Abstract
The aim of the present study was to investigate the crosstalk between resveratrol (Res)-induced autophagy and apoptosis, and the molecular pathway by which autophagy leads to apoptotic death in drug-resistant K562/ADM leukemia cells. The viability of K562/ADM cells was determined using the MTT assay. The formation of autophagic vacuoles was detected using transmission electron microscopy and monodansylcadaverine (MDC) staining. Cell apoptosis was evaluated using flow cytometry. The expression of apoptosis- or autophagy-associated proteins was measured using western blotting. The results indicated that treatment with Res inhibited cell viability in a concentration-dependent manner. Furthermore, the numbers of MDC-positive fluorescent points, autophagic vacuoles and autolysosome-engulfed cytoplasmic materials were markedly increased in Res-treated K562/ADM cells compared with untreated cells, as determined using fluorescence microscopy and transmission electron microscopy. Res-induced apoptosis was associated with increased cleaved caspase-3 and B-cell lymphoma 2 associated X protein, and decreased B-cell lymphoma 2 (Bcl-2) protein expression levels when compared with the control (P<0.05). However, the proportion of apoptotic cells decreased from 69.6 to 41.0% (40 µmol/l Res) and from 77.3 to 58.8% (80 µmol/l Res) following pre-treatment with the autophagy inhibitor 3-methyladenine (P<0.01). The protein expression levels of microtubule-associated protein 1A/1B-light chain 3 and beclin 1, two markers of autophagy, were upregulated in Res-treated cells compared with the control (P<0.05). In addition, lysosomal cathepsin D (Cath D) release increased during Res-induced autophagy and apoptosis (P<0.05). The present results demonstrated that Res-induced apoptosis of K562/ADM cells was autophagy-dependent and the released Cath D may trigger caspase-dependent cell death through the Bcl-2 family of proteins. Furthermore, the present data indicate that to enhancement of the autophagy-cathepsin-apoptosis pathway may be an effective approach for overcoming anticancer drug resistance.
Keywords: resveratrol, apoptosis, autophagy, drug resistance, cathepsin D, K562/ADM cells
Introduction
Autophagy associated with drug resistance is a major obstacle in cancer chemotherapy (1,2). It has been reported that autophagy-deficient tumors are typically more sensitive to chemotherapeutic agents than are their autophagy-proficient counterparts (3). In the previous study, it was discovered that human drug-resistant K562/ADM leukemia cells preferentially emerged with higher autophagy activity compared with the parental, sensitive K562 cells following starvation treatment (4). However, autophagy is considered to have a dual role in tumor cell resistance, since previous research has shown conflicting roles for autophagy with regard to drug resistance in different cancer cell lines; specifically, autophagy either promotes or suppresses resistance via various mechanisms (5–7).
Resveratrol (trans-3,4′,5-trihydroxystilbene; Res), a natural polyphenolic compound, has been reported to have numerous beneficial effects, including anti-oxidant and anti-inflammatory activities, and an inhibitory effect on tumor growth (8,9). Yan et al (10) reported that Res not only induced leukemia cell apoptosis and erythroid differentiation, but also autophagy. A previous report has indicated that Res induces autophagy in different cancer cell lines via either a pro-survival or pro-death mechanism (11). Additionally, a number of studies have demonstrated that Res increases the sensitivity of malignancies, including melanoma, prostate and non-small lung cancer, to chemotherapy (12,13). Although Res is known to induce apoptosis and autophagy in different types of tumor cells, few studies have explored the effects of Res-induced autophagy and its association with apoptosis in drug-resistant leukemia cells.
The molecular regulators interconnecting between autophagy and apoptosis, including B-cell lymphoma 2 (Bcl-2), Bcl-2-associated X protein (Bax) and beclin 1, have been suggested to act as switching points that are critical for the outcome of tumor cells, and lysosomes have been reported to initiate the cell death pathway in autophagic cells (14–16). It has been demonstrated that Res activates a lysosome-dependent cytotoxic pathway that results in caspase-dependent cell death in colorectal cancer cells (17). Other results have shown that lysosomal cathepsin L mediates Res-induced autophagy and apoptotic cell death in cervical cancer cells (18). However, few studies have investigated the interrelation of autophagy, apoptosis and drug resistance in leukemia. The molecular pathway by which autophagy leads to apoptosis in drug-resistant conditions was explored in the present study. Furthermore, the present study also examined the fate of K562/ADM cells during sustained Res exposure. To the best of our knowledge, the present findings are the first to show that autophagy-dependent apoptosis is mediated by lysosomal cathepsin D (Cath D).
Materials and methods
Reagents
Resveratrol (Res), MTT, dimethyl sulfoxide (DSMO), 3-methyladenine (3-MA), monodansylcadaverine (MDC), microtubule-associated protein 1A/1B-light chain 3 (LC3) antibody and RPMI-1640 were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Fetal bovine serum (FBS) was obtained from Sijiqing Biological Engineering Materials Co., Ltd. (Hangzhou, China). Antibodies against beclin 1, Bcl-2, p62, cleaved caspase-3 and Bax were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Cell culture
K562/ADM ADM-resistant cell line was kindly provided by Dr Zhang Jiwang from Hematological laboratory, Ruijin Hospital Affiliated to the Shanghai Jiao Tong University School of Medicine (Shanghai, China). Cells were grown in RPMI-1640 medium supplemented with 10% (v/v) FBS, 2 mM glutamine, 100 U/ml penicillin and 100 U/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. K562/ADM cells were stimulated with 5 mg/l Adriamycin (ADM; Shenzhen Main Luck Pharmaceuticals, Inc., Shenzhen, China) every 45 days to maintain increased drug resistance and then experiments were performed after 2 weeks of culturing without ADM. Cells in the exponential phase were used in the experiments.
