Abstract
The objective of the present study was to investigate whether deep learning could be applied successfully to the classification of images from colposcopy. For this purpose, a total of 158 patients who underwent conization were enrolled, and medical records and data from the gynecological oncology database were retrospectively reviewed. Deep learning was performed with the Keras neural network and TensorFlow libraries. Using preoperative images from colposcopy as the input data and deep learning technology, the patients were classified into three groups [severe dysplasia, carcinoma in situ (CIS) and invasive cancer (IC)]. A total of 485 images were obtained for the analysis, of which 142 images were of severe dysplasia (2.9 images/patient), 257 were of CIS (3.3 images/patient), and 86 were of IC (4.1 images/patient). Of these, 233 images were captured with a green filter, and the remaining 252 were captured without a green filter. Following the application of L2 regularization, L1 regularization, dropout and data augmentation, the accuracy of the validation dataset was ~50%. Although the present study is preliminary, the results indicated that deep learning may be applied to classify colposcopy images.
Keywords: colposcopy, cervical cancer, cervical intraepithelial neoplasia, deep learning, artificial intelligence
Introduction
In the era of big data, various types of data can be obtained and shared by all users through the internet or social network systems. One method to efficiently manage enormous amounts of data is to apply deep learning (1). One characteristic of deep learning is that this approach does not require features or representations to be selected during data input. Recently, a number of studies have focused on image classification by deep learning, and these technologies are now becoming readily available for use by corporations and individuals (2–6). For example, TensorFlow is a Google software library for machine learning that was released under an open-source license in 2015 (7). Using this technology, the present study aimed to potentially integrate deep learning into gynecological clinical practice.
Cervical cancer is a leading cause of death in women worldwide (8). Although mortality rates were drastically reduced following the introduction of the Pap smear test, determining the types of patients who should be further screened and treated as high-risk remains an important issue, particularly for avoiding overmedication (9–11). In daily practice, patient management is determined by the combined use of cytology, histology, HPV typing and colposcopy results. The present study investigated whether deep learning with a focus on colposcopy images as input could predict the postoperative diagnosis.
Colposcopy is a well-established tool for observing the cervix at up to ×10 magnification (12). Cervical intraepithelial lesions are enhanced and easily recognized when treated with acetic acid solutions. For instance, areas that turn white following acetic acid treatment (acetowhitening) and/or areas that present abnormal vascular patterns are considered for biopsy. These effects become more visible after a green filter is applied (13). Diagnoses are then evaluated by gynecologists based on the degree of staining and the underlying vascular patterns. Studies have attempted to classify images from colposcopy using neural networks (14–16). For instance, one group investigated whether neural networks could recognize the dot pattern, which represents a colposcopy finding, after learning the pattern from samples annotated by the researchers (16). The present study is distinct from the aforementioned studies because features or representations of the images, for example, the presence of this dot pattern, were not selected during data input.
For deep learning, the Keras neural network and TensorFlow libraries were used (7,17). In the present study, the classification accuracy on the validation dataset reached ~50%. While this result in itself is not satisfactory, it suggests that deep learning has the potential to classify images from colposcopy. In addition, the present study investigated methods to improve the learning rate and avoid overfitting due to the limitation of insufficient numbers of obtained images. In the process presented, L2 regularization, L1 regularization and dropout were applied, and the amount of input data was increased via data augmentation.
In the present study, the intention was not to stress the accuracy rate itself but rather to demonstrate that gynecologists, who are not specialists in artificial intelligence or machine learning, may be able to utilize deep learning in clinical practice. Furthermore, the present results suggest that relevant information from clinical practice should be appropriately stored for future use.
Materials and methods
Patients
The present study was approved by the Institutional Ethics Committee of Saitama Cancer Center (approval no. 630). Written informed consent was obtained from all the patients. Medical records and data from the gynecological oncology database were retrospectively reviewed. Patients who underwent conization at Saitama Cancer Centre (Ina, Japan) from January 2014 to December 2015 were enrolled. Conization management at the facility is determined according to the guidelines of the Japan Society of Obstetrics and Gynecology. Although each diagnosis was performed in principle according to the postoperative pathology (conization), the preoperative pathology (biopsy) was prioritized when the results were severe and used as the output (‘target’ in deep learning) for images from colposcopy.
