Abstract
The Wnt signaling pathway is aberrantly activated in the majority of human colorectal tumors. β-catenin, a key component of the Wnt signaling pathway, interacts with the T-cell factor/lymphoid enhancer-binding factor family of transcription factors and activates transcription of Wnt target genes. Sp5 is one of the Wnt target genes, and its expression is commonly upregulated in colon cancer cells. The present study demonstrates that the expression of Sp5 is not upregulated in the colon cancer cell line HCT116, in which Wnt signaling is constitutively activated. Furthermore, the results demonstrate that Sp5 has the potential to inhibit cell proliferation through upregulation of the cell cycle inhibitor p27. These findings suggest that HCT116 cells downregulate Sp5 to avoid p27-mediated growth arrest.
Keywords: Wnt signalling, β-catenin, Sp5, p27, colon cancer
Introduction
The Wnt signaling pathway plays a critical role in proliferation, cell mobility and development (1–3). Wnt signaling stabilizes cytoplasmic β-catenin, and β-catenin is subsequently translocated into the nucleus where it interacts with the T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) transcription factors to stimulate the transcription of Wnt target genes, such as such as cyclin D1, c-Myc and Axin2 (4–7). Aberrant activation of Wnt signaling has been implicated in a variety of human cancers, including colon cancer (8,9).
Specificity protein 5 (Sp5), a member of the Sp transcription factor family, is a direct target of β-catenin-TCF/LEF (10–13). Sp5 is expressed at high levels in normal and tumor tissues where Wnt signaling is activated (14), suggesting that Sp5 is a universal Wnt signaling target. Sp5 contains three conserved Cys2His2 zinc finger domains at its C-terminus, which bind the GC-box (GGGCGG) or closely associated sequences (15,16). Sp5 directly represses the transcription of the cell cycle inhibitor p21 by binding to the GC-boxes located in its promoter region, and exogenous expression promotes the growth of breast cancer MCF-7 cells, which express no detectable Sp5 protein (11,14,17). By contrast, overexpression of Sp5 inhibits cell growth and transformation mediated by oncogenic KRAS in NIH3T3 cells (18). In addition, Sp5-deficient mice do not exhibit evident abnormalities, possibly due to functional compensation by other members of the Sp family (19). Although the analysis of Sp5 could provide novel insights into the mechanisms underlying the tumorigenesis of colon cancer, the role of Sp5 in the regulation of cell proliferation has not been clearly established.
The present study demonstrates that the expression of Sp5 is not upregulated in the colon cancer cell line HCT116, in which Wnt signaling is constitutively activated. Furthermore, Sp5 has the potential to inhibit the proliferation of HCT116 cells through regulation of the expression of the cell cycle inhibitor p27.
Materials and methods
Cell culture and transfection
HCT116 cells were obtained from ATCC (Manassas, VA, USA) and cultured in McCoy's 5A medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS; Nichirei Biosciences, Tokyo, Japan). HEK293 cells were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM; Nissui, Tokyo, Japan) supplemented with 10% FBS. An siRNA targeting p27 (5′-CAAGTGGAATTTCGATTTT-3′) was obtained from Ambion (s2837; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Plasmids and siRNAs were transfected into cells using Lipofectamine LTX and RNAiMAX (both Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Silencer negative control 1 siRNA (Ambion; Thermo Fisher Scientific, Inc.) was used as a negative control.
Cloning of mouse Sp5 cDNA
Mouse embryonic fibroblasts (MEF) were sourced from C57BL/6 mice. Total RNA was isolated from MEFs using TRIsure (Bioline Reagents Limited, London, UK) and cDNA was synthesized from 500 ng of RNA using Super Script III Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) with random hexamers. A FLAG tag was added to the 5′ end of the mouse Sp5 (mSp5) cDNA by polymerase chain reaction (PCR) using KOD-plus-Neo (Toyobo Co., Ltd., Osaka, Japan) and cloned into a pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.) with the forward primer 5′-AAAGAATTCATGGCCGCTGTGGCCGTCCT-3′ and reverse primer 5′-TTTTCTAGATCATAGGTCCCGCGGATTCT-3′.
