations of the ubiquitin ligase FBXW7 also result in the stabilization of MYC and NOTCH1 proteins (O'Neil et al. 2007). Many studies with human samples and animal models have established the role of MYC as a driver oncogene in T-cell leukemia (Langenau et al. 2003; Sharma et al. 2006). One of us (Mansour et al. 2013) had reported previously that TAL1 indirectly up-regulates MYC protein expression via the induction of miR-223, which inhibits FBXW7 mRNA expression. We now provide another mechanism whereby TAL1 activates MYC through the up-regulation of ARID5B. Notably, Myc expression is up-regulated in thymocytes after DN3 stage of thymocytes in mice, while Arid5b is down-regulated at DN3 stage. Therefore, we assume that ARID5B may not be required for the expression of MYC during normal T-cell development but can positively regulate MYC when overexpressed in T-ALL.
Importantly, we showed that ARID5B is required for the growth and survival of T-ALL cells in cell lines and an animal model. Knockdown of ARID5B significantly inhibited cell growth in multiple TAL1-positive human T-ALL cell lines. This phenotype was not observed in a TAL1-negative cell line (DND-41) or a MYC-negative cell line (LOUCY), thus supporting our hypothesis that ARID5B exerts its oncogenic effects through the induction of the TAL1 and MYC oncogenes. Critically, forced expression of ARID5B in immature thymocytes resulted in thymic retention and radioresistance in zebrafish. Two ARID5B transgenic zebrafish developed a lymphoma phenotype, which was observed after a long latency period of 6 mo. Other siblings from the same founder zebrafish did not develop tumors. Therefore, we infer that ARID5B may not be a bona fide oncogene and is insufficient to induce tumors by itself. Interestingly, our data demonstrated that tumor cells in ARID5B transgenic fish showed reduced expressions of lck, tcra, cd4, or cd8. Hence, a possible explanation is that ARID5B overexpression induces differentiation arrest of thymocytes at an immature stage and promotes cell survival, which may predispose the cells to acquiring additional genetic abnormalities that can induce leukemia. This might lead to the up-regulation of endogenous myca and mycb expression in tumor samples. Follow-up studies with a large number of animals are needed to confirm this point.
In this study, we also identified the enhancer controlling ARID5B expression in T-ALL cells, which is located 135 kb upstream of the ARID5B gene and is bound by the TAL1 complex. The associated SE was present in multiple T-ALL cells but not in normal T cells. Thus, we assume that ectopic expression of TAL1 in immature thymocytes leads to aberrant activation of the −135-kb element, thereby inducing ARID5B. Notably, we also found that normal SP T cells possess a different SE within the ARID5B gene. We also observed weak H3K27ac signals at this locus in normal thymocytes. Given the expression pattern of ARID5B in normal T-cell development, this element could be regulated by different transcription factors and be responsible for the activation of ARID5B in DP and SP T cells. Notably, knockout studies reported by others have shown that a loss of Arid5b induces thymic hypoplasia in mice (Lahoud et al. 2001). One group reported recently that Arid5a, another member of the ARID family of proteins, is a key factor regulating the fate of naïve CD4+ cells (Masuda et al. 2016). Therefore, ARID5B may be induced under a different enhancer in normal mature T cells and required for T-cell development or function.
Recent genome-wide studies have reported germline genomic variations within the ARID5B genetic locus that confer increased risk of childhood B-precursor ALL and treatment outcomes (Papaemmanuil et al. 2009; Trevino et al. 2009). Among several SNPs examined, rs10821936 is strongly associated with disease susceptibility in several race groups (Xu et al. 2012). In addition, children with rs10821936 were at an increased risk of mixed lineage leukemia (MLL)-related acute leukemia (Emerenciano et al. 2014). However, investigation of SNP genotypes and ARID5B expression in B-ALL revealed no significant correlation between the ARID5B SNPs and mRNA levels (Gutierrez-Camino et al. 2013). Interestingly, in high-hyperdiploid ALL (HD-ALL), the rs7090445-C risk allele has been shown to disrupt RUNX3 binding and contribute to the reduction of ARID5B expression (Studd et al. 2017). Therefore, additional functional studies are required to elucidate the relationship between SNPs and ARID5B expression and their role in leukemogenesis.