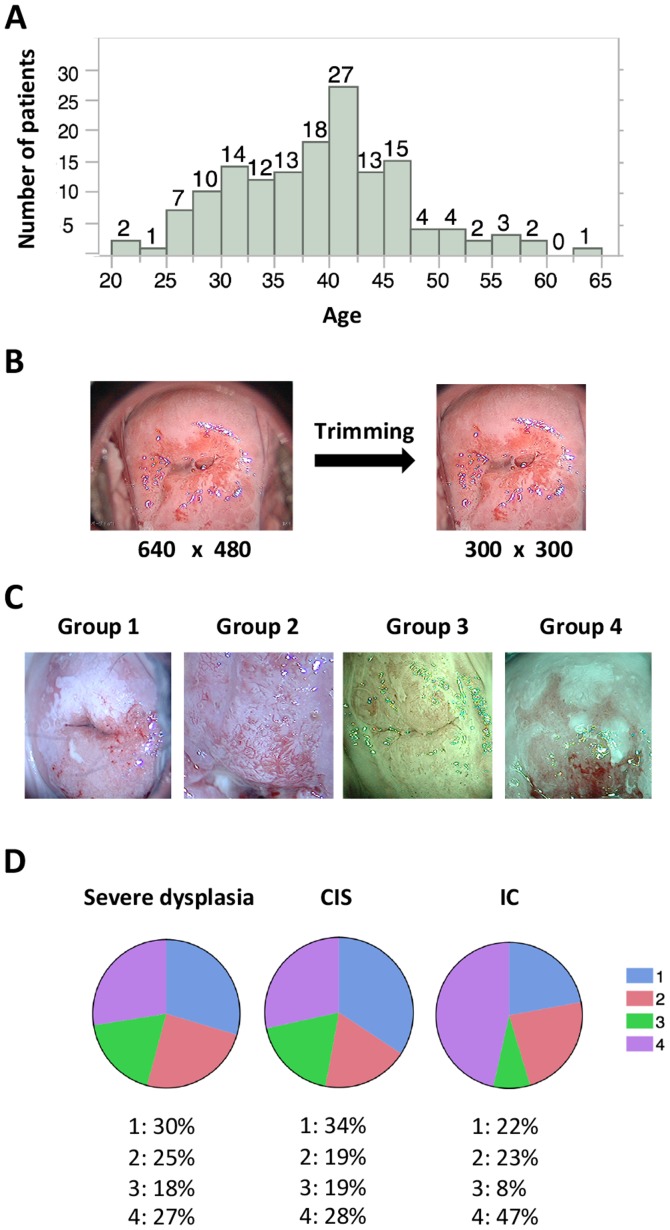
A total of 158 patients were enrolled; their median age was 39 years (range, 21–63 years; Fig. 1A). The diagnoses and corresponding patient numbers were as follows: severe dysplasia, 49; carcinoma in situ (CIS), 78; invasive cancer, (IC) 21; and others (such as adenocarcinoma in situ and invasive adenocarcinoma), 10. In the current study, patient classification was limited to three groups (severe dysplasia, CIS and IC) because of the limited number of available images.
Figure 1.
Patient characteristics and collected images. (A) Distribution of patient age. The median age of the patients was 39 years (range, 21–63 years). (B) Preprocessing of images. Images containing 640×480 pixels were trimmed to 300×300 pixels using Photoshop CC. (C) Patterns of collected images. Representative images for each group are shown. Group 1, images without a green filter that captured >two thirds of the cervix; Group 2, images without a green filter that captured <two thirds of the cervix; Group 3, images with a green filter that captured >two thirds of the cervix; and Group 4, images with a green filter that captured <two thirds of the cervix. (D) Image group distribution for each diagnosis. Images of IC tended to present lesions at greater magnification and were more likely to be captured with a green filter compared with the images of severe dysplasia and CIS. CIS, carcinoma in situ; IC, invasive cancer.