Reverse transcription (RT)-quantitative PCR (qPCR) analysis
Total RNA was isolated using TRIsure and cDNA was synthesized from 500 ng of RNA using PrimeScript RT Master Mix (Takara Bio, Inc., Otsu, Japan). Reactions were performed at 37°C for 15 min and inactivated at 85°C for 5 sec. All cDNA samples were diluted (1:9) with RNase-free water for use as templates in qPCR. qPCR was performed in duplicate using SYBR Green I (Takara Bio, Inc.), and detected on a LightCycler 480 Real-Time PCR System (Roche Diagnostics GmbH, Mannheim, Germany). qPCR was performed in a 10-µl mixture that included 5 µl 2X SYBR Premix Ex Taq (Takara Bio, Inc.), 3µ l RNase-free water containing 1.6 pmol of each primer and 2µl cDNA template. The PCR cycling conditions were as follows: A denaturation step of 30 sec at 95°C, and then 40 cycles of amplification at 95°C for 5 sec and 60°C for 30 sec. All mRNA quantification data were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression and compared as differences in ΔΔCq (20). Primer sequences were as follows: GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′; GAPDH reverse, 5′-TGGTGAAGACGCCAGTGGA-3′; Axin2 forward, 5′-GCCAATGGCCAAGTGTCTC-3′; Axin2 reverse, 5′-GGCTCTCCAACTCCAGCTTC-3′; p27 forward, 5′-GGCCTCAGAAGACGTCAAAC-3′; p27 reverse, 5′-CATGTATATCTTCCTTGCTTCATCA-3′; p21 forward, 5′-CTGGAGACTCTCAGGGTCGAAA-3′; and p21 reverse, 5′-GATTAGGGCTTCCTCTTGGAGAA-3′.
Immunoblotting analysis
Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.5 mM dithiothreitol, 1:1,000 protease inhibitor cocktail). The lysates were subjected to immunoblotting analysis with monoclonal mouse anti-FLAG (#F1804; Sigma-Aldrich; dilution, 1:1,000) or anti-tubulin (#CP06; EMD Millipore, Billerica, MA, USA; dilution, 1:1,000) primary antibodies, and horseradish peroxidase (HRP)-conjugated secondary antibodies (#405306; BioLegend, San Diego, CA, USA; dilution, 1:5,000).. Visualization was performed using Luminata Forte HRP substrate (EMD Millipore).
Cell proliferation assays
HCT116 cells were transfected with Flag-tagged mSp5 or empty vector (control) using Lipofectamine LTX. Cells were cultured for 1 day and then plated in 96-well plates (Day 0), and cell numbers were measured using the CellTiter-Glo 2.0 assay kit (Promega Corporation, Madison, WI, USA).
Luciferase assays
HEK293 cells were transfected with a luciferase-reporter plasmid and cultured for 24 h. Cells were lysed and firefly luciferase activity was measured with the Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's instructions. pRL-CMV was used as an internal control. Primer sequences were as follows: p27 promoter (−300) forward, 5′-AAACTCGAGCGGTCCTCTGGTCCAGGTCC-3′; p27 promoter (−1,000) forward, 5′-AAACTCGAGCGAGGGGAGTATTTTCACCC-3′; p27 promoter (TSS) reverse, 5′-TTTAAGCTTGGTGGCCCCGGCGCGGTTCC-3′; p27 promoter (+100) reverse, 5′-TTTAAGCTTAGAAAATGATTGACACGGCG-3′.
Chromatin immunoprecipitation (ChIP) assays. ChIP assays were performed as described previously (21). Briefly, HCT116 cells were transfected with Mock or FLAG-mSp5 expression constructs 24 h prior to the assays. DNA fragments immunoprecipitated with anti-FLAG antibody (#F1804; Sigma-Aldrich) or mouse IgG (2 µg) were analyzed by RT-qPCR using primers directed against a region containing the predicted Sp5-binding site in the p27 promoter region (p27-TSS). The promoter regions of GAPDH and p21 were used as negative and positive controls, respectively. Primer sequences were as follows: GAPDH (TSS) forward, 5′-TGCGTGCCCAGTTGAACCAG-3′; GAPDH (TSS) reverse, 5′-AACAGGAGGAGCAGAGAGCGAAGC-3′; p21 (TSS) forward, 5′-CTGGGGGAGGAGGGAAGTG-3′; p21 (TSS) reverse, 5′-CTGGCCGAGTTCCAGCAG-3′; p27 (−1,000) forward, 5′-CCTTCAGCCCTCTGAAAGCA-3′; p27 (−1,000) reverse, 5′-AGTCCAGCCGCCAAATACAA-3′; p27 (TSS) forward, 5′-ACCGCCATATTGGGCCAC-3′; and p27 (TSS) reverse, 5′-GAGTCGCAGAGCCGTGAG-3′.
