Abstract
A genetically engineered Salmonella typhimurium strain that may be applied in the medically useful therapeutic strategy of using bacterial agents to target breast cancer in a tumor-bearing nude mouse model has been previously reported. Furthermore, immune cell accumulation in breast tumor types has been observed, particularly distributed in regions surrounding the bacteria. M2 macrophages are associated with breast cancer aggressiveness, whereas M1 macrophages are prone to devouring bacteria and killing cancer cells. Therefore, this engineered tumor-targeting salmonella strain was used in an attempt to reverse the phenotype of M2 macrophages into the M1 phenotype. Subsequent to the co-culture of M2 macrophages with the bacteria for a short time, >50% of the M2 macrophages were invaded by bacteria. These M2 macrophages exhibited a decreased expression of mannose receptor (an M2 phenotypic marker) and increased expression of human leukocyte antigen-antigen D related (an M1 phenotypic marker). The results of the present study indicated that differentiated M2 macrophages may be redirected into the M1 phenotype following exposure to the engineered bacteria stimulus. This effect may be a potential mechanism by which bacteria retard tumor growth. Thus, this engineered bacterium may be a useful candidate for targeting and redirecting M2 macrophages into the M1 phenotype.
Keywords: engineered bacteria, breast cancer, macrophage, tumor-targeting therapy
Introduction
The potential use of bacteria for cancer treatment has been extensively investigated in previous years. Bacteria, including Bifidobacterium (1,2), Clostridium (3) and Salmonella have been demonstrated to preferentially target and replicate in the hypoxic and necrotic regions of a tumor, resulting in tumor repression (4–7). In a previous study, a synthetic biology approach was used to generate the novel Salmonella typhimurium strain YB1 (YB1) (8). This bacterium specifically colonizes and proliferates in the hypoxic/necrotic areas of the tumor, but avoids normal organs and retards tumor growth (8). Furthermore, a previous study reported that numerous macrophages accumulate in breast tumors and are associated with a poor prognosis (9).
Macrophages are heterogeneous cells that respond differently to various stimulating signals and display numerous different phenotypes (5). The M1 and M2 macrophage phenotypes represent the two extremes of a broad range of macrophage functional states. Fully polarized M1 (or classically activated) macrophages are stimulated by microbial agents or pro-inflammatory factors, including lipopolysaccharide (LPS), whereas M2 (or alternatively activated) macrophages respond to anti-inflammatory molecules, including interleukin-4 (IL-4) (10,11). Macrophages located in the stroma of breast cancer tissues [known as tumor-associated macrophages (TAMs)] are primarily M2 macrophages activated by IL-4-producing cluster of differentiation (CD)4+ T cells (12). TAMs are the most notable migratory hematopoietic cell type in the tumor microenvironment and promote the invasiveness of breast cancer cells (13).
Clinically, a large amount of macrophage infiltration in tumor sections from patients with breast cancer has been observed using CD68 immunohistochemical staining. TAMs are associated with breast cancer aggressiveness and promote cancer metastasis, whereas M1 macrophages are prone to killing cancer cells and devouring bacteria (14). Furthermore, studies have revealed that TAMs (which are primarily M2 macrophages activated by IL-4) exhibit a CD206high/human leukocyte antigen-antigen D related (HLA-DR)low phenotype that is associated with immune suppression (15–17). Therefore, CD206 and HLA-DR may be used as markers for M1 and M2 macrophage phenotype analysis (15). In the present study, the newly engineered tumor-targeting YB1 strain was used in order to attempt to redirect M2 macrophages into the M1 phenotype. More than half of the M2 macrophages devoured the bacteria after 2 h of co-culture. These M2 macrophages exhibited a decreased CD206 expression and an increased HLA-DR expression. Therefore, the IL-4-activated M2 macrophages switched from the CD206high/HLA-DRlow phenotype to the CD206low/HLA-DRhigh phenotype subsequent to co-culture with the engineered YB1 strain. The present study indicates that differentiated M2 macrophages may be redirected into an M1 phenotype following exposure to different stimuli. This finding may reflect a potential mechanism by which bacteria retard tumor growth. Therefore, these engineered bacteria may be used as a vector to target tumors.
Materials and methods
Patient samples and macrophage immunohistochemistry staining
All tumor samples from breast-infiltrating ductal carcinomas were obtained from female patients (mean age, 45 years; age range, 35–55 years) at the Guangdong Women and Children's Hospital (Guangdong, China). The samples were used with written informed consent and ethical approval was obtained from the Internal Review and the Ethics Boards of Guangdong Women and Children's Hospital (Guangdong, China).