In conclusion, we identified ARID5B as a critical transcriptional regulator controlled by TAL1 in T-ALL. Our data suggest that ARID5B reinforces the oncogenic transcription program induced by the oncogenes TAL1 and MYC. Together, our findings support the idea that abnormal expression of ARID5B could be oncogenic and may play a central role in the molecular pathogenesis of T-ALL.
Materials and methods
Cell culture
All T-ALL and AML cell lines were cultured in RPMI-1640 medium (BioWest) supplemented with 10% FBS (BioWest) in a CO2 incubator. The 293T cells were cultured in DMEM (BioWest) supplemented with 10% FBS in a CO2 incubator.
shRNA knockdown analysis
The shRNA sequences were designed and cloned into the lentivirus vector pLKO.1-puro according to The RNAi Consortium's recommendation (http://www.broadinstitute.org/rnai/trc). The shRNA sequences are in the Supplemental Material. Lentiviruses were produced by cotransfecting individual shRNA constructs with the packaging plasmids pMDLg/pRRE and pRSV-Rev and the envelope plasmid pMD2.G into 293T cells using FuGENE 6 transfection reagent (Roche). Supernatants containing lentivirus particles were collected and filtered through a 0.45-µm filter (Thermo). T-ALL cells were infected with lentivirus in the presence of 8 µg/mL polybrene (Millipore) by centrifugation at 1300 rcf for 1.5 h. The infected cells were selected with 0.7 µg/mL puromycin (Sigma) in RPMI-1640 medium for at least 36 h after infection.
CRISPR/Cas9 gene knockout
For genetic knockout experiments, single-guide RNA (sgRNA) was designed using the CRISPR design tool (http://crispr.mit.edu) and cloned into lentiCRISPv2 (Addgene plasmid no. 52961) or FgH1tUTG (Addgene plasmid no. 70183) lentivirus vector using BsmB1 enzyme sites. Lentiviruses were produced using the same protocol for shRNA knockdown analysis. Jurkat cells infected with the virus were selected by 0.7 µg/mL puromycin (Sigma) from day 3 to day 7. To identify genetic deletion, we isolated genomic DNA using the QIAamp DNA blood minikit (Qiagen) followed by PCR amplification using specific primers flanking the −135-kb element as follows: forward (5′-CGTCAACCACCACTGCTTTT-3′) and reverse (5′-TTCCAGTAACGTGGCAGTCC-3′).
Protein extraction and Western blotting
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 µg/mL leupeptin) (Cell Signaling Technology) with a protease inhibitor cocktail (Roche). Equivalent amounts of protein were diluted in Laemmli sample buffer (Bio-Rad) containing β-mercaptoethanol (Bio-Rad) and resolved on an SDS-PAGE gel. The proteins were subsequently transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad). The membranes were blocked with 5% nonfat milk and subjected to immunoblot analysis with specific antibodies against ARID5B (Bethyl Laboratories), TAL1, MYB (Millipore), GATA3, RUNX1, MYC, MAX, PARP, α-tubulin, and β-actin (Cell Signaling Technology). The membranes were washed with Tris-buffered saline (TBS)-Tween (0.1%), incubated with secondary antibodies, and subsequently visualized using the enhanced chemiluminescence method (Thermo).
RNA extraction and gene expression analysis
Total RNA was harvested using a NucleoSpin RNA kit (Macherey-Nagel), and 1 µg of the purified total RNA was reverse-transcribed into cDNA using QuantiTect (Qiagen). qPCR was performed on a QuantStudio 3 real-time PCR system (Thermo Fisher Scientific) using Power SYBR Green PCR Master mix (Roche) and specific primers for individual genes. The PCR primer sequences of the genes of interest are in the Supplemental Material.
RNA-seq
RNA was extracted using a miRNeasy kit (Qiagen) followed by DNase treatment (Turbo DNA-free kit, Ambion). For each sample, 10 million cells were collected and pelleted for RNA extraction. Ten microliters of ERCC RNA spike-in mix (1:10 dilution) was added to each sample. Strand-specific library construction and sequencing of paired-end 100-bp-long reads by the Illumina HiSeq 4000 were performed at BGI Biotech Solutions Co. Ltd. All RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database (GSE97514). RNA-seq data sets of two control samples and two knockdown samples for each member of the TAL1 complex were aligned to the hg19 human genome using STAR 2.5.2a with the parameter outFilterMultimapNmax set to 1. We used the tool htseq-count version 0.6.1 for the mapped reads in a .bam file to generate count tables based on the Ensembl gene annotation of hg19. From these tables, the Bioconductor package DESeq2 version 1.12.4, was used in the intersection-strict mode to analyze differential gene expression.