Images
Preoperative images from colposcopy were used as the input data for deep learning. Because this investigation was a retrospective study, there were no criteria for determining the number and type of colposcopy images to retain. Images following acetic acid treatment with or without a green filter that represented areas of biopsy and were used in the diagnoses were stored. The total number of images was 485, with 142 images for severe dysplasia (2.9 images/patient), 257 for CIS (3.3 images/patient), and 86 for IC (4.1 images/patient). Of these, 233 images were captured with a green filter, and the remaining 252 were captured without a green filter.
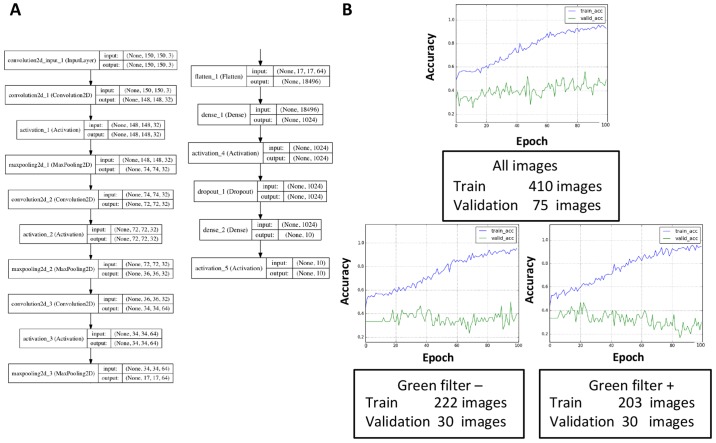
Images from colposcopy captured at our facility were stored in PNG format at a resolution of 640×480 pixels in RGB 3-channel color. These raw images often represented areas inappropriate and unwanted for deep learning, such as the Cusco speculum and vaginal wall; therefore, preprocessing was performed to focus on the cervix by trimming the images to 300×300 pixels (Fig. 1B). Trimming was performed with Photoshop CC (Adobe Systems, Inc., San Jose, CA, USA). Images without a green filter that captured >two thirds of the cervix or <two thirds of the cervix (magnified images of the lesion) were assigned to groups 1 and 2, respectively. Images with a green filter that captured >two thirds of the cervix or <two thirds of the cervix were assigned to groups 3 and 4, respectively (Fig. 1C). During deep learning, these images were re-trimmed to 150×150 pixels and then were used as input (Fig. 2A). The same procedures were performed with images containing 32×32 pixels or 300×300 pixels; however, images with 150×150 pixels were considered suitable for learning in terms of the learning efficacy and time allocated to learning (data not shown), at least in this small-scale study.
Figure 2.
Application of deep learning. (A) Neural network architecture. Tuning the dense layers appeared to improve the learning rates. (B) Influence of the number and types of images on learning rates. The validation accuracy was reduced irrespective of the presence (lower right) or absence (lower left) of a green filter. The total number of images was likely more important for increasing the validation accuracy, at least in this small-scale study (upper). Blue line, training-accuracy curve; Green line, validation-accuracy curve.
Deep learning
We used the Keras (https://keras.io) neural network library and the TensorFlow (https://www.tensorflow.org) software library. The code was frequently referred to in the Keras blog (https://blog.keras.io/building-powerful-image-classification-models-using-very-little-data.html) and the basic code was adjusted for the learning procedure of the present study. The validation dataset contained 25 randomly selected images for each diagnosis (75 images in total), and it was not used for training in the study unless otherwise mentioned.
Development environment
The development environment used in the present study was as follows: a Mac running OS X 10.11.3 (Apple, Inc., Cupertino, CA, USA); Python language v. 2.7.12; Keras 1.1.0; TensorFlow 0.8.0; and matplotlib 1.5.3.