The samples were fixed in 10% formalin for >2 h at room temperature, paraffin-embedded (3 min at 56°C) and sectioned into 5 µM-thick slices. The macrophages were visualized by immunohistochemistry staining using an anti-CD68 antibody (cat. no. M0814; dilution, 1:200; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA), and sections were treated using this antibody overnight at 4°C. For details, please refer to reference (18).
Bacterial culture
The bacterial YB1 strain was cultured in lysogeny broth medium overnight (12 to 16 h) (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with chloramphenicol and 2,3-diaminopropionic acid (Sigma-Aldrich; Merck KGaA) at 37°C (8).
Isolation and activation of human monocyte-derived macrophages
Institutional ethical approval was obtained from the Internal Review and the Ethics Boards of Guangdong Women and Children's Hospital, Guangdong, China prior to conducting the study. Human mononuclear cells were isolated from 100 ml peripheral blood of healthy donors by Ficoll density gradient centrifugation (20°C at 250 × g for 20 min), as previously described (18). The resulting monocyte-derived macrophages were activated by the addition of IL-4 (45 ng/ml) to the culture medium for 3 days (19), and LPS (20 ng/ml) was added as a control.
Bacteria and macrophage co-culture
Isolated macrophages were activated by IL-4. The bacterial YB1 strain, which carried the green fluorescent protein (GFP)-tagged plasmid, was added to the macrophage culture for a final bacterial concentration of 5×106/ml. The cultures were incubated at 37°C under hypoxic conditions as previously described (8) for 2 h. Then, the bacteria were washed away and the macrophages were harvested for further analysis.
Flow cytometry
Following the co-culture, the macrophages were fixed in paraformaldehyde (4%) at 4°C for 0.5–2 h. The macrophage phagocytic rate of YB1 (GFP) was detected using flow cytometry. CD206 or HLA-DR expression levels in the macrophages were also determined using flow cytometry. Briefly, the macrophages (105-106/ml) were collected, fixed in 4% paraformaldehyde at 4°C for 0.5–2 h, blocked with 5% bovine serum albumin for 20 min at room temperature and stained using a phycoerythrin-conjugated CD206 (dilution, 1:20; cat no. 321105; BioLegend, Inc., San Diego, CA, USA) or allophycocyanin-conjugated HLA-DR (dilution, 1:20; cat no. 307609; BioLegend, Inc.) antibody for 1 h at 4°C. The corresponding isotype control was included in each test. Then, CD206 or HLA-DR expression was analyzed using flow cytometry. FlowJo software (version 7.6.1; FlowJo LLC, Ashland, OR, USA) was used to analyze the data.
Western blotting
Cells were lysed using radioimmunoprecipitation assay lysis buffer (50 mM TrisHCl pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.1% SDS) and protease inhibitors (Sigma-Aldrich; Merck KGaA). Cells were lysed with lysis buffer for 15–20 min at 4°C. HLA-DR expression was determined according to a previously described protocol (18). The proteins (20 µl per lane) were separated on 10% SDS-PAGE gels and transferred onto nitrocellulose membranes. The membranes were blocked in 5% non-fat milk for 2 h at room temperature and then incubated with antibodies against HLA-DR (1:5,000; rabbit monoclonal IgG antibody; cat no. ab92511; Abcam, Cambridge, UK) overnight at 4°C. Subsequent to washing using 0.1% TBST (50 mM tris, 150 mM NaCl, 0.1% Tween 20, pH 7.4) three times (10 min each), the membrane was incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (GE Healthcare, Chicago, IL, USA) for 1 h at room temperature. Then, the membranes were washed again using 0.1% TBST three times (10 min each). The HRP signal was visualized using enhanced chemiluminescence (ChemiDoc MP Imaging System; Bio-Rad Laboratories, Inc., Hercules, CA, USA), and analyzed using Image Lab (version 5.2.1; Bio-Rad Laboratories, Inc.).
Results
Macrophage infiltration in breast tumor tissues
The engineered YB1 strain may specifically target and survive in a solid tumor. To confirm macrophage infiltration in the breast tumor, clinical breast cancer samples were collected and CD68 immunohistochemistry staining was used to demonstrate the macrophage distribution. As presented in Fig. 1, a large number of macrophages (CD68-positive) infiltrated the breast tumor, particularly in the tumor-associated stromal border, a result consistent with previous reports (12). This result implies that bacteria may be able to target macrophages localized in the breast tumor.