Public microarray and RNA-seq data sets
The microarray gene expression data set for T-ALL cell lines was reported previously by us (Liau et al. 2017) and has been deposited under accession number GSE90138. The microarray data set for primary T-ALL cases has been reported under accession number GSE26713 (Homminga et al. 2011) and was analyzed with R2 (http://r2.amc.nl). RNA-seq data sets for different stages of human hematopoietic cells were obtained from the NCBI GEO under accession number GSE69239 (Casero et al. 2015).
ChIP-PCR
ChIP was performed in accordance with the Agilent ChIP analysis protocol. Briefly, the cells were cross-linked in a fresh 11% formaldehyde solution followed by quenching with glycine. The cells were washed in 1× PBS, pelleted, and flash-frozen in liquid nitrogen. The cell pellets were lysed and sonicated with a Bioruptor sonicator at high power for 50 pulses of 30 sec (with a 30-sec pause between pulses). Chromatin was immunoprecipitated using the antibodies IgG (Santa Cruz Biotechnology), ARID5B (Sigma, HPA015037), and H3K27ac (Abcam) and bound to Dynabeads Protein G (Thermo Fisher Scientific) followed by elution and reverse cross-linking. qPCR was performed on a QuantStudio 3 real-time PCR system (Thermo Fisher Scientific) using Power SYBR Green PCR Master mix (Roche) and specific primers for the genomic regions of individual genes. The PCR primer sequences are in the Supplemental Material.
ChIP-seq analysis
Library construction from the ARID5B ChIP-seq samples and sequencing of paired-end 50-bp-long reads by the Illumina HiSeq 4000 were performed at BGI Biotech Solutions Co. Ltd. The ChIP-seq data for ARID5B have been deposited in the GEO database (GSE97514). The Jurkat ChIP-seq data sets for TAL1, HEB, GATA3, RUNX1, MYB, and CBP were downloaded from NCBI GEO under the superseries accession numbers GSE29181, GSE68976, GSE59657, and GSE17954 (Sanda et al. 2012; Mansour et al. 2014). H3K27ac ChIP-seq results for Jurkat (GSM1296384), RPMI-8402 (GSM1442003), CCRF-CEM (GSM2037781), MOLT-4 (GSM2037790), normal human thymus (GSM1013125), and Th17 (GSM772987) cells were also obtained from NCBI GEO (Bernstein et al. 2010; Sanda et al. 2012; Vahedi et al. 2015; Abraham et al. 2017). The ChIP-seq data sets for Th1 and Th2 can be downloaded from http://www.ncbi.nlm.nih.gov/sra/SRX290665 and http://www.ncbi.nlm.nih.gov/sra/SRX290666, respectively. All data were aligned to the human genome sequence (hg19) using the Bowtie program with parameters -e 70 -k 1 -m 1 -n 2 --best, and peaks were called using MACS2 (Model-Based Analysis of ChIP-seq). A total of 2117 peaks was identified in ARID5B ChIP-seq. Five-hundred-base-pair regions around the ARID5B ChIP-seq peaks (250 bp upstream of and 250 bp downstream from the highest point of the peak) were used as input sequences for MEME_ChIP to discover motifs. The colocalization of ARID5B with other transcription factors was analyzed by the Bioconductor package genomation version 1.6.0 with a scaled ChIP-seq signal and k-means clustering to order the heat map. The SEs were identified using ROSE (https://bitbucket.org/youngcomputation/rose).
Classification of chromatin state
The Jurkat ChIP-seq data sets for H3K27ac, H3K4me3, H3K79me2, H3K27me3, and CTCF were downloaded from NCBI GEO under accession numbers GSE50622, GSE35583, GSE23080, GSE72298, and GSE68976. The Jurkat ChIP-seq for H3K4me1 has been deposited in the GEO database (GSE97514). Chromatin states were ascertained by ChromHMM (Ernst and Kellis 2012) and classified into six active states (promoter and enhancers), one transcription-associated state, one repressed state, one low signal state, and one domain insulator state.
