In this study, Bao et al. investigated the role of the trimeric retention and splicing (RES) complex protein Snu17 and its interaction partners during spliceosome assembly and catalytic activation. Their findings reveal novel roles for the RES complex proteins during spliceosome activation as well as a potential proofreading function of the spliceosomal ATPase Prp2.
Keywords: RES complex, pre-mRNA splicing, spliceosome activation, Prp2 ATPase/RNA helicase, spliceosome disassembly
Abstract
The precise function of the trimeric retention and splicing (RES) complex in pre-mRNA splicing remains unclear. Here we dissected the role of RES during the assembly and activation of yeast spliceosomes. The efficiency of pre-mRNA splicing was significantly lower in the absence of the RES protein Snu17, and the recruitment of its binding partners, Pml1 (pre-mRNA leakage protein 1) and Bud13 (bud site selection protein 13), to the spliceosome was either abolished or substantially reduced. RES was not required for the assembly of spliceosomal B complexes, but its absence hindered efficient Bact complex formation. ΔRES spliceosomes were no longer strictly dependent on Prp2 activity for their catalytic activation, suggesting that they are structurally compromised. Addition of Prp2, Spp2, and UTP to affinity-purified ΔRES B or a mixture of B/Bact complexes formed on wild-type pre-mRNA led to their disassembly. However, no substantial disassembly was observed with ΔRES spliceosomes formed on a truncated pre-mRNA that allows Prp2 binding but blocks its activity. Thus, in the absence of RES, Prp2 appears to bind prematurely, leading to the disassembly of the ΔRES B complexes to which it binds. Our data suggest that Prp2 can dismantle B complexes with an aberrant protein composition, suggesting that it may proofread the spliceosome's RNP structure prior to activation.
The spliceosome is composed of the U1, U2, U4/U6, and U5 small nuclear RNPs (snRNPs) and numerous proteins (Wahl et al. 2009). Spliceosome assembly is initiated by interaction of the U1 snRNP with the 5′ splice site (5′ss) and the U2 snRNP with the branch site (BS) of the intron, generating the spliceosomal A complex. The U4/U6.U5 tri-snRNP, in which the U4 and U6 small nuclear RNAs (snRNAs) are extensively base-paired, subsequently interacts, forming the B complex. The latter undergoes extensive rearrangements, yielding the activated Bact complex (Supplemental Fig. S1). After the action of the Prp2 RNA helicase, the catalytically activated B* complex is formed and catalyzes step 1 of pre-mRNA splicing, yielding the cleaved 5′ exon and intron lariat 3′ exon splicing intermediates. The resulting C complex is then remodeled by the Prp16 RNA helicase, generating the C* complex. C* catalyzes step 2 of splicing, during which the intron is excised and the 5′ and 3′ exons are ligated to form mRNA. After step 2, the mRNA is released, and the intron lariat spliceosome (ILS) is disassembled by the RNA helicase Prp43 (Arenas and Abelson 1997; Martin et al. 2002; Tsai et al. 2005).
During spliceosome assembly and catalytic activation, the snRNAs and pre-mRNA form a dynamic RNA–RNA network essential for splicing catalysis (Staley and Guthrie 1998). Initially, U1 snRNA base-pairs with the 5′ss, and U2 base-pairs with the BS. Upon tri-snRNP integration, U2 and U6 snRNA base-pair, forming U2/U6 helix II. The RNA helicase Prp28 then displaces U1 snRNA from the 5′ss (Staley and Guthrie 1999), allowing the base-pairing interaction between the 5′ss and the conserved U6 ACAGA box. Subsequent unwinding of the U4/U6 helices by Brr2 (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998) enables U6 to form additional short duplexes with U2 (U2/U6 helix Ia and helix Ib) and also a catalytically important U6 internal stem–loop, which coordinates the metal ions important for splicing catalysis (Fica et al. 2013; Hang et al. 2015).
The spliceosome is a highly dynamic RNP machine that undergoes many compositional and structural rearrangements that are driven by eight conserved DEAH/D-box ATPases or RNA helicases (Staley and Guthrie 1998; Wahl et al. 2009). Several of these enzymes also promote the fidelity of the splicing process by kinetically proofreading the formation of the correct RNA/RNP structures in the spliceosome (Semlow and Staley 2012). Two of the spliceosomal ATPases/RNA helicases (namely, Brr2 and Prp2) play key roles during the transformation of the spliceosome into a catalytically active RNP enzyme. The earliest step of spliceosome activation is the unwinding of the U4/U6 snRNA duplex by Brr2. This leads to loss of U4 from the spliceosome and restructuring of U6 that is required for the formation of the catalytically active U2/U6 RNA network. U4/U6 unwinding also triggers the loss of U4/U6 snRNP proteins and U6-associated Lsm proteins and the recruitment of the nineteen complex (NTC) and NTC-related proteins plus several other Bact proteins.
Prp2 together with its cofactor, Spp2, catalyzes the remodeling of the Bact complex into a catalytically active B* complex (King and Beggs 1990; Kim and Lin 1996; Warkocki et al. 2009, 2015). Prp2 interacts transiently with the spliceosome at the time of its activation and is released after catalyzing the transformation of Bact into B* (King and Beggs 1990; Kim and Lin 1996). This implies that Prp2's binding site normally is first generated during/after Bact formation and that the structural changes that it facilitates lead to its own displacement from the spliceosome. Prp2 contacts the pre-mRNA intron downstream from the BS (Liu and Cheng 2012; Warkocki et al. 2015). Cryo-electron microscopy (cryo-EM) studies revealed that Prp2 is located on the periphery of the Bact complex, on the outer side of the U2 Hsh155 (SF3B1) HEAT domain (Rauhut et al. 2016; Yan et al. 2016). Although the precise remodeling events catalyzed by Prp2 are not completely understood, most appear to involve rearrangements in RNA–protein and protein–protein interactions. For example, Prp2 action leads to the destabilization/repositioning of the U2-associated SF3a and SF3b proteins, the pre-mRNA retention and splicing (RES) complex, and the NTC-related proteins Cwc24 and Cwc27 (Fabrizio et al. 2009; Warkocki et al. 2009; Lardelli et al. 2010; Ohrt et al. 2012). This in turn facilitates the binding of Cwc25 and, together with Yju2, promotes efficient step 1 catalysis (Liu et al. 2007; Chiu et al. 2009; Warkocki et al. 2009). Genetic data suggest that Prp2 may facilitate the remodeling of U2/U6 helix I and additionally be involved in proofreading the catalytic RNA network that is formed during activation (Wlodaver and Staley 2014). More recent biochemical studies indicate that yeast Prp2 liberates the pre-mRNA's 5′ss and BS adenosine for step 1 catalysis, primarily by catalyzing RNP rearrangements as opposed to unwinding RNA duplexes (Bao et al. 2017).
The RES complex in Saccharomyces cerevisiae, which is also present in humans, is comprised of Snu17 (also known as Ist3), Pml1 (pre-mRNA leakage protein 1), and Bud13 (bud site selection protein 13), with Snu17 acting as a binding platform for both Pml1 and Bud13 (Dziembowski et al. 2004; Brooks et al. 2009; Trowitzsch et al. 2009; Wysoczański et al. 2014). The RES complex proteins bind in a cooperative manner, which enhances the binding affinity of the individual proteins for each other and of Snu17 for RNA (Wysoczański et al. 2014). RES proteins are initially recruited at the B complex stage in yeast and become more stably associated during activation but are destabilized during the Prp2-mediated formation of B* complex (Gottschalk et al. 2001; Fabrizio et al. 2009; Ohrt et al. 2012). In the yeast Bact complex, the RES complex proteins contact the HEAT domain of the U2 Hsh155 protein, the RT domain of the U5 Prp8 protein, and the Prp45 and Cwc22 proteins (Rauhut et al. 2016; Yan et al. 2016). In addition, protein–protein cross-linking suggests that they are in close proximity to Prp2 and Spp2 in Bact (Rauhut et al. 2016). Cross-linking also revealed that the RES complex proteins interact with the intron between the BS and the 3′ss (Wysoczański et al. 2014; Schneider et al. 2015). In the absence of RES subunits, an increase in the amount of unspliced pre-mRNA in the cytoplasm was observed, indicating that RES is required in vivo for nuclear retention of unspliced pre-mRNA (Dziembowski et al. 2004).
RES subunits are not essential for yeast viability, but, in their absence, a slow growth phenotype is observed (Gottschalk et al. 2001; Dziembowski et al. 2004). Consistent with this, a marked decrease in splicing efficiency was observed both in vivo and in vitro in the absence of Snu17 and Bud13, whereas a much milder splicing defect was observed in a Δpml1 strain (Gottschalk et al. 2001; Dziembowski et al. 2004). In Δsnu17 and Δbud13 whole-cell extracts, splicing efficiency was substantially reduced, with a block observed prior to the first catalytic step. Spliceosomes formed in vitro in Δsnu17 or Δbud13 extracts exhibited aberrant migration on native gels compared with spliceosomes formed in wild-type extracts (Gottschalk et al. 2001; Dziembowski et al. 2004), suggesting that they may have an altered structure. However, the precise role of the RES complex during splicing and the stage at which it acts remain unclear.
Here we dissected the role of the RES complex proteins during the assembly and activation of the yeast spliceosome. Our data reveal that the RES complex promotes the efficient formation of the activated Bact spliceosomal complex and that complexes lacking the RES complex proteins are no longer strictly dependent on Prp2 for their catalytic activation. B complexes lacking RES allow the premature binding of Prp2, and its action leads to the disassembly of these complexes. Our data suggest that Prp2 is involved in proofreading the spliceosome's RNP structure and facilitates the disassembly of complexes with an aberrant protein composition/RNP structure. Taken together, our data reveal not only novel roles for the RES complex proteins during spliceosome activation but also a potential proofreading function of Prp2.