Figure 1.
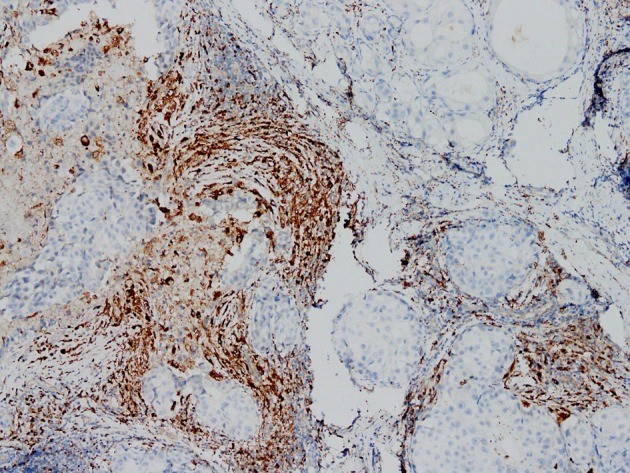
Immunohistochemical staining of breast cancer tissue revealing the mannose receptor-positive macrophages present (magnification, ×100).
YB1 invaded M2 macrophages under hypoxic conditions
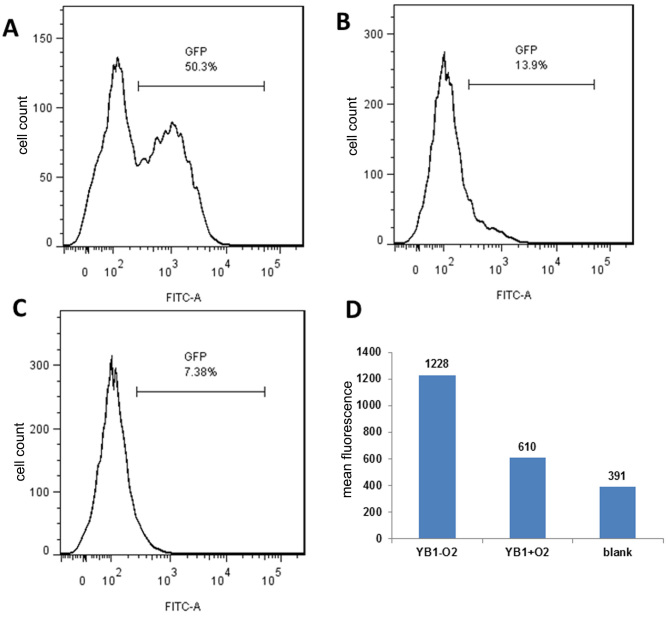
The engineered YB1 strain only survives under hypoxic conditions (8). However, whether YB1 may invade M2 macrophages under hypoxic conditions remains unknown. Thus, an anaerobic microenvironment was stimulated in vitro and co-cultured M2 macrophages with the YB1 strain with the GFP-tagged plasmid for 2 h. Flow cytometry analysis was used to determine the YB1 invasion rate. As presented in Fig. 2A, YB1 had a high invasion rate under hypoxic conditions; after 2 h, >50% of the macrophages were invaded. In contrast, the invasion rate was very low under normal conditions (21% oxygen) (Fig. 2B) as YB1 may not survive. Fig. 2C presented the results of the blank control. It was additionally identified that the mean fluorescence was substantially higher under hypoxic conditions than under normal conditions, indicating that >1 bacterium invaded each macrophage (Fig. 2D).
Figure 2.
YB1 strain invasion rate between hypoxic and normal conditions, determined by flow cytometry analysis. YB1 invasion rate under (A) hypoxic and (B) normal conditions, with (C) demonstrating the results of the blank control. (D) Mean fluorescence between hypoxic and normal conditions. YB1-O2: under hypoxic conditions; YB1+O2: under normal conditions. GFP, green fluorescent protein; FITC-A, fluorescein isothiocyanate; YB1, Salmonella typhimurium strain YB1.