Cell viability assay
Cells were incubated with various concentrations of Res and were collected for determination of the half-maximal inhibitory concentrations (IC50) of Res using an MTT assay. Briefly, K562/ADM cells were seeded at a density of 1×105 cells/ml in 96-well plates. The cells were treated with 0, 20, 40, 80, 120 and 160 µmol/l of Res for 24, 48 or 72 h at 37°C. The controls were treated with 0 µmol/l of Res. A total of 10 µl MTT (5 mg/ml) was added to each well, followed by incubation for 4 h at 37°C. 100 µl DMSO was added to each of the wells, and incubation for 20 min at 37°C to dissolve the formazan crystals. The absorbance at 490 nm was measured using a PowerWave X plate reader (Bio-Tek Instruments, Inc., Winooski, VT, USA). Moreover, an autophagic inhibitor 3-MA (5 mmol/l) was added to the cells 3 h prior to Res treatment to confirm the role of autophagy in Res-induced cell death.
Transmission electron microscopy
K562/ADM cells (treated with Res at 0, 40 or 80 µmol/l at 37°C for 48 h) were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 mol/l phosphate buffer for 90 min at room temperature, followed by post-fixation with 1% osmium tetroxide for 30 min (10). Subsequently, the cells were gradually dehydrated in 10% graded series of 50–100% ethanol and propylene oxide, and embedded in Epon 812 resin. Finally, the blocks were cut into 70-nm-thick ultrathin sections with a microtome, which were then stained with uranyl acetate for 30 min and lead citrate for 15 min at 37°C for observation. The ultrastructure of the cells was examined using a JEMI1230 transmission electron microscope (JEOL, Ltd., Tokyo, Japan). The controls were treated with 0 µmol/l of Res.
MDC staining
K562/ADM cells (treated with Res at 0, 40 or 80 µmol/l at 37°C for 24 h) were stained with 0.05 mmol/l MDC at 37°C for 30 min. The cells were washed three times with PBS and autophagic vacuoles were observed with a fluorescence inverted, phase contrast microscope (Olympus Corporation, Tokyo, Japan).
Flow cytometric analysis
K562/ADM cells were pre-incubated with or without 3-MA for 3 h and treated with Res at 0, 40, or 80 µmol/l for 48 h prior to detection of cell apoptosis using an Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). A total of 1×106 cells were collected and suspended in 100 ml binding buffer containing 0.25 mg of Annexin V-FITC. After 15 min of incubation in the dark at room temperature, cells were washed by PBS and suspended in the binding buffer with 5 mg of PI. Then, the stained cells were analyzed using a flow cytometer (EPICS XL; Beckman Coulter, Inc., Miami, FL, USA) and analyzed using Windows Multiple Document Interface for Flow Cytometry software (version 2.8; The Scripps Research Institute, La Jolla, CA, USA).
Western blotting
At the end of the treatment with Res at 0, 40, or 80 µmol/l for 24, 48 and 72 h, the cells were washed twice with PBS and lysed in ice-cold RIPA lysis buffer (Beijing Solarbio Science & Technology Co. Ltd., Beijing, China). Total protein concentration was determined using a BCA protein quantitative kit (cat no., P0010s; Beyotime Institute of Biotechnology, Haimen, China). Protein extracts (40 µg/lane) were separated by 10–12% SDS-PAGE and electrophoretically transferred onto polyacrylamide difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked in 5% non-fat milk for 1 h at room temperature, followed by overnight incubation with primary antibodies at 4°C. The primary antibodies used were as follows: Beclin 1 (cat. no. 3738), LC3 (cat. no. ABC929; EMD Millipore), Bcl-2 (cat. no. 2872), cleaved caspase-3 (cat. no. 9661), Bax (cat. no. 2774); p62 (cat. no. 5114), Cath D (cat. no. sc-10725; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) (all 1:1,000), P-gp (1:500; cat. no. BA1351-2; Boster Biological Technology, Wuhan, China) and β-actin (1:700; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.). Subsequently, the membranes were washed three times for 5 min with PBS in Tween 20 and incubated again with horseradish peroxidase-conjugated goat anti-rabbit (cat. no. ZB-2301) or goat anti-mouse (cat. no. ZB-2305) antibodies (both 1:10,000; OriGene Technologies, Inc., Beijing, China) at 37°C for 1 h. Protein bands were visualized using enhanced chemiluminescence reagents (EMD Millipore). Western blots were scanned using an Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA) and the bands were quantified using Image J software (version 1.45S; National Institutes of Health; Bethesda, MD, USA). β-actin was used as a reference protein.