Statistical analysis
JMP Pro 11 (SAS Institute, Inc., Cary, NC, USA) was used for the statistical analysis. One-way analysis of variance was used for comparing the means. The Tukey-Kramer test was used for post-hoc analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Images
Preoperative images from colposcopy were retrospectively collected as described in the Methods section. Statistical analysis suggested that a higher number of images were stored for more severe lesions (P=0.0085). The % of groups 1–4 were summarized for each diagnosis (Fig. 1D). Unlike the images of severe dysplasia and CIS, the IC images tended to include magnified lesions and were usually captured with a green filter (Fig. 1D).
The total number of images is more important for avoiding overfitting than dividing the input images according to the presence or absence of a green filter. A validation set accuracy of <33% meant that learning did not occur because the same number of images for each diagnosis was assigned to the validation dataset. The convolution layers and dense layers were tuned as described in the Methods section (Fig. 2A). In the present study, dense layers appeared to affect the learning rates, and a training accuracy that exceeded 90% in 100 epochs was obtained by tuning the dense layers. However, the validation accuracy plateaued at ~40–50%, which suggested that overfitting had occurred. Therefore, methods of avoiding overfitting to prevent discrepancies between the training curve and validation curve were explored. First, the set of collected images included images both with and without a green filter, and these images were individually used for learning because of possible learning inefficiencies caused by mixed data (Fig. 2B). However, the validation dataset was re-selected (10 images for each diagnosis, 30 in total), and the results demonstrated that the validation accuracy was reduced regardless of the presence or absence of a green filter. This result was likely related to a reduction in the total number of images. Thus, the total number of images appeared to be more important for increasing the validation accuracy than the division of input data according to the presence or absence of green a filter, at least in the present small-scale study.
L2 regularization can improve overfitting
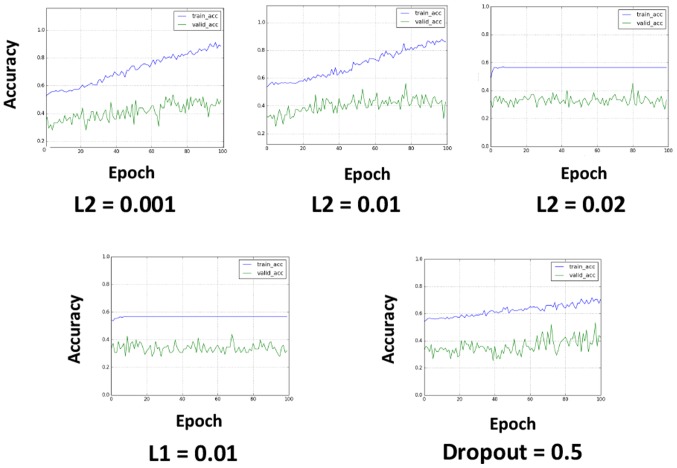
To avoid overfitting, L2 regularization, L1 regularization and dropout were applied (Fig. 3). L2 regularization and L1 regularization were applied in the first input layer, and dropout was applied to all the layers after max-pooling (the dropout rate was set to 0.5). L2 regularization appeared to be effective at avoiding overfitting when properly tuned (Fig. 3). L1 regularization caused learning failure in the investigated value set, and the application of dropout reduced the learning efficacy, although this result may have been related to the relatively short epochs.
Figure 3.
Exploring methods to avoid overfitting. L2 regularization, L1 regularization and dropout were applied. L2 regularization was thought to be somewhat effective in avoiding overfitting when set at a value of 0.001 or 0.01, but not at a value of 0.02. L1 regularization set at a value of 0.01 caused learning failure, and the application of dropout caused a lower learning efficacy; however, this result may have been caused by the relatively small epochs, or it may have represented a characteristic of applying dropout. Blue line, training-accuracy curve; Green line, validation-accuracy curve.