Results
Pre-mRNA splicing is less efficient in extracts from an S. cerevisiae Δsnu17 strain
To investigate the function of the RES complex, whose formation is dependent on Snu17, we deleted the SNU17 gene from S. cerevisiae and assayed pre-mRNA splicing in yeast total-cell extracts. Compared with the wild-type control, the amount of spliced mRNA was reduced substantially in extracts from the Δsnu17 strain (Fig. 1A), consistent with previous results (Gottschalk et al. 2001; Dziembowski et al. 2004). Splicing could be partially restored by adding a mixture of affinity-purified Snu17 and Snu17–Bud13 dimer to the Δsnu17 extract (Fig. 1A). Unspliced actin pre-mRNA accumulated in the Δsnu17 extract (Fig. 1A), indicating that the block in splicing occurs prior to the catalytic steps. We thus analyzed spliceosome formation in extracts from the wild-type and Δsnu17 strain under standard splicing conditions (2 mM ATP) using a wild-type actin pre-mRNA containing three RNA aptamers for the MS2 protein at its 5′ end. Spliceosomes were affinity purified and then subjected to glycerol gradient centrifugation. In both cases, only very small amounts of spliceosomes were detected in the 40S–50S region of the gradient (Fig. 1B), where B, Bact, B*, and C complexes normally sediment (Fabrizio et al. 2009; Warkocki et al. 2009). Most of the radiolabeled RNA sedimented in the 20S region of the gradient, where H and A complexes, the spliced mRNA, and spliceosome degradation products are typically found (Fabrizio et al. 2009). In contrast, when spliceosome assembly was blocked at the Bact complex stage by using an extract in which Prp2 was heat-inactivated (prp2-1), 45S spliceosomal complexes accumulated. In the wild-type extract, most pre-mRNA has been spliced, and thus only low levels of affinity-purified 45S spliceosomes are detected. The lack of accumulation of B, Bact, B*, and/or C complexes in the Δsnu17 extract, as compared with the wild type, suggests that there is a defect in the assembly of mature spliceosomes or, alternatively, that they are assembled but are less stable and thus subsequently dissociate.
Figure 1.
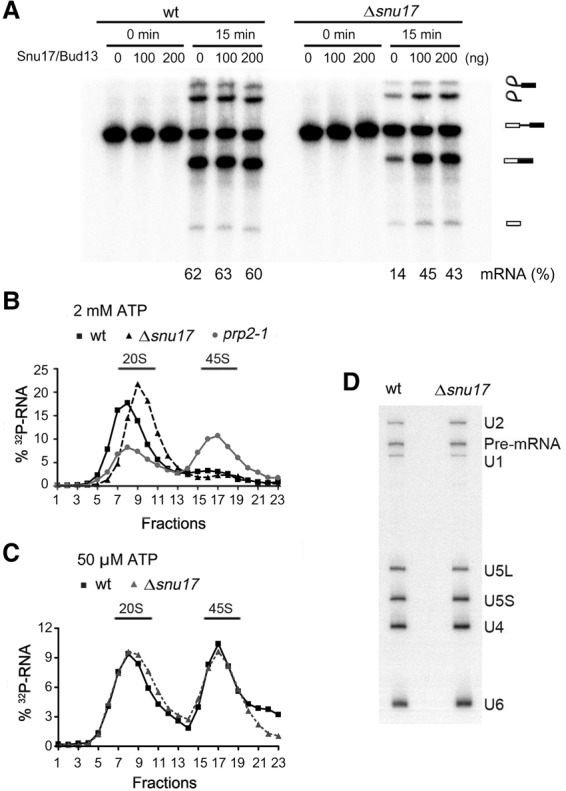
Pre-mRNA splicing, but not spliceosomal B complex formation, is less efficient in extracts from an S. cerevisiae Δsnu17 strain. (A) Splicing of 32P-labeled Actin pre-mRNA in whole-cell extracts from wild-type or Δsnu17 S. cerevisiae cells. A mixture of affinity-purified Snu17 and Snu17–Bud13 dimer (100 or 200 ng as indicated) or the corresponding buffer alone was added prior to incubation of the splicing reaction. The identities of the 32P-labeled RNA species are indicated at the right. The percentage of mRNA formed is indicated at the bottom. (B) Glycerol gradient sedimentation profile of affinity-purified spliceosomes formed on 32P-labeled Actin pre-mRNA under splicing conditions at 2 mM ATP for 60 min in extracts from wild-type, Δsnu17, and prp2-1 S. cerevisiae cells. The prp2-1 extract was preincubated at 35°C to inactivate Prp2. (C) Glycerol gradient sedimentation of affinity-purified spliceosomes formed at 50 µM ATP for 60 min in extracts from wild-type or Δsnu17 yeast cells. (D) RNA compositions of affinity-purified wild-type or Δsnu17 splicing complexes (formed at 50 µM ATP) migrating in the 45S peak fractions of the glycerol gradient. RNA was detected by Northern blotting, and the snRNAs are indicated at the right. The 32P-labeled Actin pre-mRNA was detected by autoradiography.
RES proteins are not required for B complex assembly
We next investigated whether B complex assembly is impaired in the absence of Snu17. To this end, splicing was performed in wild-type and Δsnu17 cell extracts at a low ATP concentration (50 µM as opposed to 2 mM ATP) that blocks Brr2 activity, leading to the arrest of spliceosome assembly at the B complex stage (Tarn et al. 1993; Fabrizio et al. 2009). Similar amounts of 45S spliceosomes could be affinity-purified from the wild type and the Δsnu17 splicing extracts (Fig. 1C). In both cases, these spliceosomes contained U1, U2, U4, U5, and U6 snRNAs and unspliced pre-mRNA (Fig. 1D), consistent with their designation as spliceosomal B complexes. Mass spectrometry (MS) revealed that B complexes formed in the absence of Snu17 (denoted BΔSnu17) also lacked Snu17's binding partner, Pml1, and, based on the number of peptides sequenced by MS, contained only low levels of the third RES complex protein, Bud13, compared with wild-type B complexes (Supplemental Table S1). Thus, efficient recruitment of Pml1 and Bud13 to the spliceosome is dependent on the presence of Snu17. All other proteins typically found in B complexes, including U2 snRNP, U4/U6.U5 tri-snRNP, and NTC proteins, appeared to be present at similar levels in the wild-type and BΔSnu17 complexes, based on peptide counts (Supplemental Table S1). Thus, the RES proteins are not required for the recruitment of these proteins to the B complex. Interestingly, although they normally first bind after B complex formation, there was an increase in the amount of Prp2 and Spp2 present in the BΔSnu17 versus Bwt complexes (Supplemental Table S1). However, based on peptide counts, the amount of Prp2 binding appears to be rather low. Nonetheless, this is a first indication that the RES proteins may prevent premature binding of Prp2 to the B complex. Taken together, these results show that B complex assembly is not dependent on the RES complex.
Formation of activated spliceosomes is less efficient in the absence of RES
Next we investigated whether RES affected the formation of activated Bact complexes. To stall spliceosome assembly at the Bact stage, we used a yeast strain carrying a mutant version of Prp2 (prp2-1) that is inactivated at elevated temperatures (Kim and Lin 1996). Deletion of SNU17 from the prp2-1 strain had no effect on yeast viability at 16°C, 25°C, and 30°C but led to a slower growth phenotype compared with prp2-1 containing the SNU17 gene (Supplemental Fig. S2). We incubated splicing extracts from the prp2-1 or prp2-1 Δsnu17 strains at 35°C to inactivate Prp2 and performed splicing with MS2-Act pre-mRNA at 2 mM ATP followed by MS2 affinity selection. Substantial quantities of 45S spliceosomes were affinity-purified in both the presence and absence of Snu17 (Fig. 2A). Spliceosomes formed in the prp2-1 extract contained the pre-mRNA and U2, U5, and U6 snRNAs but essentially no U4 (Fig. 2B), consistent with their designation as Bact complexes. In contrast, complexes formed in the prp2-1 Δsnu17 extract additionally contained substantial amounts of U4 but approximately half as much as that detected in B complexes formed in the prp2-1 extract at 50 µM ATP (Fig. 2B).
Figure 2.