Statistical analysis
Statistical analysis was performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). All the experiments were repeated at least three times. Data are presented as the mean ± standard deviation. One-way analysis of variance followed by Tukey's least significant difference post-hoc test was used to analyze statistical differences among groups under different conditions. P<0.05 was considered to indicate a statistically significant difference.
Results
Res induces autophagy in K562/ADM cells
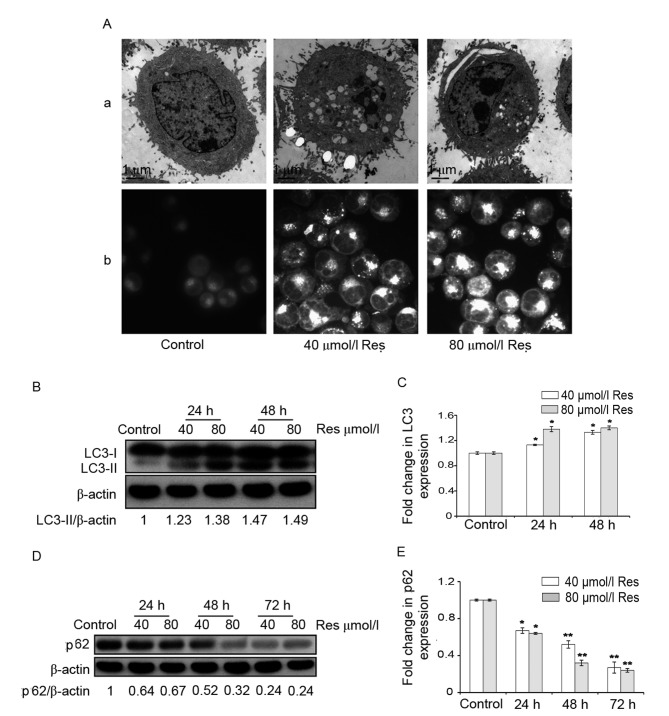
To determine whether Res induces autophagy in K562/ADM cells, autophagic bodies were investigated using transmission electron microscopy. Compared with the control group, a large number of autophagic vacuoles appeared in the cytoplasm when K562/ADM cells were treated with 40 or 80 µmol/l Res (Fig. 1A-a). Furthermore, the number of MDC-positive fluorescent points and the amount of engulfed cytoplasmic material in Res-treated cells was markedly greater compared with that in the control group, as observed under a fluorescence microscope (Fig. 1A-b).
Figure 1.
Res induced autophagy in K562/ADM cells. (A) K562/ADM cells were treated with 40 and 80 µmol/l Res for 24 h. (A-a) Morphology of autophagy was observed under a transmission electron microscope following treatment. Scale bar, 1 µm. (A-b) Cells were stained with monodansylcadaverine following treatment. The fluorescence intensity of monodansylcadaverine was examined under a fluorescence inverted phase-contrast microscope. Original magnification, ×1,000. (B) Representative image and (C) quantification of protein expression levels of LC3, detected by western blotting. (D) Representative image and (E) quantification of protein expression levels of p62, detected by western blotting β-actin served as an internal control. Data are represented as the mean ± standard deviation of triplicate experiments. The control group was treated with 0 µmol/l Res. *P<0.05 and **P<0.01 vs. control group. Res, resveratrol; LC3, microtubule-associated protein 1A/1B-light chain 3.
The expression of autophagic LC3 protein was determined using western blotting. There are two forms of LC3: LC3-I and LC3-II (3,19). During the process of autophagy, the cytoplasmic form, LC3-I, is converted into LC3-II, which is the form that is bound to the autophagosome membrane (3,19). The results revealed that, following treatment with 40 or 80 µmol/l Res, the relative ratio of LC3-II/β-actin increased in K562/ADM cells compared with the control (Fig. 1B). This difference was indicated to be statistically significant (P<0.05; Fig. 1C).
In addition to LC3, the expression level of p62 protein, which is another autophagy-specific biomarker was also explored (19,20). The p62 protein acts as a signaling hub through its involvement in the formation of autophagosomes and is degraded during autophagy (19,20). The results indicated that p62 protein expression levels were decreased by Res in a time-dependent manner, suggesting an increase in autophagic flux (Fig. 1D). Furthermore, the difference between the Res-treated cells and the control was statistically significant (P<0.05; Fig. 1E).