Statistical analysis
Statistical analysis was performed using Student's t-test with Microsoft Excel 2010 (Microsoft Corporation, Seattle, WA, USA). P<0.01 was considered to indicate a statistically significant difference.
Results
Sp5 is expressed at low levels in HCT116 cells
To examine the expression levels of Sp5 and the Wnt target gene Axin2, RT-qPCR analyses were performed on the colorectal cancer cell lines HCT116, HT-29, SW403, DLD-1, WiDr, Caco-2 and SW480 (Fig. 1A and B). HCT116 cells contain mutated β-catenin, whereas HT-29, SW403, DLD-1, WiDr, Caco-2 and SW480 cells contain a truncated mutant of the tumor suppressor adenomatous polyposis coli (22,23). In the present study, Sp5 was found to be expressed at very low levels in HCT116 cells compared with HT-29, SW403, DLD-1, WiDr, Caco-2 and SW480 cells. By contrast, Axin2 expression was not downregulated in HCT116 cells compared to the other cell lines.
Figure 1.
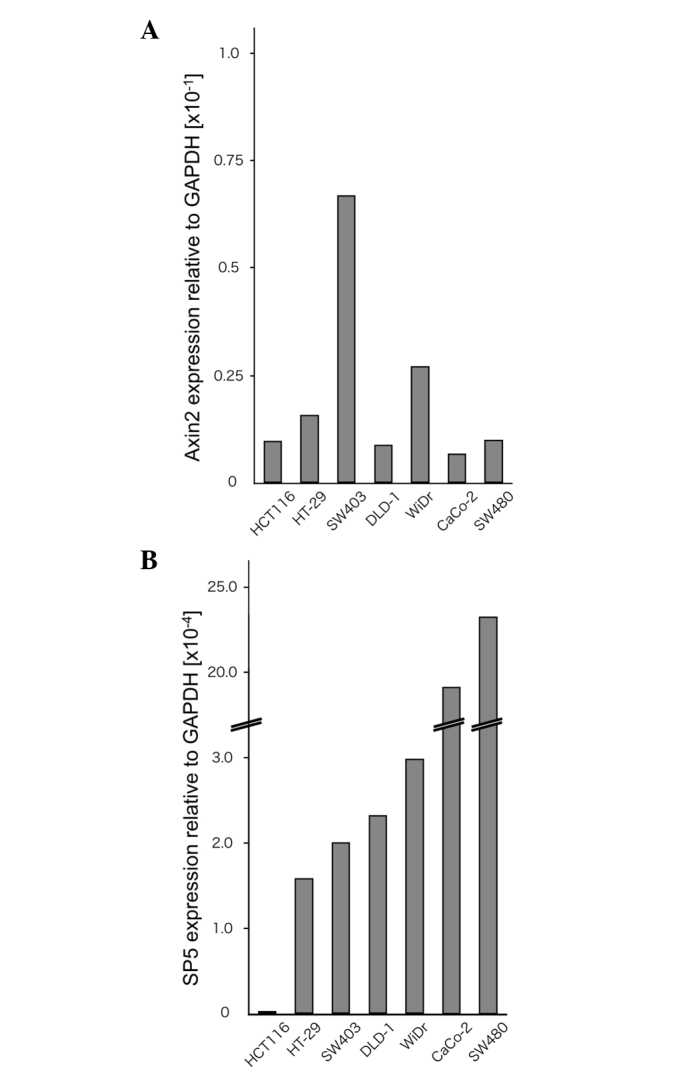
Expression levels of Axin2 and Sp5 in colon cancer cell lines determined by reverse transcription-quantitative polymerase chain reaction.(A) Axin2 and (B) Sp5 expression in HCT116, HT-29, SW403, DLD-1, WiDr, Caco-2 and SW480 cells. The values are the expression levels for each gene normalized to GAPDH. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
mSp5 inhibited the growth of HCT116 cells
To study the effect of Sp5 on the proliferation of colon cancer cells, mSp5 was overexpressed in HCT116 cells (Fig. 2A). Overexpression of mSp5 was found to result in a decrease in proliferation in these cells (Fig. 2B).