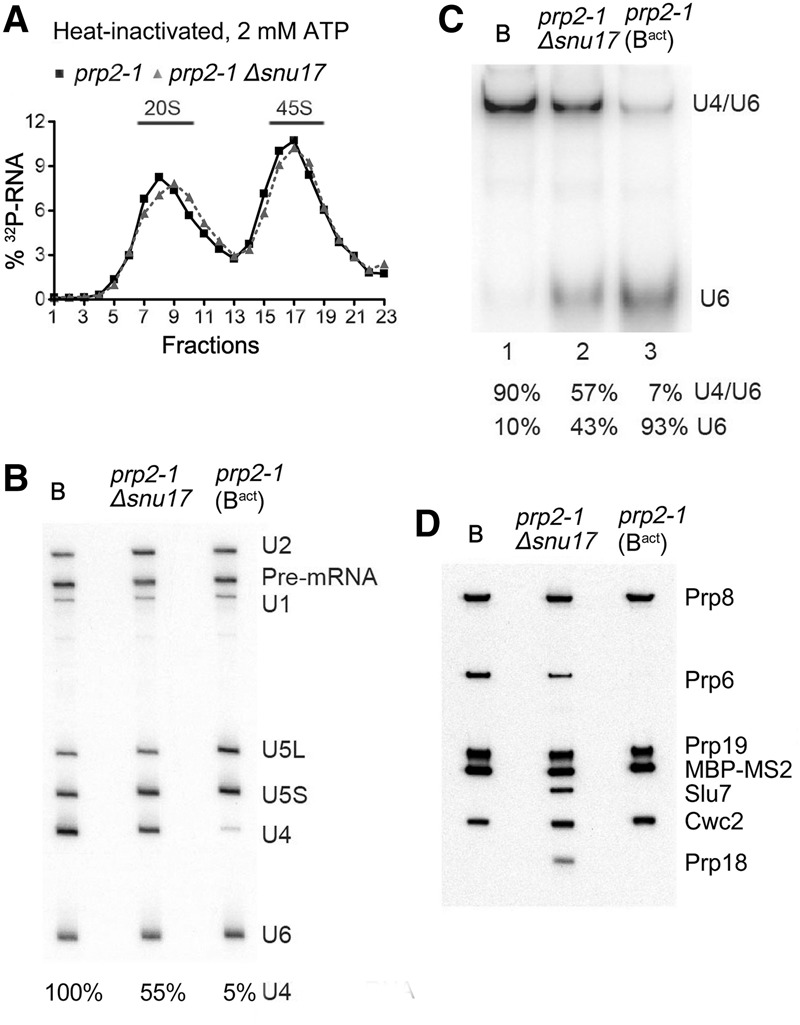
The transformation of the spliceosomal B complex into Bact is less efficient in the absence of RES complex proteins. (A) Glycerol gradient sedimentation profile of MS2-MBP affinity-purified spliceosomal complexes formed after initial heat inactivation of Prp2 on 32P-labeled Actin pre-mRNA in extracts from prp2-1 or prp2-1 Δsnu17 yeast cells after 60 min in the presence of 2 mM ATP. (B) RNA compositions of affinity-purified B complexes (formed at 50 µM ATP) or Bact or splicing complexes formed in extracts from prp2-1 Δsnu17 cells (at 2 mM ATP) sedimenting in the 45S peak fractions of the glycerol gradient. The pre-mRNA and snRNAs were separated by denaturing polyacrylamide gel electrophoresis (PAGE) and detected as described in the legend for Figure 1. The amount of U4 snRNA in each lane (shown below) was quantitated as a percentage of the pre-mRNA band (which acts as a spliceosome loading control) and then normalized to the amount of U4 in the B complex, which was set to 100%. (C) Quantitation of the amount of U4/U6 duplex and U6 snRNA in affinity-purified B, Bact, or spliceosomal complexes formed in extracts from prp2-1 Δsnu17 cells. RNAs were analyzed by native PAGE, and U6 snRNA was visualized by Northern blotting. The percent of free U6 versus U6 base-paired with U4 was quantitated with a PhosphorImager and is shown below each lane. The identity of the slower-migrating band as U4/U6 was confirmed by Northern blotting with a probe against U4 snRNA. (D) Proteins from affinity-purified B complexes (formed at 50 µM ATP) or spliceosomal complexes formed at 2 mM ATP in extracts from prp2-1 (predominantly Bact complexes) or prp2-1 Δsnu17 cells (as indicated) after heat inactivation of Prp2 were analyzed by Western blotting using antibodies against the indicated proteins. Antibodies against Prp8 were used to ensure equal loading.
To determine whether the U4 and U6 snRNAs are still base-paired in the 45S complexes containing U4, we performed native polyacrylamide gel electrophoresis (PAGE) after deproteination of RNA in the purified spliceosomes. In the prp2-1 Bact complexes, U6 is base-paired with U2 snRNA, but the U6–U2 duplexes do not survive deproteination and U6 snRNA thus migrates as a single RNA when subjected to native PAGE (Fig. 2C, lane 3). Approximately half of U6 was still base-paired with U4 in the prp2-1 Δsnu17 spliceosomes, compared with 90% in B complexes (Fig. 2C, lanes 1,2), suggesting that in the absence of Snu17, only half of the B complexes formed at 2 mM ATP undergo Brr2-mediated U4/U6 duplex unwinding compared with ∼90% in the prp2-1 control extract. Thus, in the absence of Snu17, about half of the yeast spliceosomes appear to undergo activation, while the other half are stalled at the B complex stage. As the RES proteins are missing (with the exception of low amounts of Bud13) in these purified spliceosomes (see below), we refer to these complexes as ΔRES “B/Bact-like” spliceosomes, respectively. Based on the amount of base-paired U4/U6, the ratio of ΔRES B-like to Bact-like complexes did not change significantly if the splicing reaction was incubated for 40 or 60 min (data not shown), suggesting that the mixed population of complexes generated after 60 min is not due to slower spliceosome assembly kinetics.
We next determined the protein composition of spliceosomes assembled in the prp2-1 Δsnu17 extracts at 2 mM ATP via MS. As expected, the RES complex proteins were either completely absent (Snu17 and Pml1) or strongly underrepresented (Bud13) compared with the prp2-1 (i.e., Bact control). Consistent with the presence of ∼50% each of B-like and Bact-like complexes, the ΔRES spliceosomes contained similar amounts of nearly all U2, U5, and NTC proteins compared with the prp2-1 control but substantially more of the U4/U6 proteins Prp31, Prp3, Prp4, and Snu13, which are normally lost during activation (Supplemental Table S2). Proteins typically more abundant in Bact versus B complexes were also detected in the ΔRES spliceosomes. Several of these (e.g., Ecm2, Cwc15, Bud31, Yju2, and Cwc24) were apparently less abundant (based on peptide counts) compared with the prp2-1 control, consistent with Bact-like complexes comprising only half of the ΔRES spliceosomes. Thus, in the absence of RES, most NTC-related and Bact proteins are/can be recruited during spliceosome activation. However, at present, it is not clear whether all of these Bact proteins are present in stoichiometric amounts and also whether their interactions within the spliceosome are altered in some way due to the absence of RES. Notably, similar or even lower peptide counts were observed for the Lsm proteins in the ΔRES B/Bact-like complexes versus Bact complexes. As the Lsm proteins are normally much more abundant in B complexes compared with Bact, this suggests that the ΔRES B complexes may lack the Lsm proteins. Surprisingly, the ΔRES B/Bact-like complexes also contained significant quantities of the step 1 factor Cwc25 and the step 2 splicing factors Slu7 and Prp18 as well as Prp43 and its cofactors, Ntr1/Spp382 and Ntr2 (Fig. 2D; Supplemental Table S2). In mature wild-type Bact complexes, these proteins are usually present in very low amounts, if at all, and are first incorporated at later stages of the splicing process (Supplemental Table S2; Fabrizio et al. 2009). Consistent with the MS results, immunoblotting revealed equal amounts of Prp19 and Cwc2 but higher amounts of Prp6 (which is abundant in B complexes), Slu7, and Prp18 in the prp2-1 Δsnu17 spliceosomes compared with the prp2-1 (Bact) control (Fig. 2D). Taken together, our results indicate that the RES proteins are required for the efficient transformation of the B complex into Bact.
In the absence of RES, the requirement for Prp2 activity can be bypassed in vitro
Spliceosomes formed in Prp2 heat-inactivated extracts lacking Snu17 contained a number of factors that normally first interact during the catalytic steps of splicing, suggesting that in the absence of RES, splicing catalysis can occur even if Prp2 is no longer active. We thus investigated whether prp2-1 Δsnu17 splicing extracts support pre-mRNA splicing in the absence of functional Prp2. Heat inactivation of Prp2 in prp2-1 extract led to a complete block in both steps of splicing (Fig. 3, cf. A and B, lanes 4–6). Surprisingly, with prp2-1 Δsnu17 extract, both catalytic steps were observed after heat inactivation, albeit to a lower extent than that observed in the absence of heat inactivation (i.e., 22% vs. 8% mRNA formation) (Fig. 3, cf. A and B, lane 10). Addition of a mixture of purified Snu17 and Snu17–Bud13 dimer abolished the bypass of the requirement for Prp2; that is, splicing was no longer observed in the heat-inactivated prp2-1 Δsnu17 extract (Fig. 3B, lanes 11,12). Thus, in the absence of a functional RES protein complex, some of the spliceosomes that are assembled undergo catalytic activation without Prp2-mediated remodeling. This suggests that at least a subset of the ΔRES complexes potentially has an aberrant structure that precludes the requirement for Prp2 activity to convert them into catalytically active spliceosomes.
Figure 3.