Inhibition of autophagy attenuates the Res-induced apoptosis of K562/ADM cells
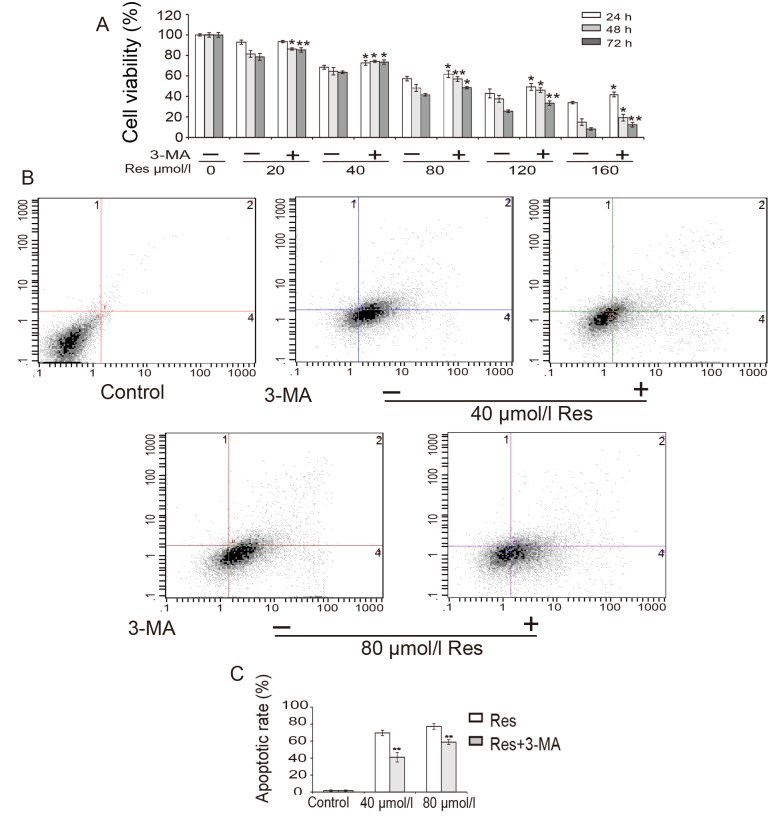
The effect of Res on the viability of K562/ADM cells was investigated. K562/ADM cells were treated with different concentrations of Res (0–160 µmol/l) for 24, 48 or 72 h, and cell viability was assessed by MTT assay. As shown in Fig. 2A, cell viability decreased in a time- and dose-dependent manner. The IC50 of Res in K562/ADM cells at 48 h was calculated to be 57.7 µmol/l. These data indicate that Res reduced the survival of K562/ADM cells. In addition, the results revealed that cell viability was restored when cells were treated with 5 mmol/l 3-MA for 3 h prior to treatment with Res (Fig. 2A).
Figure 2.
Res inhibits the viability and induces the apoptosis of K562/ADM cells. (A) Cells were pre-incubated with or without 3-MA and subsequently treated with different concentrations Res for 24, 48, and 72 h prior to detection of cell viability by MTT assays. (B) Cells were pre-incubated with or without 3-MA and treated with indicated concentrations Res for 48 h prior to detection of cell apoptosis by flow cytometry. (C) Quantification of flow cytometry data. Data represent the mean ± standard deviation of triplicate experiments. *P<0.05 and **P<0.01 vs. without 3-MA group. Res, resveratrol; 3-MA, 3-methyladenine.
To determine the association between cell death and apoptosis, a flow cytometry experiment was conducted (Fig. 2B and C). The results revealed that the proportion of apoptotic cells increased following treatment with Res in a dose-dependent manner, which suggested that Res induced apoptosis in K562/ADM cells. However, following pre-treatment with 3-MA, the proportion of apoptotic cells was significantly decreased from 69.6 to 41.0% for 40 µmol/l Res treatment and from 77.3 to 58.8% for 80 µmol/l Res treatment (P<0.01; Fig. 2C).
These data demonstrated that Res inhibited cell viability and that Res-induced apoptosis was reduced by 3-MA in K562/ADM cells. These results suggest that the Res-induced apoptosis of K562/ADM cells was autophagy-dependent.
Protein expression levels of Bax/Bcl-2 and beclin 1 increase during the Res-induced autophagy and apoptosis of K562/ADM cells
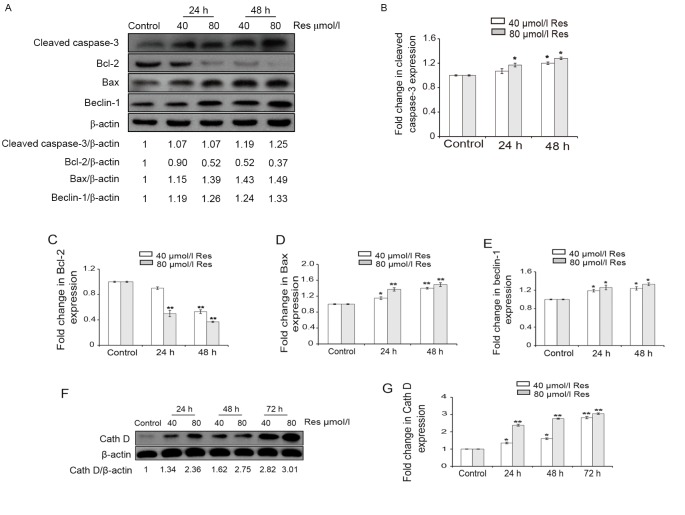
To further explore the mechanism by which Res induces apoptosis, the expression levels of cleaved caspase-3, Bcl-2, Bax, beclin 1 and Cath D proteins were determined using western blotting (Fig. 3). Following treatment with 40 or 80 µmol/l Res for 24 and 48 h, the expression levels of cleaved caspase-3 increased in a dose-dependent manner in K562/ADM cells (Fig. 3A). In addition, differences in the protein expression levels of cleaved caspase-3 between the control and Res-treated cells were statistically significant (P<0.05; Fig. 3B). The data suggested that Res significantly induced apoptosis through the activation of caspase-3.