Data augmentation slightly improves the validation accuracy and overfitting
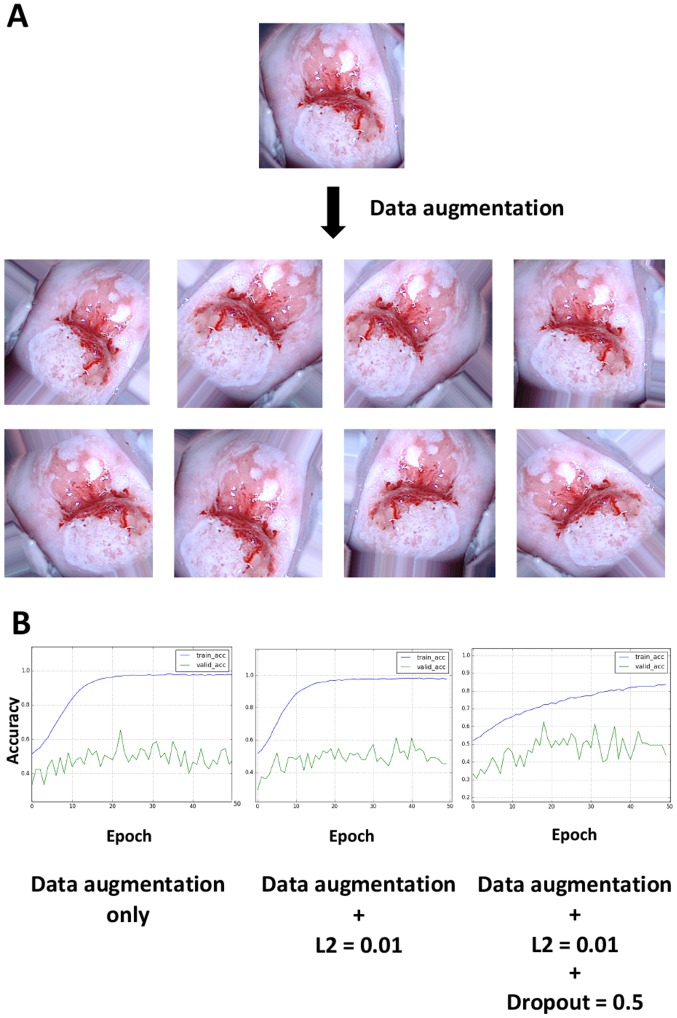
When performing deep learning, the total number of images is known to be an important factor for improving the learning accuracy and avoiding overfitting, which is generally consistent with the aforementioned results (5). Thus, the hypothesis that data augmentation could improve the accuracy rates was investigated. When viewing colposcopy images, tilted images were occasionally encountered because the angles of the cervix relative to the camera posture varied; however, differences in the angle should not be important for recognizing lesions. To test this hypothesis, 20 images were obtained from a single image by randomly rotating it, applying different zoom magnifications and horizontally flipping the image and the resulting images were then used as input. Thus, one image was converted into ~20 images by data augmentation. Examples of the augmentation results are illustrated in Fig. 4A. Data augmentation appeared to worsen the overfitting limitation; however, the application of L2 regularization and dropout slightly improved overfitting (Fig. 4B). The validation accuracy ultimately reached a stable level of ~50%.
Figure 4.
Data augmentation. (A) Example of data-augmented images. A total of 20 images were obtained from a single image by randomly rotating, zooming in/out and horizontally flipping the image. Therefore, one image was increased to ~20 images by data augmentation. (B) Combination of data augmentation, L2 regularization and dropout. Data augmentation worsened the overfitting; however, the application of L2 regularization and dropout slightly improved the overfitting. The final validation accuracy reached a stable value of ~50%. Blue line, training-accuracy curve; Green line, validation-accuracy curve.
Discussion
The present study investigated whether deep learning could be applied to the classification of images from colposcopy. Various types of data are increasingly available through the Internet, and inexpensive high-end smartphones are more readily available for the general public, facilitating the uploading and sharing of information, such as pictures. The same is true for data processing. High-performance personal computers are affordable for individuals, and statistical analyses or machine learning can be performed without supercomputers if the information volume is limited. Furthermore, deep learning technologies are becoming more accessible for corporations and individuals. For example, the Google software library for machine learning, TensorFlow, was released under an open-source license in 2015 (7). Based on these trends, the present study aimed to apply deep learning to gynecological clinical practice.