M2 macrophages exhibited increased HLA-DR and decreased CD206 expression
Subsequent to co-culturing M2 macrophages with YB1, the macrophages were collected and flow cytometry was used to determine the HLA-DR expression levels (Fig. 3A). LPS activated the macrophages to express higher levels of HLA-DR compared with those in the IL-4-activated macrophages. Notably, the YB1 alone strain induced the highest HLA-DR expression in macrophages, suggesting the high efficiency of YB1 in activating the M1 macrophage phenotype. When the macrophages were activated in advance with IL-4 and then co-cultured with YB1, an increase in HLA-DR expression was observed compared with LPS or IL-4 alone activated macrophages. Western blotting confirmed these results (Fig. 3B). Next, the M2 macrophage phenotype marker CD206 was examined (Fig. 3C) and it was identified that IL-4-activated macrophages expressed higher CD206 levels compared with LPS-activated macrophages. The YB1 strain alone reduced the CD206 expression levels compared with every other group. Notably, the YB1 strain reduced CD206 expression in the IL-4-activated M2 macrophages.
Figure 3.
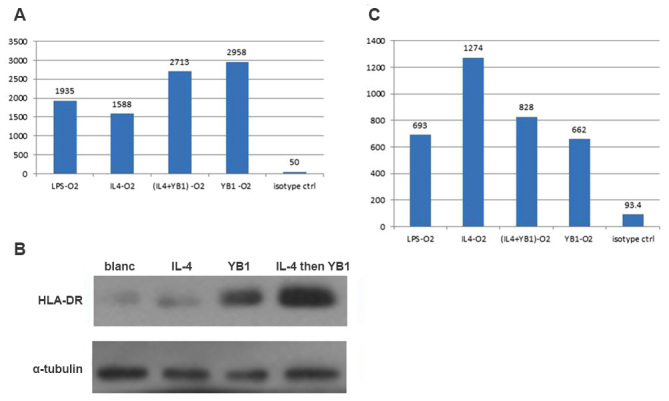
M2 macrophages exhibited increased HLA-DR and decreased CD206 expression levels. (A) HLA-DR expression levels in M2 macrophages activated using LPS, IL-4, YB1, a combination of YB1 and IL-4 or an isotype. (B) HLA-DR expression confirmed using western blotting. (C) CD206 expression levels in M2 macrophages activated using LPS, IL-4, YB1, a combination of YB1 and IL-4 or an isotype. HLA-DR, human leukocyte antigen-antigen D related; CD206, mannose receptor; LPS, lipopolysaccharide; IL, interleukin; YB1, Salmonella typhimurium strain YB1.
Discussion
Although a broad range of macrophage subsets have been identified, the two major macrophage populations are the M1 (classically activated) macrophages and M2 (alternatively activated) macrophages (20). These two different types of macrophages have different effects on tumor progression. As early as 1980, studies have demonstrated that bacterial LPS activates macrophages (21–23) to specifically kill tumor cells including breast cancer cells but has no effect on normal cells (24,25). This effect on macrophages requires LPS for maintenance (26). Additionally, macrophages themselves possess a phagocytic ability. When a tumor occurs, macrophages migrate to the tumor location, as directed by the action of chemokines, and devour the tumor cells (27). Macrophages may kill tumor cells, though it remains unknown why this killing effect halts tumor growth and distant metastasis (28). Beyond the tumor immune escape mechanism (29), studies have identified that macrophages are induced by tumor cells in the tumor microenvironment and develop tumor-promoting properties (M2 type, otherwise known as TAMs) (30,31). Statistical data analyses have revealed that the proportion of TAMs in solid tumor tissues may be as high as 80% (29). Likewise, numerous macrophages have been detected in clinical breast cancer samples and, furthermore, macrophage infiltration and breast cancer metastasis are associated (32–34). However, although M1 and M2 macrophages serve different functions in tumor progression, there is no absolute boundary between the two types of macrophages. In the tumor microenvironment, factors including the MHC expression level in tumor cells and the oxygen pressure in the microenvironment affect the macrophage phenotype (29). Therefore, the phenotype of differentiated macrophages may change. This finding indicates a novel target of tumor treatment: Macrophages in the tumor microenvironment. If M2 macrophages may be directed to become the M1 type, one of the drivers of tumor progression would be eliminated, and result in the gain of one more helper to kill tumor cells.
Deng et al (35) has demonstrated that suppression of heme oxygenase-1 in TAMs in a breast cancer mouse model alternatively activates the switching of the M2 macrophage type to the classically activated M1 macrophage type. However, the ‘weapons’ used to specifically target macrophages in breast cancer are still lacking.
In the present study, a novel engineered Salmonella strain (YB1) was reported to induce increased HLA-DR expression and decreased CD206 expression in differentiated M2 macrophages. These M2 macrophages changed from the CD206high/HLA-DRlow phenotype to the CD206low/HLA-DRhigh phenotype, indicating an M2 to M1-type switch. This result suggests a potential use for the engineered tumor-targeting bacteria YB1 in redirecting M2 type macrophages into the M1 type and thus suppressing tumor growth.