Figure 3.
Protein expression levels of cleaved caspase-3, Bcl-2, Bax, beclin 1 and Cath D were detected by western blotting. Cells were treated with 40 and 80 µmol/l Res for 24, 48, or 72 h. β-actin served as an internal control. (A) Blot images of cleaved caspase-3, Bcl-2, Bax and beclin 1 protein expression. Quantification of (B) cleaved caspase-3, (C) Bcl-2, (D) Bax and (E) beclin 1 protein expression levels. (F) Blot image of Cath D protein expression. (G) Quantification of Cath D protein expression levels. Data are represented as the mean ± standard deviation of triplicate experiments. The control group was treated with 0 µmol/l Res. *P<0.05 and **P<0.01 vs. control group. Res, resveratrol; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2 associated X protein; Cath D, cathepsin D.
The Bcl-2 family is a crucial integrator of cellular survival and death signals (14,16). Bcl-2 is an anti-apoptotic protein that has the ability to inhibit autophagy and apoptosis through interactions with beclin 1 and Bax/Bak, respectively, which are mediated through their Bcl-2-homology-3-binding pockets (14,16). The western blotting results indicated that the expression levels of Bcl-2 were decreased by Res (Fig. 3A and C); this was accompanied by increased expression levels of Bax protein in a dose-dependent manner (Fig. 3A and D). Statistically significant differences were detected in the protein expression levels of Bax and Bcl-2 in Res-treated cells compared with the control (P<0.05; Fig. 3C and D). Therefore, the increased expression levels of Bax and decreased expression levels of Bcl-2 may be important in the apoptotic process in K562/ADM cells following treatment with Res.
Beclin 1, another hallmark of autophagosomes, is a well-known protein with respect to the formation of autophagic vesicles (1,5). Western blotting results in the present study demonstrated that the expression level of beclin 1 protein was increased by 1.19- to 1.33-fold following treatment with Res, further corroborating the hypothesis that autophagy is induced by Res treatment (Fig. 3A and E). In addition, the differences in beclin 1 protein expression levels between the Res-treated cells and the control were statistically significant (P<0.05; Fig. 3E). These results suggest that complex interactions of the anti-apoptotic Bcl-2 and pro-autophagic beclin 1 proteins co-regulate the ultimate fate of Res-induced K562/ADM cells with respect to apoptosis.
Cath D is activated during res-induced autophagy and apoptosis in K562/ADM cells
Lysosomal proteases, also known as cathepsins, participate in the activation of the intrinsic programmed cell death pathway (17,19). Cath D, a major intracellular aspartic protease, is typically involved in apoptosis through its release from lysosomes (17,19). For this reason, whether Cath D is a possible mediator of Res-induced cytotoxicity was investigated in the present study. The western blotting results indicated that Cath D expression levels were increased by res in a time- and concentration-dependent manner, with an increase of ~3-fold in the 80 µmol/l Res-treated group at 72 h compared with the control group (Fig. 3F and G). Furthermore, the differences in Cath D protein expression levels between the Res-treated cells and the control were statistically significant (P<0.05; Fig. 3G). This suggests that lysosomal Cath D may be involved in the process of Res-induced autophagy and the subsequent progression to apoptotic death in K562/ADM cells treated with Res.
Discussion
Res has been shown to induce cell death through various mechanisms in different cancer cell lines (2,11,12,19). Various antitumor effects of Res have been described previously, including the inhibition of DNA polymerase, protein kinase C, cyclooxygenase-2 or interleukin-6 activities, and the suppression of transcription factors such as nuclear factor-κB (4,11,12,21,22). The present study revealed that Res increased autophagosome formation, decreased cell viability and induced apoptotic death in drug-resistant K562/ADM cells. It was hypothesize that the mediation of these effects by Res may involve the permeation of Res through the lysosomal membrane, resulting in the release of Cath D from lysosomes into the cytosol. In addition, the released Cath D may trigger caspase-dependent cell death through interaction with Bcl-2 and beclin 1 and the activation of Bax.
Yan et al (10) reported that Res inhibited the proliferation of K562 leukemia cells and induced apoptosis and autophagy, and demonstrated the contribution of Res-induced autophagy to erythroid differentiation in K562 cells. However, the present results indicated that autophagy preceded apoptosis in K562/ADM cells following Res treatment. To investigate the role of autophagy in Res-induced cell death, 3-MA, an autophagic inhibitor (4,15), was used to block autophagy and resulted in the marked attenuation of Res-induced apoptosis. These data indicated that Res-induced apoptosis may be autophagy-dependent in K562/ADM cells.
In the present study, autophagic responses increased significantly following Res treatment for 24 h in K562/ADM cells. As observed under a transmission electron microscope, a large number of autophagic bodies appeared in K562/ADM cells following Res treatment. This finding was further supported by evaluation of the expression levels of LC3 using western blotting. LC3 is considered to be a specific marker of autophagic activity (3,19). The LC3-II/β-actin ratio was significantly increased in K562/ADM cells treated with 40 or 80 µmol/l Res for 24 or 48 h. Previous studies have indicated that the LC3-II turnover assay is suitable for monitoring autophagic flux in the presence of a lysosomal inhibitor (23,24). To distinguish whether autophagosome accumulation was due to enhanced autophagic activity or reduced autophagosome degradation, the protein expression levels of p62 were determined in the present study. p62, an autophagy-related protein, is involved in the formation of autophagosomes and degraded through autophagy (19,20). The present results showed that the protein expression level of p62 was significantly decreased in Res-treated cells compared with the control. These data therefore indicated that Res induced a high level of LC3-II protein by the induction of autophagic activity rather than by blocking autophagosome degradation in K562/ADM cells.