Preoperative images from colposcopy were retrospectively collected. A total of 485 images were obtained, with 142 images for severe dysplasia (2.9 images/patient), 257 for CIS (3.3 images/patient), and 86 for IC (4.1 images/patient). These results indicate that more images were stored when the lesions were more severe (P=0.0085), because gynecologists tend to capture a higher number of images in lesions in order to record important findings. Accordingly, the IC images tended to include lesions under greater magnification. Furthermore, these images were more frequently captured with a green filter compared with the severe dysplasia and CIS images (Fig. 1D).
One of the greatest challenges associated with machine learning, including deep learning, is the prevention of overfitting (5). Overfitting is a condition in which the model cannot be applied to unknown data because it has been overly adjusted to the training data. In the present study, the large discrepancy between the training curve and validation curve suggests that overfitting occurred (Figs. 2–4), most likely due to the small number of included images. Ordinarily, 500–1,000 images are prepared for each class during image classification with deep learning (2). The present study explored methods to improve the learning rate and avoid overfitting under the limitation of an insufficient number of included images. In addition, L2 regularization, L1 regularization and dropout were applied, and the amount of input data was increased by data augmentation.
In clinical practice, it would be of interest for clinicians to distinguish CIN1, CIN2 and CIN3, or to distinguish CIN1 (or low-grade squamous intraepithelial lesions) from CIN2 (or high-grade squamous intraepithelial lesions). Furthermore, classification might not be clinically necessary for severe dysplasia and carcinoma in situ (CIS), because there is little difference in diagnosis and treatment between these two conditions. However, due to technical reasons, the present preliminary study used images from CIN3 and invasive cancer patients. In terms of deep learning, the output (i.e., the ground-truth classification result of an image) is very important. For instance, what is considered CIN1 may not always be ‘genuine’ CIN1 because only a biopsy is performed in most cases. Ideally, conization should be performed to provide the true answer. In addition, providing ground-truth classifications such as ‘white lesion’ or ‘glandular opening’ would not construct a reliable model, because those answers are subject to human perception: There is no strict definition for these terms. As such, for this initial study, images from CIN3 and invasive cancer patients were used, because pathological diagnoses of the conization samples were readily obtained. For clinicians, an improved solution would be to use the ‘patient's prognosis’ as an output. A clinical application designed to screen a target group not in need of invasive testing, such as biopsy and conization could be desirable as well. Therefore, although the clinical significance of the classification into three groups (severe dysplasia, CIS and IC) is currently limited, the present study demonstrated that deep learning, by inputting only images, could be used to classify colposcopy images.
The final validation accuracy was ~50%, which is better than a random result (33%). To the best of our knowledge, no report using ‘deep learning’ for classification of images from colposcopy exists to date. Although previous studies have used automation diagnosis, deep learning was not employed and the patient cohort was different than the current study (CIN3 and invasive cancer) (14–16). As such, the present study cannot be directly compared to the previous literature, in order to evaluate whether the 50% accuracy result was good or poor. However, although this result may not be satisfactory in terms of deep-learning tasks, it may be sufficient in the clinic considering the difficulty of distinguishing among severe dysplasia, CIS and IC even among experts.
The present study aimed, not to stress the accuracy rate itself, but rather to demonstrate that gynecologists, who are not specialists in artificial intelligence or machine learning, can utilize deep learning in clinical practice. The barriers to using artificial intelligence and deep learning will likely be decreased in the near future. Thus, as much relevant clinical information as possible should be stored appropriately for future use. For instance, the images used in this study contained as few as 150×150 pixels and 3 RGB channels. Images of this size could be obtained by most users, even those using smart phones. Our facility is a cancer center, and the patient population could have been biased in terms of disease conditions because those who require operations or intensive observation tend to be referred to our hospital. Consequently, the collected images could also be biased. Furthermore, as mentioned above, there were many more images taken of lesions considered to be more severe. Therefore, the same proportion of images is likely stored at many other facilities, regardless of the diagnosis and the apparent severity of the lesion.
The current study investigated a method for applying deep learning to colposcopy image classification. The accuracy on the final validation dataset reached ~50%. Although this result is preliminary, it suggests that clinicians and researchers, who are not specialists in artificial intelligence or machine learning, can utilize deep learning. Furthermore, these findings suggest that as much relevant clinical practice information as possible, including colposcopy data and images, should be stored for future use.