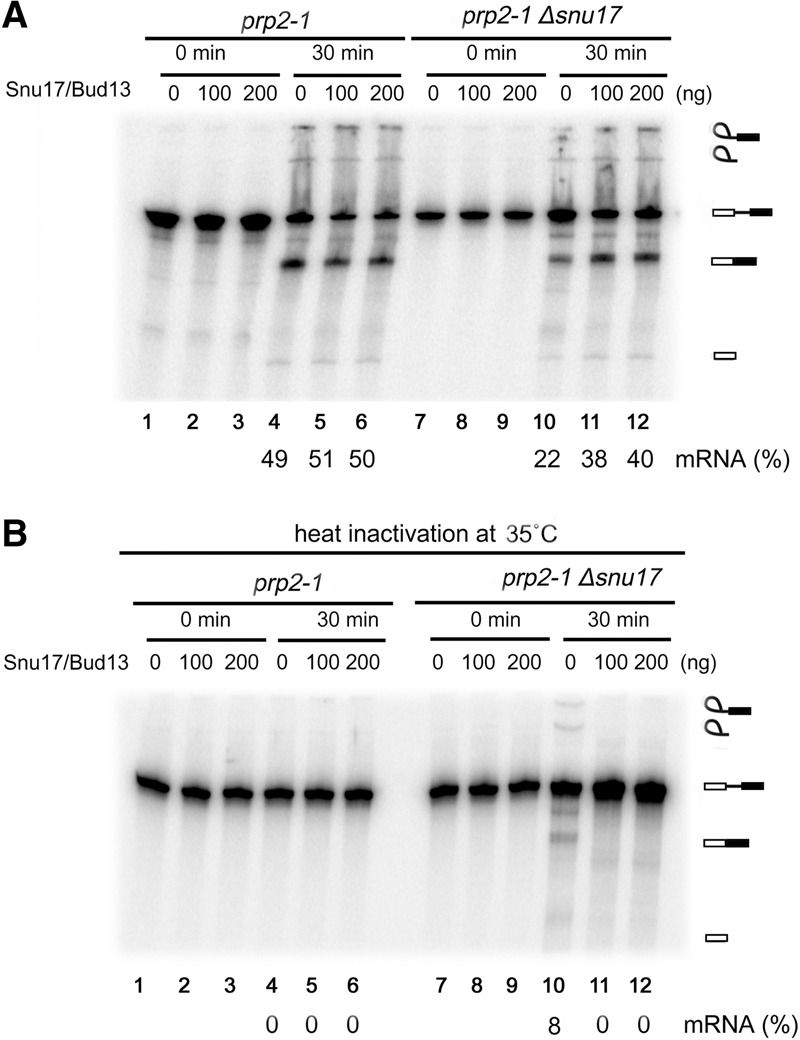
The requirement for Prp2 activity can be bypassed in the absence of Snu17. Pre-mRNA splicing in extracts from prp2-1 or prp2-1 Δsnu17 yeast cells without heat inactivation of Prp2 (A) or after heat inactivation of Prp2 at 35°C (B). Splicing was performed without or after addition of affinity-purified Snu17/Bud13 (100 or 200 ng as indicated). RNA was separated by denaturing PAGE and visualized using a PhosphorImager. The identities of the 32P-labeled RNA species are indicated at the right. The percentage of mRNA formed after 30 min is indicated below.
Brr2 unwinds U4/U6 in affinity-purified B complexes in a RES-independent manner
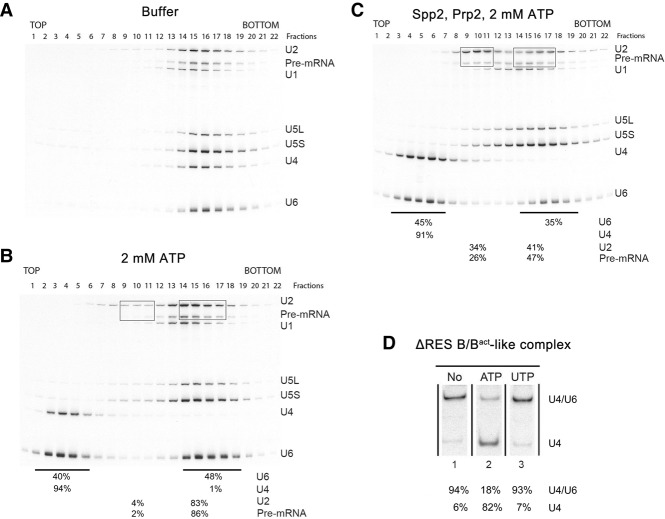
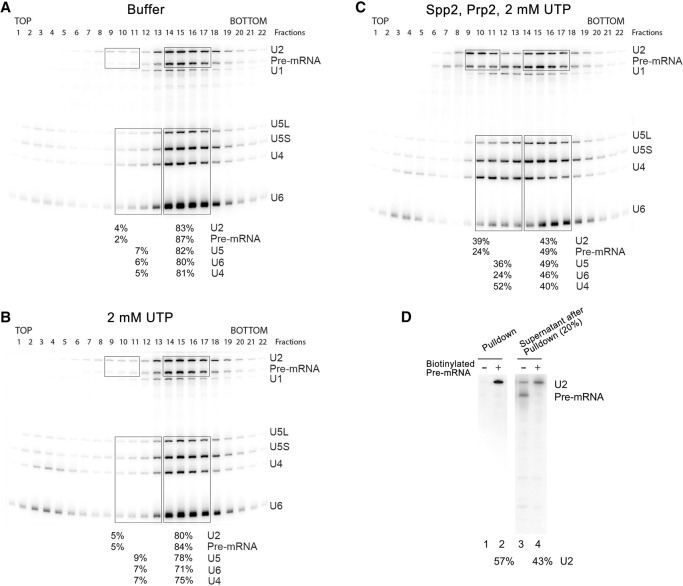
In Δsnu17 splicing extracts incubated with 2 mM ATP, spliceosomes appear to accumulate only in the absence of functional Prp2 RNA helicase (cf. Figs. 1B and 2A), suggesting that Prp2 may play an active part in the destabilization of ΔRES spliceosomes. Thus, we next investigated whether ΔRES B/Bact-like spliceosomes can be dissociated by Prp2. For this purpose, we incubated affinity-purified ΔRES B/Bact-like spliceosomes with ATP in the absence and presence of recombinant Prp2 and Spp2 and then subjected them to glycerol gradient centrifugation. Incubation with ATP alone led to the quantitative release of U4 and a substantial amount of U6 (Fig. 4A,B). These two snRNAs comigrated in fractions 3–6 (around 10S) of the gradient, whereas the pre-mRNA peaked together with U2, U5, and the remainder of the U6 snRNA in fractions 14–18 (around 40S–45S). Native PAGE revealed that addition of 2 mM ATP leads to the unwinding of a substantial amount (82%) of the U4/U6 duplex (Fig. 4D), suggesting that Brr2 is active under these conditions. To test whether Brr2 helicase activity is responsible for the observed disruption of the U4/U6 snRNP, we incubated the purified ΔRES B/Bact-like spliceosomes with UTP instead of ATP; in contrast to Prp2, Brr2 activity is not stimulated in the presence of UTP (Santos et al. 2012). Indeed, the U4/U6 snRNA duplex was not unwound in the presence of UTP (Fig. 4D), and the majority of U4/U6 snRNPs remained bound to the ΔRES spliceosomes (Fig. 5, cf. A and B). We thus conclude that Brr2, as opposed to Prp2, is responsible for the release of both U4 and U6 from the isolated ΔRES B-like complexes when incubated with ATP. We also analyzed the effect of ATP or UTP addition on affinity-purified B or BΔRES complexes formed at 50 µM ATP. A similar amount of U4/U6 unwinding (i.e., 87%) (Supplemental Fig. S3) and release of both U4 and U6, as evidenced by gradient centrifugation (Supplemental Fig. S3), was observed with both B and BΔRES affinity-purified complexes upon incubation with 2 mM ATP, whereas addition of UTP had little or no effect (Supplemental Fig. S4). These data indicate that Brr2 unwinds the U4/U6 snRNPs in affinity-purified B complexes in a RES-independent manner. Thus, affinity-purified B complexes formed at 50 µM ATP may contain Brr2 that is no longer subject to negative regulation (see the Discussion).
Figure 4.
Affinity-purified ΔRES spliceosomes are partially disassembled upon addition of Prp2, Spp2, and ATP. Spliceosomal complexes formed in extracts from prp2-1 Δsnu17 yeast cells after initial heat inactivation of Prp2 (designated ΔRES B/Bact-like complexes) were affinity-purified, and 45S complexes were subjected to a second glycerol gradient after incubation with buffer (A), 2 mM ATP (B), or ATP, Prp2, and Spp2 (C). RNA was isolated from each gradient fraction, separated by denaturing PAGE, and detected by Northern blotting with 32P-labeled probes against the snRNAs indicated at the right. The 32P-labeled Actin pre-mRNA was detected by autoradiography. The percentage of the pre-mRNA or the indicated snRNAs in the boxed or underlined fractions is shown below the gel in B and C. (D) RNA was isolated from ΔRES B/Bact-like complexes and analyzed by native PAGE. Free U4 snRNA or U4 base-paired with U6 was subsequently visualized by Northern blotting with a 32P probe complementary to U4 snRNA. The percentage of free U4 versus U4 base-paired with U6 was quantitated with a PhosphorImager and is shown below each lane.
Figure 5.
The Prp2 RNA helicase facilitates the disassembly of ΔRES spliceosomes. ΔRES B/Bact-like complexes were affinity-purified, and 45S complexes were subjected to a second glycerol gradient after incubation with buffer (A), 2 mM UTP (B), or UTP, Prp2, and Spp2 (C). RNA was analyzed as described in the legend for Figure 4. The percentage of the pre-mRNA or the indicated snRNAs in the boxed or underlined fractions is shown below each panel. (D) Affinity-purified 45S spliceosomes formed on nonbiotinylated or biotinylated Actin pre-mRNA in prp2-1 Δsnu17 extract (after heat inactivation of Prp2) were incubated with UTP, Prp2, and Spp2 and then analyzed on a second glycerol gradient. RNP complexes in gradient fractions 8–10 were subjected to streptavidin agarose pull-downs. RNA precipitated together with the biotinylated pre-mRNA (which was not released from the streptavidin beads) as well as RNA remaining in the supernatant were analyzed by denaturing PAGE followed by Northern blotting (to visualize the U2 snRNA) or autoradiography (to detect the radiolabeled pre-mRNA). The percentage of U2 in the pulled-down fraction versus the supernatant is shown below the gel.