Beclin 1 is required for the initiation stage in the formation of the autophagosome (1,5). In the present study, an increase in beclin1 protein expression levels was observed in Res-treated cells compared with the control, suggesting an enhanced induction of autophagy. Beclin 1 was originally identified as a Bcl-2-interacting protein. Previous studies have demonstrated that beclin 1 and Bcl-2 interact in a multiprotein complex on the membrane of the endoplasmic reticulum; this observation in turn suggests a mechanism by which these proteins assist in mediating the intricate relationship between apoptosis and autophagy (1,16). It is known that beclin 1 generates a pro-apoptotic protein fragment and also interacts with the anti-apoptotic Bcl-2 family protein to inhibit autophagy and initiate mitochondrial apoptosis (22). Prabhu et al (25) reported that the silencing of key regulators of autophagy, such as beclin 1 and autophagy protein (ATG)5, significantly enhanced Res-induced caspase activation. In the present study, western blotting results revealed that Res significantly increased the expression levels of Bax and decreased those of Bcl-2. The interaction of beclin 1 with Bcl-2 in the cytosolic compartment may increase efficient Bax translocation to mitochondria, leading to permeabilization of the mitochondrial membrane, cytochrome c release and activation of a caspase cascade, and finally, initiation of caspase-dependent cell death (2,10,11).
Lysosomes contain numerous hydrolases, which degrade intracellular and extracellular delivered materials, and have been implicated in the regulation of cell death (17,19). Various stimuli may induce lysosomal membrane permeabilization, thereby releasing a class of hydrolytic enzymes, the cathepsins, from the lysosomal compartment into the cytosol, which contributes to apoptosis (17,18). Cath D has been suggested to be a key regulator of apoptosis via its catalytic activity, and there is also evidence to support the hypothesis that cathepsins act in concert with caspases in apoptotic cell death (17). However, little is known regarding the mechanism by which Cath D mediates the cytotoxic agent-induced autophagic death of resistant leukemia cells. In the present study, a significantly higher protein expression level of Cath D was observed during the autophagy and apoptosis of Res-induced K562/ADM cells, and this indicated that Cath D may have a key role in Res-induced autophagy-dependent apoptotic death in these cells. Trincheri et al (17) considered Bax to be the probable target of Cath D in Res-mediated cytotoxicity. Furthermore, Yelamanchili et al (26) observed that the upregulation of Cath D correlated with cellular damage leading to typical caspase-dependent activation of apoptosis. The present study data revealed that cleaved caspase-3 and Cath D protein expression levels were significantly increased following Res treatment and, therefore, may be involved in mediating the apoptotic effect of Res in K562/ADM cells. Specifically, the death pathway may be initiated by the promotion of lysosomal membrane permeabilization and release of lysosomal Cath D to the cytosol to trigger the activation of Bax and the selective mitochondrial release of apoptosis-inducing factors, eventually resulting in activation of the caspase proteolytic cascade and apoptosis (17,19,24).
Autophagy has a cytoprotective function under stress conditions, such as during cytotoxic drug administration, and may result in increased drug resistance (1). It has been reported that autophagy increases the resistance of colorectal cancer stem-like cells to photodynamic therapy-induced apoptosis (1,27,28). Similarly, cisplatin-activated autophagy, involving the activation of ATG7, has been indicated to increase liver cancer cell resistance to cisplatin (29). Eum et al (1) suggested that the inhibition of autophagy may resensitize resistant tumor cells to anticancer therapy. Previous observations by our group highlighted the elevation of autophagic activity in drug-resistant K562/ADM cells (15). These findings suggest that cancer cells may activate the autophagic process to fight against cytotoxin-induced cell injury by recycling damaged organelles and proteins, resulting in drug resistance. However, under sustained exposure to cytotoxic agents, autophagic machinery could be recruited to trigger both caspase-dependent and -independent lethal pathways (17,19). In addition, P-glycoprotein serves a significant role in causing multidrug resistance (30). The expression of P-glycoprotein was also examined, but its expression did not change, indicating that it did not participate in the autophagy cell death induced by resveratrol (data not shown). Therefore, it was hypothesize that ongoing exposure to Res drives the autophagic K562/ADM cells to a higher degree of autophagy and activates Bax, a downstream effector of Cath D (17,19,26). The activation of Bax promotes lysosomal membrane permeabilization with release of Cath D from lysosomes (17,18). In turn, Cath D contributes to apoptosis through the clearance of dying cells, activation of Bax and subsequent amplification of apoptotic signaling.