Overall, the results suggest that the engineered bacterial YB1 strain may be a good candidate for targeting and redirecting M2 macrophages into the M1 type. In addition to its tumor targeting ability, these bacteria may survive and proliferate in the tumor microenvironment; therefore, the effects would be long-lasting, and the activation of the M1 type would be sustained. Furthermore, for safety, this engineered Salmonella YB1 strain is controllable and may be eliminated by antibiotics. Finally, the genetic background of YB1 is clear and may be engineered to carry further ‘weapons’ in order to kill cancer cells.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81202076) and the Guangzhou Science and Technology Program (grant no. 2014J2200007). The authors would like to thank Dr Jian-dong Huang (Hong Kong University) for supplying the bacteria strain.
References
- 1.Yazawa K, Fujimori M, Nakamura T, Sasaki T, Amano J, Kano Y, Taniguchi S. Bifidobacterium longum as a delivery system for gene therapy of chemically induced rat mammary tumors. Breast Cancer Res Treat. 2001;66:165–170. doi: 10.1023/A:1010644217648. [DOI] [PubMed] [Google Scholar]
- 2.Fujimori M. Anaerobic bacteria as a gene delivery system for breast cancer therapy. Nihon Rinsho. 2008;66:1211–1218. (In Japanese) [PubMed] [Google Scholar]
- 3.Liu SC, Ahn GO, Kioi M, Dorie MJ, Patterson AV, Brown JM. Optimized clostridium-directed enzyme prodrug therapy improves the antitumor activity of the novel DNA cross-linking agent PR-104. Cancer Res. 2008;68:7995–8003. doi: 10.1158/0008-5472.CAN-08-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao M, Yang M, Ma H, Li X, Tan X, Li S, Yang Z, Hoffman RM. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer Res. 2006;66:7647–7652. doi: 10.1158/0008-5472.CAN-06-0716. [DOI] [PubMed] [Google Scholar]
- 5.Low KB, Ittensohn M, Le T, Platt J, Sodi S, Amoss M, Ash O, Carmichael E, Chakraborty A, Fischer J, et al. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol. 1999;17:37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- 6.Felgner S, Kocijancic D, Frahm M, Curtiss R, III, Erhardt M, Weiss S. Optimizing salmonella enterica serovar typhimurium for bacteria-mediated tumor therapy. Gut microbes. 2016;7:171–177. doi: 10.1080/19490976.2016.1155021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frahm M, Felgner S, Kocijancic D, Rohde M, Hensel M, Curtiss R, III, Erhardt M, Weiss S. Efficiency of conditionally attenuated Salmonella enterica serovar Typhimurium in bacterium-mediated tumor therapy. mBio. 2015;6:pii: e00254–15. doi: 10.1128/mBio.00254-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu B, Yang M, Shi L, Yao Y, Jiang Q, Li X, Tang LH, Zheng BJ, Yuen KY, Smith DK, et al. Explicit hypoxia targeting with tumor suppression by creating an ‘obligate’ anaerobic Salmonella Typhimurium strain. Sci Rep. 2012;2:436. doi: 10.1038/srep00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013;332:3–10. doi: 10.1016/j.canlet.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 10.Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, Markowitz D, Wu W, Liu C, Reisfeld RA, Xiang R. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest. 2006;116:2132–2141. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leek RD, Lewis CE, Whitehouse R, Greenall M, Clarke J, Harris AL. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56:4625–4629. [PubMed] [Google Scholar]
- 12.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Gao L, Cai Y, Liu H, Gao D, Lai J, Jia B, Wang F, Liu Z. Inhibition of tumor growth and metastasis by photoimmunotherapy targeting tumor-associated macrophage in a sorafenib-resistant tumor model. Biomaterials. 2016;84:1–12. doi: 10.1016/j.biomaterials.2016.01.027. [DOI] [PubMed] [Google Scholar]