The Prp2 RNA helicase facilitates the disassembly of ΔRES spliceosomes
When affinity-purified ΔRES B/Bact-like spliceosomes were incubated with Prp2, Spp2, and 2 mM ATP and fractionated by glycerol gradient centrifugation, a shift in not only U4 and U6 snRNA but also a substantial portion (∼30%) of the U2 snRNA (Fig. 4C) was observed. The latter peaked in fractions 9–11 of the gradient (around 20S), whereas the remaining fraction of the U2 snRNA comigrated with the pre-mRNA, U5 snRNA, and U6 snRNA at 40S–45S (i.e., fractions 14–18) (Fig. 4C). In contrast to the ATP-only control, a significant fraction of the pre-mRNA and U5 also now sedimented in fractions 9–13 (Fig. 4C). To exclude any effects of the Brr2 helicase activity, we incubated the ΔRES B/Bact-like spliceosomes with Prp2/Spp2 and UTP. Prp2 hydrolyzes UTP and activates spliceosomes in the presence of UTP with an efficiency similar to that observed with ATP (Kim et al. 1992; Warkocki et al. 2015). Notably, incubation of the ΔRES B/Bact-like spliceosomes with Prp2/Spp2 and UTP led to a shift of 39% of the U2 snRNPs together with 25% of the pre-mRNA to fractions 9–11 (Fig. 5, cf. B and C), while the remainder migrated together with the 40S–45S spliceosomes (fractions 14–17). In contrast, only 5% of the U2 and pre-mRNA sedimented in these fractions after incubation with solely UTP. A shift of U5 as well as U4 and U6 to fractions 10–13 was also observed upon addition of Prp2/Spp2 and UTP but not with UTP alone. In contrast, addition of Prp2, Spp2, and 2 mM UTP to Bact complexes containing RES proteins led to the release of only small amounts of U2 (Supplemental Fig. S5).
A major fraction of the U2-associated proteins (e.g., SF3a and SF3b proteins) peaked together with U2 snRNA in fractions 9–11, suggesting that intact 18S U2 snRNPs are present in these fractions (cf. Supplemental Tables S3 and S4). Part of the pre-mRNA comigrated with the U2 snRNPs in fractions 9–11, suggesting that U2 may still be bound to the former (Fig. 5C). To test this, we assembled spliceosomes on biotinylated actin pre-mRNA in heat-treated splicing extracts from the prp2–1 Δsnu17 strain, affinity-purified the resulting ΔRES B/Bact-like complexes, incubated them with Prp2/Spp2 and UTP, and subjected them to glycerol gradient centrifugation. Pull-down experiments with streptavidin beads and the 20S gradient fractions containing U2 and the pre-mRNA indicated that approximately half of the U2 in these fractions were bound to the pre-mRNA, as evidenced by the presence of U2 in the pulled-down fraction but ∼50% of U2 and essentially none of the pre-mRNA remaining in the post-pull-down supernatant (Fig. 5D).
Some of the U4 snRNA migrated in the intact spliceosome peak in fractions 14–17, while a substantial amount of U4 also sedimented together with U5 and U6 snRNAs in fractions 10–13 (25S–30S) (Fig. 5C). This suggests that the majority of the U4/U6.U5 tri-snRNPs is released from the ΔRES B-like complexes. Consistent with this, yeast tri-snRNP proteins such as Prp38 and Snu66 were enriched in gradient fractions 11–14 after addition of Prp2/Spp2 and UTP to the ΔRES spliceosomes (cf. Supplemental Table S3 and S4). Moreover, native gel electrophoresis of the deproteinated RNAs along the gradient showed that the greater part of the U4 snRNA present in fractions 10–12 and 14–17 was base-paired with the U6 snRNA (Supplemental Fig. S6). Finally, immunoprecipitation experiments with antibodies against the U5 snRNP Snu114 protein and the various gradient fractions containing the ΔRES complexes generated after incubation with Prp2, Spp2, and UTP indicated that most of the U5 snRNP is indeed complexed with U4/U6 in fractions 10–13 (Supplemental Fig. S7). In summary, these results indicate that in the absence of RES, Prp2 facilitates spliceosome disassembly, leading to primarily tri-snRNP release, while at least part of the U2 snRNP remains bound to the pre-mRNA.
As incubation of the mixture of ΔRES B/Bact-like spliceosomes with Prp2, Spp2, and UTP led to the release of predominantly U4/U6.U5 tri-snRNPs, B-like spliceosomes appear to be the major target for dissociation by Prp2. We thus investigated whether ΔRES B complexes assembled in the presence of 50 µM ATP could also be dismantled by Prp2. When ΔRES B complexes were incubated with UTP plus Prp2 and Spp2, an increase in the amount of U2 and pre-mRNA in fractions 8–10 was observed compared with UTP alone (Supplemental Fig. S4E,F); approximately six times as much U2 and pre-mRNA migrate in this region (3% vs. 19% for U2 and 2% vs. 16% for the pre-mRNA). In contrast, with wild-type B complexes, this difference was substantially less (Supplemental Fig. S4B,C). Thus, these results show that in the absence of RES, Prp2 facilitates the disassembly of B complexes formed at 50 µM ATP.
Disassembly of ΔRES spliceosomes is dependent on Prp2 activity
ΔRES B/Bact-like spliceosomes also contain the helicases Prp22 and Prp43, both of which hydrolyze UTP efficiently and are involved in spliceosome disassembly (Arenas and Abelson 1997; Martin et al. 2002; Tanaka and Schwer 2005; Tsai et al. 2005; Fourmann et al. 2013). However, addition of UTP alone did not lead to substantial disassembly of the ΔRES spliceosomes, indicating that these helicases do not contribute significantly to U2 and tri-snRNP release under these conditions. To provide additional evidence that the observed disassembly is dependent on Prp2 action, we used the ActinΔ6 (ActΔ6) pre-mRNA that contains only 6 nucleotides (nt) downstream from the BS (Fabrizio et al. 2009). Prp2 activity requires the presence of at least 25–30 intron nucleotides downstream from the BS (Fabrizio et al. 2009; Liu and Cheng 2012; Warkocki et al. 2015). Thus, with the ActΔ6 pre-mRNA, Bact complexes assemble efficiently, and Prp2 still binds, but the Prp2-induced remodeling that yields the B* complex no longer occurs (Warkocki et al. 2009, 2015). As observed with the wild-type Act pre-mRNA, a mixture of 45S B-like and Bact-like complexes appear to form on ActΔ6 pre-mRNA in the absence of Snu17, based on (1) the higher amount of U4 snRNA present (compared with the complexes formed on ActΔ6 in the presence of Snu17) and (2) the increased amounts of U4/U6 and tri-snRNP-specific proteins detected by MS (Supplemental Fig. S8; Supplemental Table S5). ActΔ6 spliceosomes formed in the absence of Snu17 also lacked Pml1 and contained only low amounts of Bud13. The protein compositions of the ΔRES spliceosomes were largely the same irrespective of whether they were assembled on ActΔ6 or Act wild-type pre-mRNA, except that Prp2 was present in the ActΔ6 spliceosomes (Supplemental Tables S2, S5). Incubation of the ΔRES ActΔ6 B/Bact-like spliceosomes with UTP and Prp2/Spp2 did not lead to an increase in the amount of U2 and pre-mRNA in the 20S region of the gradient compared with the UTP-only control (Fig. 6A–C). Thus, disruption of the ΔRES spliceosomes requires the presence of >6 nt downstream from the BS, demonstrating that disassembly is dependent on Prp2 activity and not solely on Prp2 binding per se.
Figure 6.
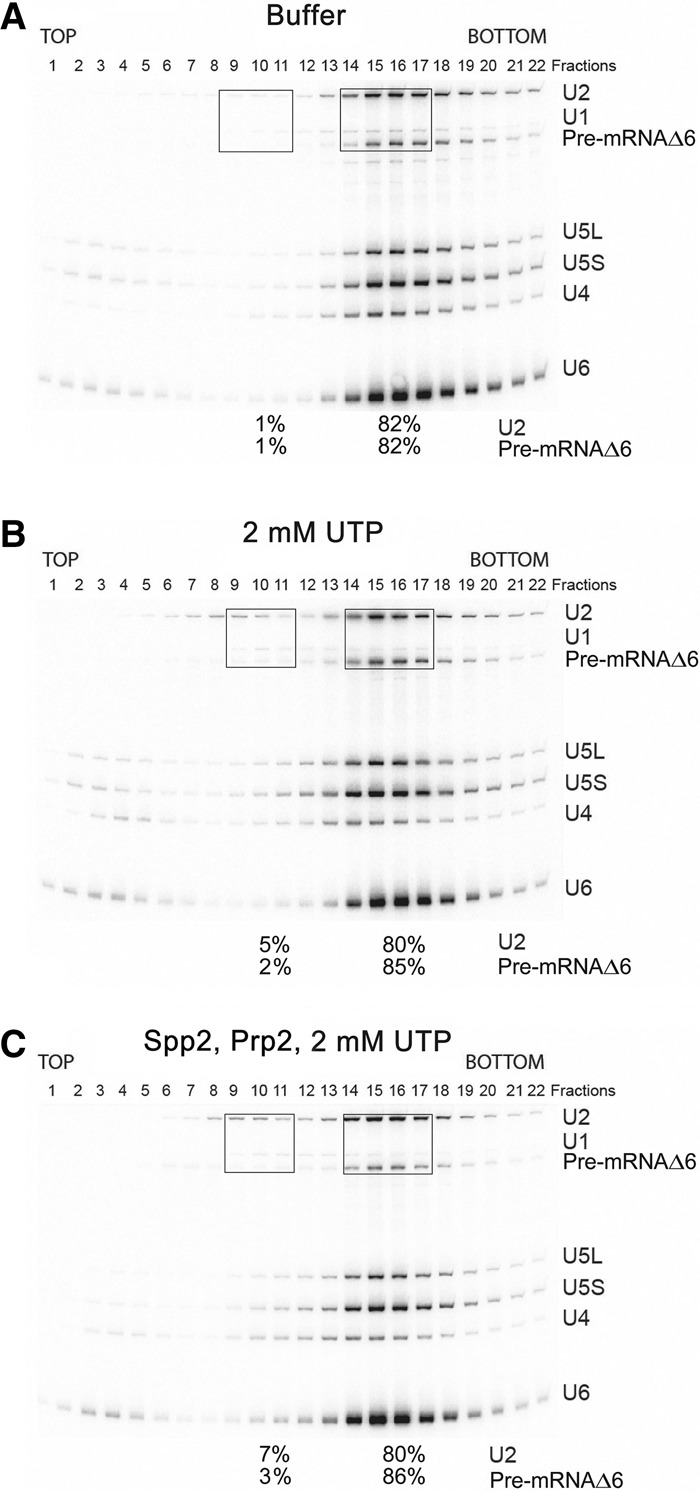
ΔRES spliceosomes formed on truncated ActΔ6 pre-mRNA are not disassembled upon addition of UTP, Prp2, and Spp2. Affinity-purified ΔRES B/Bact-like spliceosomes formed on truncated ActΔ6 pre-mRNA at 2 mM ATP were subjected to a second glycerol gradient after incubation with buffer (A), 2 mM UTP (B), or UTP, Prp2, and Spp2 (C). RNA was analyzed as described in the legend for Figure 4. The percentage of U2 and the pre-mRNA in the boxed fractions is shown below the gel in each panel.