In summary, the present study revealed that Res promoted autophagy in leukemia-drug-resistant K562/ADM cells and that this enhanced autophagy was involved in the Res-induced apoptosis of K562/ADM cells, with apoptosis occurring via a series of events that potentially involves the interaction between Bcl-2 and beclin 1, the activation of Bax and the release of lysosomal Cath D. Res cytotoxicity was associated with the up-regulation of Cath D expression, suggesting that Cath D may be the key to triggering Res-induced cell death (17). Based on the present data, it was hypothesize that K562/ADM cells increase their autophagic activity to counteract cytotoxin-induced cellular injury and sustain drug resistance under physiological conditions. However, with ongoing exposure to Res or other anticarcinogens, the drug-resistant cells are forced to maintain high levels of autophagy for a long period of time, resulting in injury to the cells due to the activation of a Bcl-2/beclin1-Cath D/Bax loop. This loop may promote a series of changes: Lysosome permeabilization and the release of Cath D, Bax-mediated mitochondrial permeabilization with cytochrome c release, activation of the caspase cascade and apoptosis. Therefore, manipulating the balance of autophagy and apoptosis via activation of the Cath D-Bax loop may have great therapeutic potential for overcoming drug resistance in leukemia.
Acknowledgements
The present study was supported by the Fundamental Research Funds for the Central Universities (grant no. lzujbky-2014-143) and National Natural Science Foundation of China (grant no. 81541025).
References
- 1.Eum KH, Lee M. Targeting the autophagy pathway using ectopic expression of Beclin1 in combination with rapamycin in drug-resistant v-Ha-ras-transformed NIH 3T3 cells. Mol Cells. 2011;31:231–238. doi: 10.1007/s10059-011-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin CJ, Lee CC, Shih YL, Lin TY, Wang SH, Lin YF, Shih CM. Resveratrol enhances the therapeutic effect of temozolomide against malignant glioma in vitro and in vivo by inhibiting autophagy. Free Radic Biol Med. 2012;52:377–379. doi: 10.1016/j.freeradbiomed.2011.10.487. [DOI] [PubMed] [Google Scholar]
- 3.Burada F, Nicoli ER, Ciurea ME, Uscatu DC, Ioana M, Gheonea DI. Autophagy in colorectal cancer: An important switch from physiology to pathology. World J Gastrointest Oncol. 2015;7:271–284. doi: 10.4251/wjgo.v7.i11.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Wei HL, Chen J, Xie B. Antitumor effect of arsenic trioxide in human K562 and K562 ADM cells by autophagy. Toxicol Mech Methods. 2012;22:512–519. doi: 10.3109/15376516.2012.686534. [DOI] [PubMed] [Google Scholar]
- 5.Shuhua W, Chenbo S, Yangyang L, Xiangqian G, Shuang H, Tangyue L, Dong T. Autophagy-related genes raptor, rictor and beclin 1 expression and relationship with multidrug resistance in colorectal carcinoma. Hum Pathol. 2015;46:1752–1759. doi: 10.1016/j.humpath.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 6.Panda PK, Mukhopadhyay S, Das DN, Sinha N, Naik PP, Bhutia SK. Mechanism of autophagic regulation in carcinogenesis and cancer therapeutics. Semin Cell Dev Biol. 2015;39:43–55. doi: 10.1016/j.semcdb.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao XY, Li GY, Liu Y, Chai LM, Chen JX, Zhang Y, Du ZM, Lu YJ, Yang BF. Resveratrol protects against arsenic trioxide-induced cardiotoxicity in vitro and in vivo. Br J Pharmacol. 2008;154:105–113. doi: 10.1038/bjp.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barjot C, Tournaire M, Castagnino C, Vigor C, Vercauteren J, Rossi JF. Evaluation of antitumor effects of two vine stalk oligomers of resveratrol on a panel of lymphoid and myeloid cell lines: Comparison with resveratrol. Life Sci. 2007;81:1565–1574. doi: 10.1016/j.lfs.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 10.Yan HW, Hu WX, Zhang JY, Wang Y, Xia K, Peng MY, Liu J. Resveratrol induces human K562 cell apoptosis, erythroid differentiation and autophagy. Tumour Biol. 2014;35:5381–5388. doi: 10.1007/s13277-014-1701-y. [DOI] [PubMed] [Google Scholar]