Figure 2.
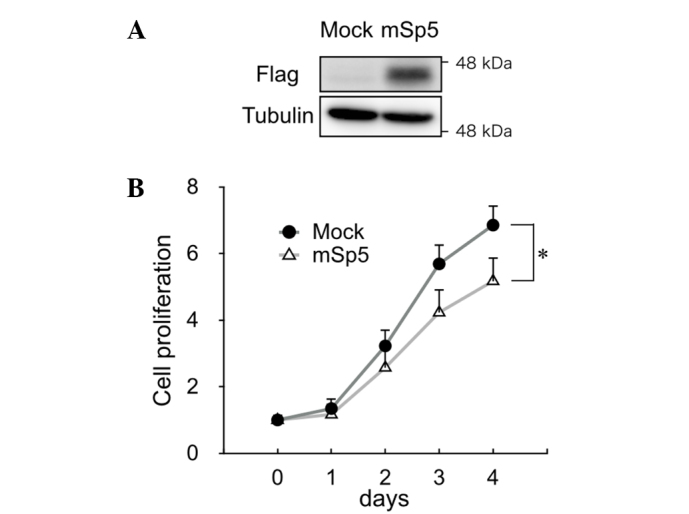
Overexpression of mSp5 inhibited the growth of HCT116 cells. (A) Lysates from HCT116 cells transfected with FLAG-tagged mSp5 were subjected to immunoblotting analysis with an anti-FLAG antibody. Tubulin was used as a loading control. (B) Growth of HCT116 cells transfected with mock or mSp5 expression vectors was measured by CellTiter-Glo assay. Results are shown as mean ± standard deviation of 3 wells. *P=0.009, Student's t-test. mSp5, mouse specificity protein 5.
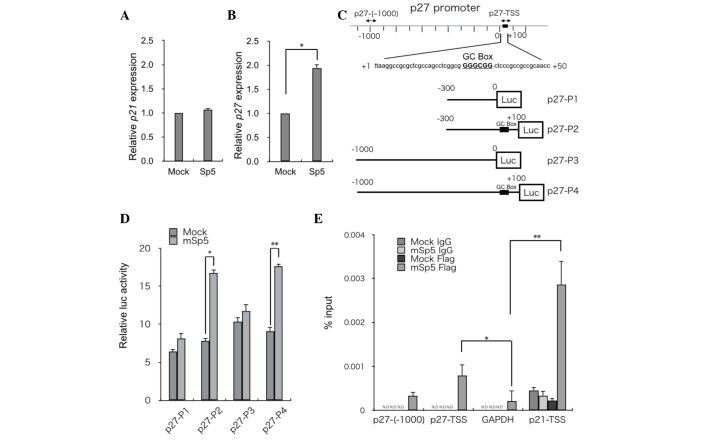
mSp5 activates p27 transcription
It has been reported that overexpression of Sp5 suppresses p21 expression (11,14,17). However, in the present study, RT-qPCR analysis revealed that overexpression of mSp5 did not affect the expression levels of p21 in HCT116 cells (Fig. 3A). By contrast, the overexpression of mSp5 resulted in the upregulation of p27 (Fig. 3B). To clarify the mechanisms underlying Sp5-mediated upregulation of p27, reporter assays were performed using constructs in which various fragments of the p27 promoter region were inserted upstream of the luciferase gene (p27-P1, -P2, -P3 and -P4 in Fig. 3C). Overexpression of mSp5 in HEK293 cells enhanced the activities of the p27-P2 and -P4, but not the p27-P1 and -P3 reporters (Fig. 3D). To examine whether Sp5 transactivates p27 directly, ChIP assays were performed on HCT116 cells transfected with FLAG-tagged mSp5 using an anti-FLAG antibody. mSp5 binding to a DNA fragment (p27-TSS; shown in Fig. 3C), which contains the GC-box (Fig. 3E) was detected. The promoter regions of GAPDH and p21 were used as negative and positive controls, respectively. These results suggest that Sp5 directly upregulates the transcription of p27 by binding to the GC-box in its promoter region.