References
- 1.Schmidhuber J. Deep learning in neural networks: An overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajkomar A, Lingam S, Taylor AG, Blum M, Mongan J. High-throughput classification of radiographs using deep convolutional neural networks. J Digit Imaging. 2017;30:95–101. doi: 10.1007/s10278-016-9914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu L, Chen H, Dou Q, Qin J, Heng PA. Automated melanoma recognition in dermoscopy images via very deep residual networks. Ieee Trans Med Imaging. 2016;36:994–1004. doi: 10.1109/TMI.2016.2642839. [DOI] [PubMed] [Google Scholar]
- 6.Zhang YC, Kagen AC. Machine learning interface for medical image analysis. J Digit Imaging. 2017;30:615–621. doi: 10.1007/s10278-016-9910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rampasek L, Goldenberg A. Tensorflow: Biology's gateway to deep learning? Cell Syst. 2016;2:12–14. doi: 10.1016/j.cels.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Ginsburg O, Bray F, Coleman MP, Vanderpuye V, Eniu A, Kotha SR, Sarker M, Huong TT, Allemani C, Dvaladze A, et al. The global burden of women's cancers: A grand challenge in global health. Lancet. 2017;389:847–860. doi: 10.1016/S0140-6736(16)31392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook DA, Smith LW, Law J, Mei W, van Niekerk DJ, Ceballos K, Gondara L, Franco EL, Coldman AJ, Ogilvie GS, et al. Aptima HPV assay versus hybrid capture® 2 HPV test for primary cervical cancer screening in the HPV FOCAL trial. J Clin Virol. 2017;87:23–29. doi: 10.1016/j.jcv.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Coste J, Cochand-Priollet B, de Cremoux P, Le Galès C, Cartier I, Molinié V, Labbé S, Vacher-Lavenu MC, Vielh P, French Society of Clinical Cytology Study Group Cross sectional study of conventional cervical smear, monolayer cytology and human papillomavirus DNA testing for cervical cancer screening. BMJ. 2003;326:733. doi: 10.1136/bmj.326.7392.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Boon JA, Pyeon D, Wang SS, Horswill M, Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal oestrogen receptor signalling; Proc Natl Acad Sci USA; 2015; pp. E3255–E3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Arteaga JD, Kybic J, Li W. Automatic colposcopy video tissue classification using higher order entropy-based image registration. Comput Biology Med. 2011;41:960–970. doi: 10.1016/j.compbiomed.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Balas C. A novel optical imaging method for the early detection, quantitative grading and mapping of cancerous and precancerous lesions of cervix. IEEE Trans Biomed Eng. 2001;48:96–104. doi: 10.1109/10.900259. [DOI] [PubMed] [Google Scholar]
- 14.Acosta-Mesa HG, Cruz-Ramirez N, Hernández-Jiménez R. Aceto-white temporal pattern classification using k-NN to identify precancerous cervical lesion in colposcopic images. Comput Biol Med. 2009;39:778–784. doi: 10.1016/j.compbiomed.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Acosta-Mesa HG, Rechy-Ramirez F, Mezura-Montes E, Cruz-Ramirez N, Hernández Jiménez R. Application of time series discretization using evolutionary programming for classification of precancerous cervical lesions. J Biomed Inform. 2014;49:73–83. doi: 10.1016/j.jbi.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Simoes PW, Izumi NB, Casagrande RS, Venson R, Veronezi CD, Moretti GP, da Rocha EL, Cechinel C, Ceretta LB, Comunello E, et al. Classification of images acquired with colposcopy using artificial neural networks. Cancer Infor. 2014;13:119–124. doi: 10.4137/CIN.S17948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medved D, Nugues P, Nilsson J. Predicting the outcome for patients in a heart transplantation queue using deep learning; Conf Proc IEEE Eng Med Biol Soc; 2017; pp. 74–77. [DOI] [PubMed] [Google Scholar]