Discussion
Here we investigated the role of the RES complex in yeast pre-mRNA splicing. Splicing efficiency was substantially lower in extract from the Δsnu17 strain, and the recruitment of Pml1 and Bud13 to B and Bact spliceosomal complexes was either abolished or substantially reduced. As Snu17 acts as the binding platform for both Bud13 and Pml1, RES complex formation will be abolished in the extract from the Δsnu17 strain. Furthermore, depletion of Snu17 might additionally affect the stability or expression of the other two RES components. RES was not required for the assembly of spliceosomal B complexes at low ATP. However, its absence hindered the efficient formation of Bact complexes at 2 mM ATP. Although B-like and Bact-like spliceosomes were generated in the absence of the RES protein complex, some of these ΔRES spliceosomes were no longer dependent on Prp2 activity for their catalytic activation, consistent with the idea that they are structurally compromised. Incubation of wild-type and ΔRES spliceosomes with ATP led to U4/U6 duplex unwinding and the release of both U4 and U6 from the spliceosome, indicating that affinity-purified B complexes formed at 50 µM ATP as well as ΔRES B complexes formed at 2 mM ATP contain Brr2 that is no longer subject to negative regulation. Incubation of the ΔRES spliceosomes with Prp2, Spp2, and UTP led to the release of U2 snRNPs and U4/U6.U5 tri-snRNPs, indicating that Prp2 is involved in their disassembly. This raises the interesting possibility that Prp2 proofreads the RNP structure of the spliceosome and facilitates the dismantling of those complexes with an aberrant protein composition or RNP structure.
ΔRES spliceosomes allow premature binding of Prp2
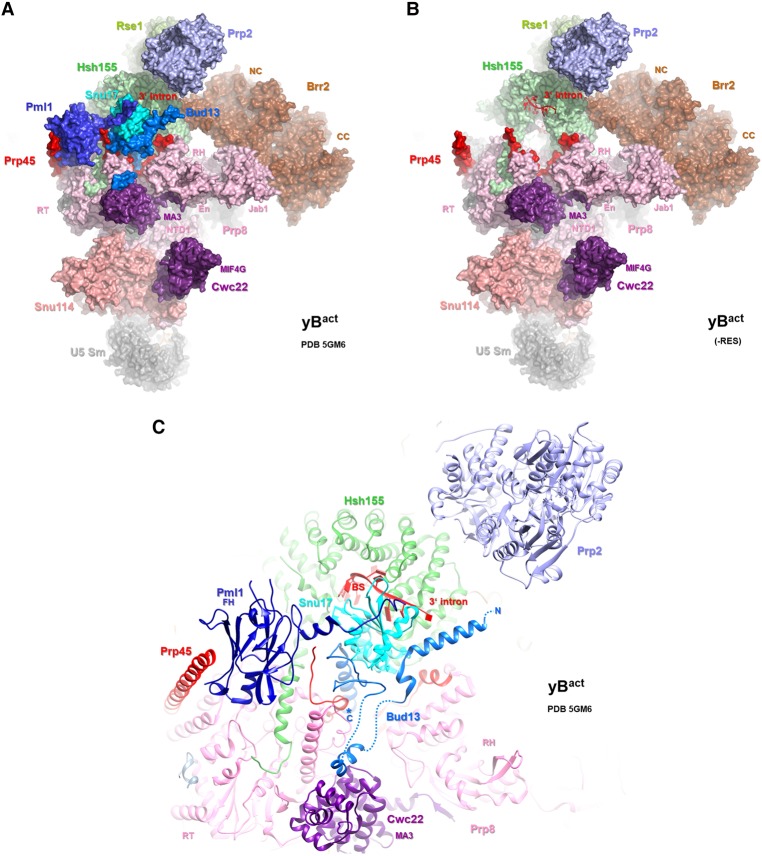
Our affinity-purified ΔRES B and B/Bact-like complexes did not exhibit significantly different S-values on glycerol gradients compared with B and Bact complexes containing the RES complex, which would be an indication of a very large structural change. Nonetheless, the ΔRES spliceosomes could potentially still be structurally compromised, as the absence of Snu17 and Pml1 and, in most complexes, also Bud13 (Supplemental Tables S1, S2) could affect the interaction of other spliceosomal proteins with which they interact, including Hsh155, Prp8, Prp45, and Cwc22 (Fig. 7). However, essentially all other proteins typically found in the yeast B and/or Bact complexes were detected in affinity-purified ΔRES B-like and Bact-like complexes (Supplemental Table S1, S2). This suggests that RES does not affect the recruitment of these spliceosomal proteins. However, it is unclear whether they all still bind in a productive/functional way in the absence of RES.
Figure 7.
The location of the RES complex proteins in the yeast Bact complex. (A,B) Overview of the location of selected proteins, shown with space-filling models, in the yeast Bact structure (Rauhut et al. 2016). In B, Snu17, Pml1, and Bud13 have been removed. (C) Close-up of the RES proteins and other spliceosomal proteins that they contact or that are in close proximity in the yeast Bact complex. Proteins are depicted as ribbon diagrams.
Unexpectedly, the absence of RES appeared to lead to enhanced Prp2 binding to the B complex, to which it normally does not bind (Supplemental Table S1). Based on our MS results, the endogeneous Prp2 is likely present in low amounts and thus in only a very small number of the purified ΔRES B complexes. After heat inactivation, the Prp2 temperature-sensitive mutant does not associate with the stalled Bact complex or the ΔRES B/Bact-like complexes formed at 2 mM ATP (Supplemental Table S2). However, the fact that the ΔRES B-like complexes formed at 2 mM ATP could be disassembled upon addition of Prp2, Spp2, and UTP is consistent with the idea that the exogenously added Prp2 readily binds prematurely to these ΔRES complexes as well. This suggests that the RES complex either directly or indirectly prevents Prp2 from associating prior to formation of mature Bact complexes, at least in yeast extracts. That the RES complex may influence the correct temporal association and/or productive interaction of Prp2 is consistent with the facts (1) that protein–protein cross-linking indicated that the RES proteins and Prp2/Spp2 are close to one another in the yeast Bact complex (Rauhut et al. 2016) and (2) that both Prp2 and Snu17 bind to the U2 Hsh155 protein, albeit on opposite sides of the Hsh155 HEAT domain (Fig. 7). The Hsh155 HEAT repeats are organized as a relatively relaxed right-handed superhelix whose overall structure can be modulated by protein interactions; thus, the binding of a protein on one side of the HEAT domain could potentially affect the interaction of proteins on the other.
RES is required for efficient formation of Bact complexes in splicing extracts
The transformation of ΔRES B complexes into Bact complexes was substantially hindered (Fig. 2), revealing a role for Snu17 and/or Pml1 and Bud13 in the activation process. The initial step of activation (namely, unwinding of the U4/U6 duplex by Brr2) was hindered in ∼50% of the complexes when the RES complex was absent. This suggests that the efficient triggering of Brr2 activity is compromised in about half of the ΔRES B-like complexes formed at 2 mM ATP. The precise mechanism whereby Brr2 activity is normally triggered is not clear, but our data suggest that the RES complex may directly or indirectly aid in activating Brr2 during the B-to-Bact transition. While the recent cryo-EM structure of the yeast B complex did not reveal the location of the RES complex proteins, the latter do not appear to contact Brr2 in the yeast Bact complex (Rauhut et al. 2016; Yan et al. 2016; Plaschka et al. 2017); however, only the stable regions of the RES proteins and other spliceosomal proteins could be localized in the Bact structure. The absence of RES may hinder Brr2 helicase activity by altering the spliceosome's structure, for example, such that Brr2 no longer can productively engage its U4/U6 RNA substrate. During the B-to-Bact transition, there is a large exchange of spliceosomal proteins, which likely occurs stepwise, with the loss of certain proteins being a prerequisite for the recruitment of others. Interestingly, the Lsm proteins appeared to dissociate from the ΔRES B complexes (Supplemental Table S2), suggesting that the absence of RES destabilizes other spliceosomal proteins and disturbs the normal exchange of proteins during the B-to-Bact transition. In this context, it should be noted that the Bact-protein Prp45 interacts with RES and several other Bact-proteins (Fig. 7; Rauhut et al. 2016; Yan et al. 2016); thus, in the absence of RES, Prp45 might no longer coordinate/stabilize the incorporation of these Bact proteins during activation.