- 14.Khabbazi S, Goumon Y, Parat MO. Morphine modulates interleukin-4- or breast cancer cell-induced pro-metastatic activation of macrophages. Sci Rep. 2015;5:11389. doi: 10.1038/srep11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su S, Liu Q, Chen J, Chen J, Chen F, He C, Huang D, Wu W, Lin L, Huang W, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer cell. 2014;25:605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Dangaj D, Abbott KL, Mookerjee A, Zhao A, Kirby PS, Sandaltzopoulos R, Powell DJ, Jr, Lamazière A, Siegel DL, Wolf C, Scholler N. Mannose receptor (MR) engagement by mesothelin GPI anchor polarizes tumor-associated macrophages and is blocked by anti-MR human recombinant antibody. PLoS One. 2011;6:e28386. doi: 10.1371/journal.pone.0028386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuang DM, Wu Y, Chen N, Cheng J, Zhuang SM, Zheng L. Tumor-derived hyaluronan induces formation of immunosuppressive macrophages through transient early activation of monocytes. Blood. 2007;110:587–595. doi: 10.1182/blood-2007-01-068031. [DOI] [PubMed] [Google Scholar]
- 18.Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, Liu B, Deng H, Wang F, Lin L, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castoldi A, Naffah de Souza C, Câmara NO, Moraes-Vieira PM. The macrophage switch in obesity development. Front Immunol. 2016;6:637. doi: 10.3389/fimmu.2015.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamaidia M, Staumont B, Duysinx B, Louis R, Willems L. Improvement of malignant pleural mesothelioma immunotherapy by epigenetic modulators. Curr Top Med Chem. 2016;16:777–787. doi: 10.2174/1568026615666150825141152. [DOI] [PubMed] [Google Scholar]
- 22.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hambleton J, Weinstein SL, Lem L, DeFranco AL. Activation of c-Jun N-terminal kinase in bacterial lipopolysaccharide-stimulated macrophages; Proc Natl Acad Sci USA; 1996; pp. 2774–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chimal-Ramírez GK, Espinoza-Sánchez NA, Chávez-Sánchez L, Arriaga-Pizano L, Fuentes-Pananá EM. Monocyte differentiation towards protumor activity does not correlate with M1 or M2 phenotypes. J Immunol Res. 2016;2016:6031486. doi: 10.1155/2016/6031486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, Liu Y, Gu J, Wang Y, Liu L, Zhang P, Li Y. Endostatin inhibits the growth and migration of 4T1 mouse breast cancer cells by skewing macrophage polarity toward the M1 phenotype. Cancer Immunol Immunother. 2016;65:677–688. doi: 10.1007/s00262-016-1824-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron DJ, Churchill WH. Cytotoxicity of human macrophages for tumor cells: Enhancement by bacterial lipopolysaccharides (LPS) J Immunol. 1980;124:708–712. [PubMed] [Google Scholar]
- 27.Sinha P, Clements VK, Miller S, Ostrand-Rosenberg S. Tumor immunity: A balancing act between T cell activation, macrophage activation and tumor-induced immune suppression. Cancer Immunol Immunother. 2005;54:1137–1142. doi: 10.1007/s00262-005-0703-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills CD, Lenz LL, Harris RA. A breakthrough: Macrophage-directed cancer immunotherapy. Cancer Res. 2016;76:513–516. doi: 10.1158/0008-5472.CAN-15-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitra R, Singh S, Khar A. Antitumour immune responses. Expert Rev Mol Med. 2003;5:1–19. doi: 10.1017/S1462399403005623. [DOI] [PubMed] [Google Scholar]
- 30.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 31.Wahl LM, Kleinman HK. Tumor-associated macrophages as targets for cancer therapy. J Natl Cancer Inst. 1998;90:1583–1584. doi: 10.1093/jnci/90.21.1583. [DOI] [PubMed] [Google Scholar]
- 32.Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K, Koike M, Inadera H, Matsushima K. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res. 2000;6:3282–3289. [PubMed] [Google Scholar]
- 33.Leek RD, Harris AL. Tumor-associated macrophages in breast cancer. J Mammary Gland Biol Neoplasia. 2002;7:177–189. doi: 10.1023/A:1020304003704. [DOI] [PubMed] [Google Scholar]
- 34.Valković T, Dobrila F, Melato M, Sasso F, Rizzardi C, Jonjić N. Correlation between vascular endothelial growth factor, angiogenesis, and tumor-associated macrophages in invasive ductal breast carcinoma. Virchows Arch. 2002;440:583–588. doi: 10.1007/s004280100458. [DOI] [PubMed] [Google Scholar]
- 35.Deng R, Wang SM, Yin T, Ye TH, Shen GB, Li L, Zhao JY, Sang YX, Duan XG, Wei YQ. Inhibition of tumor growth and alteration of associated macrophage cell type by an HO-1 inhibitor in breast carcinoma-bearing mice. Oncol Res. 2013;20:473–482. doi: 10.3727/096504013X13715991125684. [DOI] [PubMed] [Google Scholar]