TR calculation
The Jurkat ChIP-seq data set for RNAPII was downloaded from NCBI GEO under accession number GSM1224784. TR was calculated as the ratio between RNAPII density in the promoter region and in the gene body region (Rahl et al. 2010). The promoter region was defined as the region from −30 to +300 relative to the TSS of the gene, and the gene body was defined as the remaining length of the gene. “Active” genes in Jurkat were defined as those bound by RNAPII and H3K4me3 within ±2.5 kb of the TSS and bound by H3K79me2 in the first 5 kb of the gene.
GSEA and GO analysis
GSEA (Subramanian et al. 2005) was performed for the normalized genes using the ratio of classes metric to rank genes. The TAL1 target genes that were identified by ChIP-seq analysis as bound by TAL1 and by RNA-seq analysis as down-regulated by knockdown of TAL1 were first defined and used as a gene set for GSEA. A heat map was created using GSEA or the Gene Pattern program (Broad Institute). The GO analysis and ChEA were performed with the Enrichr tool (http://amp.pharm.mssm.edu/Enrichr; Chen et al. 2013). This program generates statistics determining which GO terms/pathways/phenotypes are overrepresented in the analyzed list of genes. The combined score, which was calculated by multiplying the P-value computed using Fisher's exact test by the z-score of the deviation from the expected rank, was used to rank ontology terms.
Cell viability assay
The viability of the cells was measured using a CellTiter Glo kit (Promega). Briefly, T-ALL cells were seeded in 96-well plates after lentivirus infection. Cell viability was then measured according to the manufacturer's instructions using the Tecan Infinite 200 Pro plate reader (Tecan).
Annexin V-FITC early apoptosis detection
Cells transduced with lentivirus expressing shRNA were harvested after 3 d and subjected to analysis using an annexin V-FITC early apoptosis detection kit (Cell Signaling Technology, 6592). Approximately 10,000 cells were analyzed with a BD LSR II flow cytometer using BD FACSDiva software. Annexin V-FITC-positive staining indicated cell apoptosis, whereas propidium iodide-positive DNA staining indicated necrosis.
Luciferase reporter assay
A luciferase reporter construct (pGL4.27 luc2/minP/hygro) (Promega) containing the human MYC enhancer (1524 bp in size: chromosome 8: 130,179,809–130,180,997, hg19) was cloned and transfected into Jurkat cells using a Neon transfection system (Life Technologies). The cells were selected using 400 µg/mL hygromycin B (Sigma-Aldrich). Cells expressing the ALDH1A2 enhancer reporter construct have been established previously (Tan et al. 2016). The cells were infected with lentiviruses encoding shRNAs targeting ARID5B or a control (shGFP). The luciferase activity was then determined using the One-Glo luciferase assay on a Tecan Infinite 200 Pro plate reader (Tecan). The fluorescence (cell viability) was also measured. The relative luciferase activity of each sample was then calculated by normalizing the luminescence readings to the fluorescence readings.
Mouse study
Mouse studies strictly adhered to the recommendations of the Institutional Animal Care and Use Committee, and all protocols were approved by the Committee at the National University of Singapore. Thymus specimens from 8- to 10-wk-old NOD-Rag1null IL2rgnull (NRG) mice were harvested and sorted by fluorescence-activated cell sorting (FACS) into DN1 (CD44+CD25−), DN2 (CD44+CD25+), and DN3 (CD44−CD25+) populations. Total RNA was harvested using an RNeasy Plus minikit (Qiagen) and reverse-transcribed into cDNA using SuperScript IV reverse transcriptase (Thermo Fisher Scientific). qPCR was performed on a QuantStudio 3 real-time PCR system (Thermo Fisher Scientific) using Power SYBR Green PCR Master mix (Roche) and specific primers for individual genes.