Brr2-negative regulation appears to be lost after affinity purification of the B complex
Surprisingly, while Brr2 activity appeared to be hindered in B complexes in the presence of cell extracts and 2 mM ATP, addition of 2 mM ATP to affinity-purified B complexes (both containing or lacking the RES complex) or the mixture of ΔRES B/Bact-like complexes formed in extract at 2 mM ATP led to the quantitative unwinding of U4/U6 present in these spliceosomes (Fig. 4; Supplemental Fig. S3). Thus, Brr2 in affinity-purified B complexes appears to be no longer subject to negative regulation, and, in a purified splicing system, the RES proteins are not required for Brr2 activation. Thus, one or more proteins that inhibit Brr2 presumably are removed from the spliceosome during the affinity purification step. Notably, in contrast to the situation in purified human B complexes, which do not disassemble when incubated with ATP, Brr2 appears to be prematurely engaged with its U4 RNA substrate in purified yeast B complexes prior to the formation of the U6 ACAGA/5′ss helix (Bertram et al. 2017; Plaschka et al. 2017). The Sad1 protein plays an essential part in maintaining the stability of U4/U6.U5 tri-snRNPs in S. cerevisiae splicing extracts in the presence of high concentrations of ATP (Huang et al. 2014). In the absence of Sad1, the tri-snRNPs dissociate into U5 and U4/U6 snRNPs in the presence of ATP and thus probably are dependent on Brr2 action, while, in its presence, they remain stable (Huang et al. 2014). It is conceivable that the lack of Sad1 in the purified yeast B complexes may therefore be responsible for the Brr2-mediated destabilization/dissociation of U4/U6 snRNPs. In the splicing reaction, on the other hand, Sad1 could potentially directly or indirectly suppress the activity of Brr2.
Prp2 action is not a prerequisite for catalytic activation when RES is not present
In the absence of RES, spliceosomes that can undergo catalytic activation and thus catalyze both steps of splicing without prior Prp2-mediated remodeling are generated in vitro (Fig. 3). Interestingly, Prp2 action is thought to lead to the destabilization/restructuring of the SF3a/b proteins and the RES complex, which allows the subsequent binding of Cwc25, which together with Yju2 promotes efficient step 1 catalysis. Destabilization of RES proteins within the spliceosome could be a key initial event that leads to the subsequent destabilization of SF3a/b and the exchange of spliceosomal proteins needed for step 1 of splicing. Thus, the absence of RES would preclude the requirement for its destabilization/displacement by Prp2, which in turn could allow subsequent protein rearrangements without Prp2 activity. In Bact, the Hsh155 HEAT repeats form a tight clamp that sequesters the U2/BS helix, including the nucleophile for the first step of splicing: the BS adenosine (Rauhut et al. 2016; Yan et al. 2016). During catalytic activation, the HEAT repeats must be restructured to liberate the U2/BS helix for its docking into the catalytic center, and this remodeling has been proposed to be facilitated by Prp2, which binds to the Hsh155 HEAT domain (Fig. 7). As the RES proteins also contact the HEAT domain and stabilize it by bridging it to several other proteins, including Prp8 (Fig. 7), it is conceivable that in their absence, the Hsh155 clamp may no longer be as tight, allowing release of the U2/BS helix without the action of Prp2. This is reminiscent of studies showing that when the U1-C protein, which stabilizes the U1/5′ss interaction, is altered, the requirement for Prp28 helicase activity to disrupt the latter interaction can be bypassed (Chen et al. 2001).
Prp2 action leads to the disassembly of ΔRES spliceosomes
Addition of Prp2, Spp2, and UTP to affinity-purified ΔRES spliceosomes formed at either 50 µM ATP or 2 mM ATP led to their disassembly. That this is dependent on Prp2 action as opposed to Prp43 is supported by the facts (1) that addition of UTP alone had little effect on the stability of the ΔRES spliceosomes even though they contain significant amounts of Prp43 and (2) that ΔRES spliceosomes formed on the ActΔ6 pre-mRNA, which lacks intron nucleotides required for Prp2 activity, were not disassembled (Fig. 6). It is conceivable that in the absence of RES, Prp2 action leads to a conformational change and that this Prp2-mediated rearrangement earmarks spliceosomes for their subsequent disassembly by Prp43. However, when RES is absent, our affinity-purified B complexes (in contrast to the ΔRES B/Bact-like complexes formed at 2 mM ATP) appear to contain very little Prp43 (cf. Supplemental Tables S1 and S2), and, more importantly, neither of its essential cofactors (Ntr1/Spp382 and Ntr2) was detected by MS (Supplemental Table S1). Thus, it is highly unlikely that Prp43 is active in our purified ΔRES B complexes. Nonetheless, addition of Prp2, Spp2, and UTP led to their dissociation, consistent with the idea that Prp2 alone is directly responsible for their disassembly.
It is also likely that Prp2 facilitates the disassembly of ΔRES spliceosomes in splicing extracts where Prp2 is active; that is, in Δsnu17 yeast cell extracts, B complexes did not accumulate at 2 mM ATP (Fig. 1), consistent with the idea that they are subject to destabilization/disassembly by Prp2 if the latter is present in an active form. Both U2 snRNP and U2 bound to the pre-mRNA plus predominantly the U4/U6.U5 tri-snRNP were released from the ΔRES spliceosomes, indicating that ΔRES B complexes are the main disassembly target (Fig. 5). However, as it is difficult to resolve the U5 snRNP from the tri-snRNP on our gradients, ΔRES Bact complexes may also be disassembled. This suggests that Prp2 facilitates the release of the tri-snRNP (or U5 in the case of Bact) from the spliceosome when the RES complex is absent—leaving U2 bound to the pre-mRNA—and that a portion of U2 subsequently dissociates from the pre-mRNA potentially during gradient centrifugation or first during the streptavidin agarose pull-down experiment. Alternatively, Prp2 could potentially also facilitate the release of U2, and, as a consequence, the tri-snRNP may subsequently dissociate.
The RES complex first binds during B complex formation in yeast, indicating that its binding site is created upon the stable tri-snRNP association with the spliceosome. Although the location of the RES proteins in the yeast B complex is currently unknown (Plaschka et al. 2017), the cryo-EM structure of the yeast Bact complex revealed that RES complex proteins contact both U2 and U4/U6.U5 components at this stage. Specifically, Pml1's N-terminal forkhead-associated domain and also Bud13's C-terminal helix contact the RT domain of the U5 protein Prp8, whereas Snu17's RRM contacts HEAT repeats of the U2 Hsh155 protein (Fig. 7). Thus, the RES proteins are at the interface between U2 and the U4/U6.U5 tri-snRNP or U5 snRNP interaction within the spliceosome and may normally help to stabilize their interaction in the spliceosome. Prp2 contacts the U2 Hsh155 protein in the Bact complex (Fig. 7), but it is not clear where it binds in the ΔRES B complex. Given that, in the Bact complex, Prp2 normally facilitates the destabilization of the U2 SF3a and b proteins, which stabilize the U2/BS interaction, it is tempting to speculate that, at least in some instances, disassembly of the ΔRES B complex involves destabilization of the U2/pre-mRNA interaction by Prp2. However, release of U2 from the ΔRES B complexes does not appear to involve the dismantling of the U2 snRNP, as the U2 snRNPs that are released still contain the SF3a/b proteins (Supplemental Tables S3, S4). Future studies should help to clarify the precise targets of Prp2 in the ΔRES B complexes.
Proofreading the protein composition of the spliceosome by Prp2?
The DEAH-box ATPases/RNA helicases Prp2, Prp16, and Prp22 bind transiently and in a defined temporal and mutually exclusive manner to the spliceosome. This ensures that these enzymes act at a well-defined stage of the splicing process. As discussed above, in the absence of RES, Prp2 appears to be able to bind the spliceosome prematurely (i.e., prior to the activation stage), which in turn leads to disassembly of the spliceosomal B complexes lacking RES. This suggests that Prp2 is involved in proofreading the proper protein composition and/or RNP structure of the precatalytic spliceosome. Complexes lacking RES or other proteins may allow premature binding of Prp2 and thereby trigger their disassembly/discard, preventing the progression of improperly assembled spliceosomes toward the activation stage. At least five of the spliceosomal ATPases/RNA helicases promote splicing fidelity by proofreading the pre-mRNA splice sites and BS that are selected and then earmarking spliceosomes assembled on suboptimal sites for the discard pathway involving Prp43 (Semlow and Staley 2012). More recently, Prp2 was proposed to proofread the catalytic RNA network of the activated spliceosome (Wlodaver and Staley 2014). It is not clear from our data whether Prp2 inspects changes in the spliceosome's protein composition directly, or instead the absence of RES affects the structure of the spliceosome, and Prp2 senses this indirectly via its interactions with RNA. Taken together, our studies suggest that in addition to proofreading the structure and sequence of RNA elements in the spliceosome, Prp2 and potentially other ATPases/RNA helicases can also proofread whether spliceosomes with an aberrant protein composition and/or RNP structure are formed.