- 11.Naponelli V, Modernelli A, Bettuzzi S, Rizzi F. Roles of autophagy induced by natural compounds in prostate cancer. Biomed Res Int. 2015;2015:121826. doi: 10.1155/2015/121826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y, DeMarco VG, Nicholl MB. Resveratrol enhances radiation sensitivity in prostate cancer by inhibiting cell proliferation and promoting cell senescence and apoptosis. Cancer Sci. 2012;103:1090–1098. doi: 10.1111/j.1349-7006.2012.02272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Bradley MJ, Cook KM, Herrick EJ, Nicholl MB. A potential role for resveratrol as a radiation sensitizer for melanoma treatment. J Surg Res. 2013;183:645–653. doi: 10.1016/j.jss.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 14.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibits Beclin1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Chen J, Xie B, Wei HL. Acquired multidrug resistance in human K562/ADM cells is associated with enhanced autophagy. Toxicol Mech Methods. 2013;23:678–683. doi: 10.3109/15376516.2013.843105. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Yao S. JNK-Bcl-xL-Bax/Bak pathway mediates the crosstalk between matrine-induced autophagy and apoptosis via interplay with Beclin1. Int J Mol Sci. 2015;16:25744–25758. doi: 10.3390/ijms161025744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trincheri NF, Nicotra G, Follo C, Castino R, Isidoro C. Resveratrol induces cell in colorectal cancer cells by a novel pathway involving lysosomal cathepsin D. Carcinogenesis. 2007;28:922–931. doi: 10.1093/carcin/bgl223. [DOI] [PubMed] [Google Scholar]
- 18.Hsu KF, Wu CL, Huang SC, Wu CM, Hsiao JR, Yo YT, Chen YH, Shiau AL, Chou CY. Cathepsin L mediates resveratrol-induced autophagy and apoptotic cell death in cervical cancer cells. Autophagy. 2009;5:451–460. doi: 10.4161/auto.5.4.7666. [DOI] [PubMed] [Google Scholar]
- 19.Jang BG, Choi BY, Kim JK, Kim MJ, Sohn M, Suh SW. Impairment of autophagic flux promotes glucose reperfusion-induced neuro2A cell death after glucose deprivation. PloS One. 2013;8:e76466. doi: 10.1371/journal.pone.0076466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Ma K, Qi T, Zhang Q, Li G, Chiu JF. P62 regulates resveratrol-mediated Fas/Cav-1 complex formation and transition from autophagy to apoptosis. Oncotarget. 2015;6:789–801. doi: 10.18632/oncotarget.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Xu W, Su J, Chu M, Jin H, Li G, Tan C, Wang X, Wang C. The prosurvival role of autophagy in resveratrol-induced cytotoxicity in GH3 cells. Int J Mol Med. 2014;33:987–993. doi: 10.3892/ijmm.2014.1660. [DOI] [PubMed] [Google Scholar]
- 22.Bhardwaj A, Sehthi G, Vadhan-Raj S, Bueso-Ramos C, Takada Y, Gaur U, Nair AS, Shishodia S, Aggarwal BB. Resveratrol inhibits proliferation, induces apoptosis and overcomes chemoresistance through down-regulation of STAT3 and nuclear factor-kappaB-regulated antiapoptotic and cell survival gene products in human multiple myeloma cell. Blood. 2007;109:2293–2302. doi: 10.1182/blood-2006-02-003988. [DOI] [PubMed] [Google Scholar]
- 23.Liu YN, Wang YX, Liu XF, Jiang LP, Yang G, Sun XC, Geng CY, Li QJ, Chen M, Yao XF. Citreoviridin induces ROS-dependent autophagic cell death in human liver HepG2 cells. Toxicon. 2015;95:30–37. doi: 10.1016/j.toxicon.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Chen QY, Shi JG, Yao QH, Jiao DM, Wang YY, Hu HZ, Wu YQ, Song J, Yan J, Wu LJ. Lysosomal member permeabilization is involved in curcumin-induced apoptosis of A549 lung carcinoma cells. Mol Cell Biochem. 2012;359:389–398. doi: 10.1007/s11010-011-1033-9. [DOI] [PubMed] [Google Scholar]
- 25.Prabhu V, Srivastava P, Yadav N, Amadori M, Schneider A, Seshadri A, Pitarresi J, Scott R, Zhang H, Koochekpour S, et al. Resveratrol depletes mitochondrial DNA and inhibition of autophagy enhances resveratrol-induced caspase activation. Mitochondrion. 2013;13:493–499. doi: 10.1016/j.mito.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yelamanchili SV, Chaudhuri AD, Flynn CT, Fox HS. Upregulation of cathepsin D in the caudate nucleus of primates with experimental parkinsonism. Mol Neurodegener. 2011;6:52. doi: 10.1186/1750-1326-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei MF, Chen MW, Chen KC, Lou PJ, Lin SY, Hung SC, Hsiao M, Yao CJ, Shieh MJ. Autophagy promotes resistance to photodynamic therapy-induced apoptosis selectively in colorectal cancer stem-like cells. Autophagy. 2014;10:1179–1192. doi: 10.4161/auto.28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DH, Lee NY, Sung WJ, Baek JH, Kim JG, Sohn SK, Suh JS, Lee KS, Lee KB. Multidrug resistance as a potential prognostic indicator in acute myeloid leukemia with normal karyotype. Acta Haematol. 2005;114:78–83. doi: 10.1159/000086579. [DOI] [PubMed] [Google Scholar]
- 29.Xu N, Zhang J, Shen C, Xia L, Xue F, Xia Q. Cisplatin-induced down regulation of mir-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Cimmun. 2012;423:826–831. doi: 10.1016/j.bbrc.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 30.Xue C, Wang C, Liu Q, Meng Q, Sun H, Huo X, Ma X, Liu Z, Ma X, Peng J, Liu K. Targeting P-glycoprotein expression and cancer cell energy metabolism: Combination of metformin and 2-deoxyglucose reverses the multidrug resistance of K562/Dox cells to doxorubicin. Tumor Biol. 2016;37:8587–8597. doi: 10.1007/s13277-015-4478-8. [DOI] [PubMed] [Google Scholar]