Zebrafish transgenic study
Zebrafish studies strictly adhered to the recommendations of the Institutional Animal Care and Use Committee, and all protocols were approved by the Committee at the National University of Singapore. The human ARID5B gene was cloned downstream from the zebrafish rag2 promoter in a construct containing the I-SceI meganuclease sequences (“rag2-ARID5B”). A reporter plasmid (“rag2-mCherry”) was created by introducing the fluorescent protein mCherry into the same backbone. The rag2-ARID5B and rag2-mCherry constructs were resuspended in buffer and linearized by the enzyme I-SceI (New England Biolabs). The mixture was injected into zebrafish embryos at the one-cell stage of development. The rag2-mCherry construct was used alone as a control.
Monitoring of the thymus in zebrafish
Bright-field and fluorescence microscopy images from 10 rag2-ARID5B and rag2-mCherry transgenic zebrafish were captured every 2 wk from week 4 to week 16. The thymic area was calculated as a percentage of the size of the zebrafish head using the image processing program ImageJ. For T-cell ablation assays, 8-wk-old rag2-mCherry and rag2-ARID5B transgenic zebrafish were given a whole-body dose of 20 Gy of γ irradiation and analyzed for mCherry expression within the thymic area at 1, 2, 4, and 8 d after treatment. Loss of thymic cells was measured based on the decrease in mCherry expression in the thymic area after γ irradiation.
Statistical analysis
Significant values (P-values) were determined with Student's t-test in GraphPad Prism software. A P-value of <0.05 was considered statistically significant. Spearman's correlation was used to analyze the correlation between variables.
Supplementary Material
Acknowledgments
We thank A. Thomas Look, Takashi Okamoto, Toshio Suda, Motomi Osato, and members of the Sanda laboratory for discussions and critical reviews. We thank Marc Mansour and Shuning He for advice on imaging of zebrafish. We thank Shing Leng Chan for providing the NRG mice, and Tze King Tan for help with bioinformatics analysis. We are grateful to the National University of Singapore Zebrafish Facility for zebrafish analyses. This research is supported by the National Research Foundation, Prime Minister's Office, Singapore, under its Competitive Research Programme (NRF-NRFF2013-02); the RNA Biology Center at Cancer Science Institute of Singapore, National University of Singapore for funding by the Singapore Ministry of Education's Tier 3 grants (MOE2014-T3-1-006); and the US National Cancer Institute (1K99CA157951). The research is also supported by the National Research Foundation of Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.302646.117.
References
- Abraham BJ, Hnisz D, Weintraub AS, Kwiatkowski N, Li CH, Li Z, Weichert-Leahey N, Rahman S, Liu Y, Etchin J, et al. 2017. Small genomic insertions form enhancers that misregulate oncogenes. Nat Commun 8: 14385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aifantis I, Raetz E, Buonamici S. 2008. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol 8: 380–390. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Look AT. 2005. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol 23: 6306–6315. [DOI] [PubMed] [Google Scholar]
- Baba A, Ohtake F, Okuno Y, Yokota K, Okada M, Imai Y, Ni M, Meyer CA, Igarashi K, Kanno J, et al. 2011. PKA-dependent regulation of the histone lysine demethylase complex PHF2–ARID5B. Nat Cell Biol 13: 668–675. [DOI] [PubMed] [Google Scholar]
- Begley CG, Aplan PD, Davey MP, Nakahara K, Tchorz K, Kurtzberg J, Hershfield MS, Haynes BF, Cohen DI, Waldmann TA, et al. 1989. Chromosomal translocation in a human leukemic stem-cell line disrupts the T-cell antigen receptor δ-chain diversity region and results in a previously unreported fusion transcript. Proc Natl Acad Sci 86: 2031–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belver L, Ferrando A. 2016. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer 16: 494–507. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Cheng JT, Chen Q, Siciliano MJ, Crist W, Buchanan G, Baer R. 1990. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. EMBO J 9: 3343–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, Luong A, Parekh C, Crooks GM. 2015. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat Immunol 16: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Cheng JT, Tasi LH, Schneider N, Buchanan G, Carroll A, Crist W, Ozanne B, Siciliano MJ, Baer R. 1990. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix–loop–helix protein. EMBO J 9: 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. 2007. RBP2 belongs to a family of demethylases, specific for tri- and dimethylated lysine 4 on histone 3. Cell 128: 1063–1076. [DOI] [PubMed] [Google Scholar]