Materials and methods
Yeast growth and whole-cell extract preparation
The S. cerevisiae 3.2.AID/CRL2101 strain (MATα, prp2-1, ade2, his3, lys2-801, ura3) was kindly provided by Dr. Ren-Jang Lin. This Prp2 mutation renders it temperature-sensitive at 35°C and, after heat inactivation, prevents the mutant protein from binding and stalls spliceosome assembly at the Bact complex stage. Yeast strains (wild-type or prp2-1 background) lacking Snu17 were generated by one-step PCR transformation by using the pFA6A-KanMX6 plasmid DNA template (Longtine et al. 1998). Whole-cell yeast extracts used for in vitro splicing was prepared as described previously (Rauhut et al. 2016).
Yeast pre-mRNA splicing in vitro
Uniformly 32P-labeled MS2-Actin pre-mRNA bearing three tandem phage R17 MS2-binding sites was produced by in vitro transcription with T7 RNA polymerase. To prepare uniformly biotinylated Actin pre-mRNA, biotin-16-UTP (Jena Bioscience) was additionally included in the transcription reaction. Dialyzed prp2-1 or prp2-1 Δsnu17 extract was incubated for 30 min at 35°C to heat-inactivate Prp2. A standard splicing reaction contained 60 mM K-phosphate buffer (pH 7.25); 0.3% (w/v) PEG8000; 2.5 mM MgCl2; 2 mM spermidine; 2 mM ATP; 40% of dialyzed wild-type, prp2-1, or prp2-1 Δsnu17 extract; and 1.8 nM 32P-labeled Actin pre-mRNA and was incubated for the indicated times (15, 30, 40, or 60 min) at 23°C. RNA was isolated and analyzed by denaturing PAGE, and radiolabeled unspliced pre-mRNA or splicing intermediates and products were visualized with a PhosphorImager (GE Healthcare). For Snu17 and Snu17–Bud13 add-back experiments, His10-TEV-tagged-Snu17 and Bud13-S tag were cloned into a petDuet-1 vector and coexpressed in Escherichia coli BL21 (DE3) cells (Novagen) overnight at 20°C. Cells were lysed in 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), and 10 mM imidazole using a microfluidizer, and Snu17 and Snu17–Bud13 dimer (in an ∼3:1 ratio) were copurified from the cleared soluble fraction by a standard Ni-NTA agarose purification method (Qiagen).
Affinity purification of yeast spliceosomes
To affinity-purify spliceosomes, 32P-labeled Actin pre-mRNA was preincubated with a 30-fold molar excess of the MS2-MBP fusion protein for 30 min at 4°C. The pre-mRNA bound by the MS2-MBP protein was then incubated with yeast splicing extracts under standard in vitro splicing conditions with either 2 mM ATP to isolate Bact complexes or 50 µM ATP to isolate stalled B complexes. The reaction was loaded onto an amylose–agarose column (New England Biolabs) that was pre-equilibrated with GK75 buffer (20 mM HEPES at pH 7.25, 75 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA at pH 8.0). The column was washed with GK75, and spliceosomes were eluted with 100 mM maltose in GK75, loaded onto a linear 10%–30% (v/v) glycerol gradient containing GK75 buffer, and centrifuged at 60,000 rpm for 1 h and 47 min in a Sorvall TH660 rotor. The distribution of 32P-labeled RNA was determined by Cherenkov counting. Peak fractions containing spliceosomes were pooled and used for reconstitution assays. Approximately 0.5 pmol of purified B, Bact from prp2-1, or a mixture of complexes from the prp2-1 Δsnu17 splicing extract was incubated with a 20-fold excess of recombinant purified Prp2 and Spp2 and 2 mM UTP and subsequently incubated for 1 h at 23°C. The complexes were then subjected to gradient centrifugation, and the gradient was fractioned as described above. RNA isolated from the gradient fractions was separated by 8% denaturing PAGE or, to analyze the extent of U4/U6 duplex formation, by native 9% PAGE. snRNAs were detected by Northern blotting (Boon et al. 2006).
MS and Western blotting
Proteins in gradient fractions were separated by SDS-PAGE and subsequently identified by MS as described previously (Fabrizio et al. 2009). For Western blotting, proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Protran, Whatman), which was incubated with the indicated antibodies. Bound antibody was detected using an ECL detection kit (GE Healthcare).
Streptavidin agarose pull-downs
For pull-down experiments, spliceosomal complexes were formed on biotinylated Actin pre-mRNA in extracts from prp2-1 Δsnu17 yeast cells (after heat inactivation of Prp2) and subsequently affinity-purified and subjected to glycerol gradient centrifugation. Spliceosomes in the 45S gradient peak (ΔRES B/Bact-like complexes) were incubated with UTP, Prp2, and Spp2 and then analyzed on a second glycerol gradient. RNP complexes in gradient fractions 8–10 (which contain mainly U2 and the pre-mRNA) were incubated with streptavidin agarose for 1 h on a rotating wheel. Beads were pelleted by centrifugation, and the supernatant was transferred to a new tube. The beads were incubated with NTN buffer (150 mM NaCl, 50 mM Tris-HCl at pH 7.5, 0.1% [v/v] nonidet P40] for 1 min at 95°C, and the eluted material was transferred to a new tube. RNA was recovered from the post-beads supernatant and eluate and subsequently detected by Northern blotting.
Supplementary Material
Acknowledgments
We thank Monika Raabe and Thomas Conrad for excellent technical assistance, and Jana Schmitzova and Constantin Cretu for providing Prp2 and Spp2 recombinant proteins. We are grateful to Berthold Kastner for helpful discussions and help in preparing Figure 7. This work was supported by a grant from the Deutsche Forschungsgemeinschaft SFB 860 to R.L.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.308163.117.
References
- Arenas JE, Abelson JN. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci 94: 11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao P, Höbartner C, Hartmuth K, Lührmann R. 2017. Yeast Prp2 liberates the 5′ splice site and the branch site adenosine for catalysis of pre-mRNA splicing. RNA 23: 1770–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Lührmann R, Stark H. 2017. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170: 701–713. [DOI] [PubMed] [Google Scholar]
- Boon KL, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, Beggs JD. 2006. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol Cell Biol 26: 6016–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks MA, Dziembowski A, Quevillon-Cheruel S, Henriot V, Faux C, van Tilbeurgh H, Séraphin B. 2009. Structure of the yeast Pml1 splicing factor and its integration into the RES complex. Nucleic Acids Res. 37: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Stands L, Staley JP, Jackups RR Jr, Latus LJ, Chang TH. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell 7: 227–232. [DOI] [PubMed] [Google Scholar]
- Chiu YF, Liu YC, Chiang TW, Yeh TC, Tseng CK, Wu NY, Cheng SC. 2009. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol 29: 5671–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprévote O, Séraphin B. 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 23: 4847–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608. [DOI] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. 2013. RNA catalyses nuclear pre-mRNA splicing. Nature 503: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmann JB, Schmitzová J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Lührmann R. 2013. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 27: 413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Bartels C, Neubauer G, Lührmann R, Fabrizio P. 2001. A novel yeast U2 snRNP protein, Snu17p, is required for the first catalytic step of splicing and for progression of spliceosome assembly. Mol Cell Biol 21: 3037–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang J, Wan R, Yan C, Shi Y. 2015. Structural basis of pre-mRNA splicing. Science 349: 1191–1198. [DOI] [PubMed] [Google Scholar]
- Huang YH, Chung CS, Kao DI, Kao TC, Cheng SC. 2014. Sad1 counteracts Brr2-mediated dissociation of U4/U6.U5 in tri-snRNP homeostasis. Mol Cell Biol 34: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin RJ. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol 16: 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Smith J, Claude A, Lin RJ. 1992. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J 11: 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DS, Beggs JD. 1990. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res 18: 6559–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Lührmann R. 1998. The human U5–200kD DEXH-box protein unwinds U4/U6 RNA duplices. Proc Natl Acad Sci 95: 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR III, Stevens SW. 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Cheng SC. 2012. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol Cell Biol 32: 5056–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Chen HC, Wu NY, Cheng SC. 2007. A novel splicing factor, Yju2, is associated with NTC and acts after prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol 27: 5403–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 277: 17743–17750. [DOI] [PubMed] [Google Scholar]
- Ohrt T, Prior M, Dannenberg J, Odenwalder P, Dybkov O, Rasche N, Schmitzova J, Gregor I, Fabrizio P, Enderlein J, et al. 2012. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA 18: 1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin P-C, Nagai K. 2017. Structure of a pre-catalytic spliceosome. Nature 546: 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. 1998. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 279: 857–860. [DOI] [PubMed] [Google Scholar]
- Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee C-T, Urlaub H, Kastner B, et al. 2016. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Santos KF, Jovin SM, Weber G, Pena V, Lührmann R, Wahl MC. 2012. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci 109: 17418–17423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Agafonov DE, Schmitzova J, Hartmuth K, Fabrizio P, Luhrmann R. 2015. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3′ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLoS Genet 11: e1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlow DR, Staley JP. 2012. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci 37: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Schwer B. 2005. Characterization of the NTPase, RNA-binding and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry 44: 9795–9803. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Lee KR, Cheng SC. 1993. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc Natl Acad Sci 90: 10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowitzsch S, Weber G, Lührmann R, Wahl MC. 2009. Crystal structure of the Pml1p subunit of the yeast precursor mRNA retention and splicing complex. J Mol Biol. 385: 531–541. [DOI] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243. [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Schneider C, Mozaffari-Jovin S, Schmitzová J, Höbartner C, Fabrizio P, Lührmann R. 2015. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev 29: 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodaver AM, Staley JP. 2014. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA 20: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysoczański P, Schneider C, Xiang S, Munari F, Trowitzsch S, Wahl MC, Lührmann R, Becker S, Zweckstetter M. 2014. Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex. Nat Struct Mol Biol 21: 911–918. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2016. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353: 904–911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.