Figure 3.
mSp5 binds to the promoter region of p27 and activates its transcription. Reverse transcription-quantitative polymerase chain reaction analysis of (A) p27 and (B) p21 in HCT116 cells transfected with mSp5. Results are expressed as the means ± SD of three independent experiments (*P=0.008, Student's t-test). (C) Schematic diagrams of reporter constructs used for luciferase assays. Fragments of the p27 promoter were cloned upstream of the luciferase (Luc) gene: p27-P1 (from −300 to 0), p27-P2 (from −300 to +100), p27-P3 (from −1000 to 0), p27-P4 (from −1000 to +100). The potential Sp5 binding site [GC-box (GGGCGG)] is indicated by black boxes. The positions of the primers used for ChIP assays are indicated by arrows. (D) HEK293 cells were transfected with the indicated p27 reporter constructs along with mSp5 and were subjected to luciferase assays. The pRL-CMV vector was used as an internal control. Results are expressed as the means ± SD of three independent experiments (*P=0.014, **P=0.013, Student's t-test). (E) ChIP assays were performed on HCT116 cells that had been transfected with mSp5 using anti-Flag antibody or control IgG. The promoter regions of p27 [p27-(−1000) and p27-TSS] were amplified. The promoter regions of GAPDH and p21-TSS were used as negative and positive controls, respectively. Results are expressed as the means ± SD of three independent experiments (*P<0.042, **P=0.036, Student's t-test). mSp5, mouse specificity protein 5; SD, standard deviation; ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; N.D, not detected; TSS, transcription start site.
Knockdown of p27 partially restores the growth of HCT116 cells overexpressing mSp5
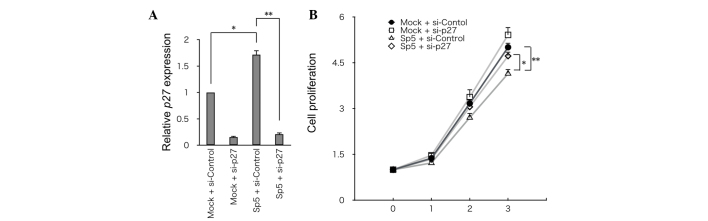
To address the significance of p27 in Sp5-mediated suppression of cell growth, p27 was knocked down using siRNA in HCT116 cells that had been transfected with mSp5 (Fig. 4A). Knockdown of p27 partially restored the growth of HCT116 cells overexpressing mSp5 (Fig. 4B). Thus, Sp5-induced upregulation of p27 may be required for inhibition of the growth of HCT116 cells.
Figure 4.
Knockdown of p27 partially restores the growth of HCT116 cells overexpressing mSp5. (A) Reverse transcription-quantitative polymerase chain reaction analysis of p27 expression in HCT116 cells transfected with the indicated expression constructs (*P=0.028,**P=0.007, Student's t-test). (B) Cell proliferation in HCT116 cells transfected with the indicated constructs. Growth was quantified by CellTiter-Glo assay. Results are expressed as the means ± standard deviation of three independent experiments (*P=0.04, **P=0.03, Student's t-test. mSp5, mouse specificity protein 5; si, small interfering RNA.
Discussion
It has been shown that Sp5 is a direct target of Wnt signaling and that its expression is commonly upregulated in several colon cancer cell lines (24). The present study showed that the expression of Sp5 is not upregulated in HCT116 cells, in which Wnt signaling is constitutively activated. Although other studies have reported that Sp5 represses the expression of p21 and promotes the growth of MCF-7 cells (11,14,17), no significant changes could be detected in the p21 expression in the HCT116 cells transfected with Sp5 in the present study. This discrepancy may be due to the differences in cell types and the different expression levels of the Sp family of proteins. Furthermore, the present results indicate that Sp5 has the potential to inhibit cell proliferation by regulating the cell cycle inhibitor p27 in HCT116 cells. We speculate that HCT116 cells downregulate Sp5 in order to escape p27-mediated growth arrest. The molecular mechanisms underlying the downregulation of Sp5 in HCT116 cells remain to be elucidated.