- Dang CV. 2013. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 3: a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerenciano M, Barbosa TC, Lopes BA, Blunck CB, Faro A, Andrade C, Meyer C, Marschalek R, Pombo-de-Oliveira MS. 2014. ARID5B polymorphism confers an increased risk to acquire specific MLL rearrangements in early childhood leukemia. BMC Cancer 14: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Look AT. 2000. Clinical implications of recurring chromosomal and associated molecular abnormalities in acute lymphoblastic leukemia. Semin Hematol 37: 381–395. [DOI] [PubMed] [Google Scholar]
- Gabay M, Li Y, Felsher DW. 2014. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 4: a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Mao TL, Panuganti PK, Kuhn E, Kurman RJ, Maeda D, Chen E, Jeng YM, Wang TL, Shih Ie M. 2011. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol 35: 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Gao M, Wu CH, Wang TL, Shih Ie M. 2012. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia 14: 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Camino A, Lopez-Lopez E, Martin-Guerrero I, Sanchez-Toledo J, Garcia de Andoin N, Carbone Baneres A, Garcia-Miguel P, Navajas A, Garcia-Orad A. 2013. Intron 3 of the ARID5B gene: a hot spot for acute lymphoblastic leukemia susceptibility. J Cancer Res Clin Oncol 139: 1879–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Takashima R, Amano K, Ono K, Nakanishi M, Yoshida M, Wakabayashi M, Matsuda A, Maeda Y, Suzuki Y, et al. 2013. Arid5b facilitates chondrogenesis by recruiting the histone demethylase Phf2 to Sox9-regulated genes. Nat Commun 4: 2850. [DOI] [PubMed] [Google Scholar]
- Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. 2000. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A–HEB function and pre-Tα chain expression. Nat Immunol 1: 138–144. [DOI] [PubMed] [Google Scholar]
- Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, Xu L, Castillo-Martin M, Llobet-Navas D, Cordon-Cardo C, et al. 2014. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 20: 1130–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351: 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, Vuerhard M, Buijs-Gladdines J, Kooi C, Klous P, et al. 2011. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 19: 484–497. [DOI] [PubMed] [Google Scholar]
- Hsu HL, Cheng JT, Chen Q, Baer R. 1991. Enhancer-binding activity of the tal-1 oncoprotein in association with the E47/E12 helix–loop–helix proteins. Mol Cell Biol 11: 3037–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba H, Greaves M, Mullighan CG. 2013. Acute lymphoblastic leukaemia. Lancet 381: 1943–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen JW, Ludwig WD, Sterry W, Bartram CR. 1993. SIL–TAL1 deletion in T-cell acute lymphoblastic leukemia. Leukemia 7: 1204–1210. [PubMed] [Google Scholar]
- Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. 2013. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol 9: 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusy S, Gerby B, Goardon N, Gault N, Ferri F, Gerard D, Armstrong F, Ballerini P, Cayuela JM, Baruchel A, et al. 2010. NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia. J Exp Med 207: 2141–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahoud MH, Ristevski S, Venter DJ, Jermiin LS, Bertoncello I, Zavarsek S, Hasthorpe S, Drago J, de Kretser D, Hertzog PJ, et al. 2001. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res 11: 1327–1334. [DOI] [PubMed] [Google Scholar]
- Landry JR, Kinston S, Knezevic K, de Bruijn MF, Wilson N, Nottingham WT, Peitz M, Edenhofer F, Pimanda JE, Ottersbach K, et al. 2008. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood 111: 3005–3014. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, et al. 2003. Myc-induced T cell leukemia in transgenic zebrafish. Science 299: 887–890. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T. 2004. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol 32: 11–24. [DOI] [PubMed] [Google Scholar]
- Liau WS, Tan SH, Ngoc PCT, Wang CQ, Tergaonkar V, Feng H, Gong Z, Osato M, Look AT, Sanda T. 2017. Aberrant activation of the GIMAP enhancer by oncogenic transcription factors in T-cell acute lymphoblastic leukemia. Leukemia 31: 1798–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Song W, Bi X, Zhao J, Huang Z, Li Z, Zhou J, Cai J, Zhao H. 2014. Recent advances in the ARID family: focusing on roles in human cancer. Onco Targets Ther 7: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look AT. 1997. Oncogenic transcription factors in the human acute leukemias. Science 278: 1059–1064. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Sanda T, Lawton LN, Li X, Kreslavsky T, Novina CD, Brand M, Gutierrez A, Kelliher MA, Jamieson CH, et al. 2013. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J Exp Med 210: 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. 2014. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346: 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ripley B, Nyati KK, Dubey PK, Zaman MM, Hanieh H, Higa M, Yamashita K, Standley DM, Mashima T, et al. 2016. Arid5a regulates naive CD4+ T cell fate through selective stabilization of Stat3 mRNA. J Exp Med 213: 605–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl NG Jr, Patsialou A, Haines DS, Dallas PB, Beck GR Jr, Moran E. 2005. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res 65: 9236–9244. [DOI] [PubMed] [Google Scholar]
- Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng JF, Prabhakar S, Rubin EM, Li PS, Sloane-Stanley J, Kong ASJ, et al. 2007. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110: 4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil J, Tchinda J, Gutierrez A, Moreau L, Maser RS, Wong KK, Li W, McKenna K, Liu XS, Feng B, et al. 2007. Alu elements mediate MYB gene tandem duplication in human T-ALL. J Exp Med 204: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Fukuhara N, Yoshie O. 1998. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol Cell Biol 18: 6939–6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, Kinsey SE, Lightfoot T, Roman E, Irving JA, et al. 2009. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet 41: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Robison LL, Look AT. 2008. Acute lymphoblastic leukaemia. Lancet 371: 1030–1043. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH. 1998. LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev 12: 2651–2657. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, Ma W, Tatarek J, Ahn Y, Kelliher MA, et al. 2012. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell 22: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J, Sahoo D, Rossi DJ, Bhattacharya D, Serwold T, Inlay MA, Ehrlich LI, Fathman JW, Dill DL, Weissman IL. 2012. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One 7: e40321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. 2006. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol 26: 8022–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studd JB, Vijayakrishnan J, Yang M, Migliorini G, Paulsson K, Houlston RS. 2017. Genetic and regulatory mechanism of susceptibility to high-hyperdiploid acute lymphoblastic leukaemia at 10p21.2. Nat Commun 8: 14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SH, Yam AW, Lawton LN, Wong RW, Young RA, Look AT, Sanda T. 2016. TRIB2 reinforces the oncogenic transcriptional program controlled by the TAL1 complex in T-cell acute lymphoblastic leukemia. Leukemia 30: 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosello V, Ferrando AA. 2013. The NOTCH signaling pathway: role in the pathogenesis of T-cell acute lymphoblastic leukemia and implication for therapy. Ther Adv Hematol 4: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, Willman C, Neale G, Downing J, Raimondi SC, et al. 2009. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet 41: 1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SC, Erdos MR, Davis SR, Roychoudhuri R, Restifo NP, Gadina M, et al. 2015. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520: 558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, Stubbs A, Cools J, Nagata K, Fornerod M, et al. 2008. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 111: 4668–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutven LJ, Onen E, Velthuizen SC, van Drunen E, von Bergh AR, van den Heuvel-Eibrink MM, Veronese A, Mecucci C, Negrini M, de Greef GE, et al. 2006. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer 45: 437–446. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grütz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J 16: 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Nagl NG Jr, Flowers S, Zweitzig D, Dallas PB, Moran E. 2004. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int J Cancer 112: 636. [DOI] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. 2009. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 20: 2096–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitson RH, Huang T, Itakura K. 1999. The novel Mrf-2 DNA-binding domain recognizes a five-base core sequence through major and minor-groove contacts. Biochem Biophys Res Commun 258: 326–331. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson AC, Gottgens B. 2013. Transcriptional regulation of haematopoietic stem cells. Adv Exp Med Biol 786: 187–212. [DOI] [PubMed] [Google Scholar]
- Wilsker D, Patsialou A, Dallas PB, Moran E. 2002. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ 13: 95–106. [PubMed] [Google Scholar]
- Wu JN, Roberts CW. 2013. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov 3: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Wang TL, Shih Ie M. 2014. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther 15: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Cheng C, Devidas M, Pei D, Fan Y, Yang W, Neale G, Scheet P, Burchard EG, Torgerson DG, et al. 2012. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J Clin Oncol 30: 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.