Acknowledgements
This study was supported by the Research Program of Innovative Cell Biology by Innovative Technology (Integrated Systems Analysis of Cellular Oncogenic Signaling Networks); Grants-in-Aid for Scientific Research on Innovative Areas (Integrative Research on Cancer Microenvironment Network); Project for Development of Innovative Research on Cancer Therapeutics; Takeda Science Foundation; and in part by the Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) and MEXT (Ministry of Education, Culture, Sports, Science and Technology), Japan.
References
- 1.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Eastman Q, Grosschedl R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/S0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 5.Beier F, Lee RJ, Taylor AC, Pestell RG, LuValle P. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc Natl Acad Sci USA. 1999;96:1433–1438. doi: 10.1073/pnas.96.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 7.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 9.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 10.Takahashi M, Nakamura Y, Obama K, Furukawa Y. Identification of SP5 as a downstream gene of the beta-catenin/Tcf pathway and its enhanced expression in human colon cancer. Int J Oncol. 2005;27:1483–1487. [PubMed] [Google Scholar]
- 11.Fujimura N, Vacik T, Machon O, Vlcek C, Scalabrin S, Speth M, Diep D, Krauss S, Kozmik Z. Wnt-mediated down-regulation of Sp1 target genes by a transcriptional repressor Sp5. J Biol Chem. 2007;282:1225–1237. doi: 10.1074/jbc.M605851200. [DOI] [PubMed] [Google Scholar]
- 12.Wallmen B, Schrempp M, Hecht A. Intrinsic properties of Tcf1 and Tcf4 splice variants determine cell-type-specific Wnt/β-catenin target gene expression. Nucleic Acids Res. 2012;40:9455–9469. doi: 10.1093/nar/gks690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fancy SP, Harrington EP, Baranzini SE, Silbereis JC, Shiow LR, Yuen TJ, Huang EJ, Lomvardas S, Rowitch DH. Parallel states of pathological Wnt signaling in neonatal brain injury and colon cancer. Nat Neurosci. 2014;17:506–512. doi: 10.1038/nn1214-1841a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Guo Y, Ge X, Itoh H, Watanabe A, Fujiwara T, Kodama T, Aburatani H. Elevated expression and potential roles of human Sp5, a member of Sp transcription factor family, in human cancers. Biochem Biophys Res Commun. 2006;340:758–766. doi: 10.1016/j.bbrc.2005.12.068. [DOI] [PubMed] [Google Scholar]
- 15.Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 16.Kadonaga JT, Courey AJ, Ladika J, Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 17.Hoverter NP, Ting JH, Sundaresh S, Baldi P, Waterman ML. A WNT/p21 circuit directed by the C-clamp, a sequence-specific DNA binding domain in TCFs. Mol Cell Biol. 2012;32:3648–3662. doi: 10.1128/MCB.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Zapico ME, Lomberk GA, Tsuji S, DeMars CJ, Bardsley MR, Lin YH, Almada LL, Han JJ, Mukhopadhyay D, Ordog T, et al. A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth. Biochem J. 2011;435:529–537. doi: 10.1042/BJ20100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison SM, Houzelstein D, Dunwoodie SL, Beddington RS. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with Brachyury. Dev Biol. 2000;227:358–372. doi: 10.1006/dbio.2000.9878. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Matsuura K, Jigami T, Taniue K, Morishita Y, Adachi S, Senda T, Nonaka A, Aburatani H, Nakamura T, Akiyama T. Identification of a link between Wnt/β-catenin signalling and the cell fusion pathway. Nat Commun. 2011;2:548. doi: 10.1038/ncomms1551. [DOI] [PubMed] [Google Scholar]
- 22.Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 23.Ilyas M, Tomlinson IP, Rowan A, Pignatelli M, Bodmer WF. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Göke B, Kolligs FT. Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC Genomics. 2014;15:74. doi: 10.1186/1471-2164-15-74. [DOI] [PMC free article] [PubMed] [Google Scholar]