Abstract
Ovarian cancer is the seventh most common type of cancer and the eighth most common cause of cancer-associated mortality among women. A number of studies have hypothesized that the expression status of certain genes may be used to predict prognosis in ovarian cancer. In the present study, the RNA expression data from next-generation sequencing and the clinical information of 413 patients from The Cancer Genome Atlas dataset was downloaded to identify the association between gene-expression level and the survival time of the patients with ovarian serous cystadenocarcinoma. A five-gene model was predicted to be significantly associated with patient survival in ovarian serous cystadenocarcinoma by using random survival forests variable hunting algorithm and Cox analysis. A total of two genes, mesencephalic astrocyte-derived neurotrophic factor and dedicator of cytokinesis 11, of the predicted five genes demonstrated positive expression in the ovarian serous cystadenocarcinoma cancer tissues by polymerase chain reaction analysis. Kaplan-Meier and Receiver Operating Characteristic analysis confirmed that the model of the two genes exhibited high sensitivity and specificity to predict the prognostic survival of patients. In conclusion, the expression of the two genes in the two-gene model was associated with the prognostic outcomes of patients with ovarian serous cystadenocarcinoma; the model demonstrated potential as a novel prognostic indicator, which may have important clinical significance.
Keywords: ovarian serous cystadenocarcinoma, gene expression, survival, random survival forests variable hunting algorithm, biomarker
Introduction
Ovarian cancer is the seventh most common type of cancer and the eighth most common cause of cancer-associated mortality among females until 2012 (1). During 2012, ~238,700 female ovarian cancer cases were diagnosed, and globally 151,900 females succumbed to this disease (1). The majority of the female patients who developed ovarian cancer were not aware of the condition, or received diagnoses until an advanced stage, which were primary causes of recurrence and early mortality (2,3). Despite advances in imaging diagnosis, preoperative and postoperative care, and chemotherapy delivery, there has been little improvement in 5-year overall survival (4–6).
Gene expression assays have been introduced in daily clinical treatments for the care of patients with numerous conditions, for example, patients with newly diagnosed breast cancer (7). The Oncotype DX assay (Genomic Health, Inc., Redwood City, CA, USA) is a 21-gene assay that is designed to quantify risk of distant recurrence at 10 years for a group of women with early stage breast cancer. The assay includes genes associated with cell proliferation (Ki-67, STK15, survivin, cyclin B1, MYBL2), invasion (stromelysin 3, cathepsin L2), HER2, estrogen (ER, PR, Bcl2, SCUBE2), in addition to GSTM1, CD68, BAG1, and several reference genes (β-actin, GAPDH, RPLP0, GUS, and TFRC) (7). Zhan et al (8) identified a five-gene (cytoskeleton associated protein 4, Solute carrier family 40 member 1, otoferlin, mannosidase-α class 2A member 2, isoprenoid synthase domain containing) panel that was significantly associated with patient survival in those with renal clear cell carcinoma from The Cancer Genome Atlas (TCGA) database. Using a publicly available microarray database, an inverse association between elevated SHANK-associated RH domain interactor gene expression with reduced patient survival in PR+ or ER+ breast cancer was identified by De Melo and Tang (9).
In the study of ovarian cancer, gene expression profiling has been utilized extensively. Previous studies have focused on differential gene expression between the tissue of normal and tumors (10), characterizing between histologic subtypes (11,12) and marking differences between invasive and tumors with low malignancy potential (13,14). Several studies have attempted to target the gene expression signatures that correlate with clinical data, to identify genes that are determinative of survival, to generate predictive biomarkers (15,16). The present study aimed to identify genes that were associated with the overall survival time of patients with ovarian cancer by analyzing high-throughput RNA sequencing data downloaded from TCGA using the random survival forests variable hunting (RSFVH) algorithm (17). Multiple genes were selected to predict the survival time of patients following fitting, and then used to verify the expression of the predicted genes in fresh ovarian cancer tissue by using polymerase chain reaction (PCR) analysis, and evaluate the prognostic value, sensitivity and specificity of the model.
Materials and methods
Materials and kits
A total of two ovarian serous cystadenocarcinoma fresh tissues were obtained during surgery from patients undergoing surgical treatment in Hubei Maternal and Child Health Hospital (Wuhan, China). Patients provided written informed consent.
Reverse transcription PCR (RT-PCR)
For RT-PCR experiments, tissue RNA was extracted using TRIzol (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. cDNA was synthesized using random hexamers (Takara Biotechnology Co., Ltd., Dalian, China). Briefly, and synthesized according to the following conditions: 96°C for 5 min; 96°C for 20 sec; 55°C for 30 sec and 72°C for 1 min for 30 cycles. RT-PCR was performed with the RT-PCR kit KOD HOT Start polymerase chain Reaction (EMD Millipore, Billerica, MA, USA). The PCR cycling conditions were as follows: 95°C, for 5 sec, 95°C for 15 sec, 50°C for 5 sec and 60°C for 90 sec, for 35 cycles. With the use of ethidium bromide, amplified products were visualized on 1.5% agarose gels. Finally, a UV-IV UV analyzer instrument (Beyotime Institute of Biotechnology, Haimen, China) was used to capture images. The following primer pairs were used: Forward primer, 5′-ACCTTGGTCTGCGTTTG-3′; and reverse 5′-GCACATCTGGGTCTTGG-3′ for clathrin heavy chain-like 1 (CLTCL1); forward, 5′-ATCCTGGAGGCTGTGCT-3′; and reverse, 5′-CTGAACGCTGGAACTGG-3′ for calcium/calmodulin dependent protein kinase 11α (CAMK2A); forward 5′-ACCAGGACCTCAAAGACAGA-3′ and reverse, 5′-GGGCATATTTAGGCATCAGT-3′ for mesencephalic astrocyte-derived neurotrophic factor (MANF); forward primer, 5′-GAGCAGGAAATGGAGGA-3′; and reverse, 5′-TGGTTGTGATGCGAGAC-3′ for dedicator of cytokinesis 11 (DOCK11); forward, 5′-CAGCAGCCTCGGCAGTA-3′; and reverse, 5′-CCGCAGGGTTTCTTTCAT-3′ for dehydrogenase/reductase 4-like 1 (DHRS4L1).
Ovarian cancer gene expression data from TCGA
The mRNA level 3 expression data of 413 patients with ovarian cancer were downloaded from the TCGA database via the data portal (https://cancergenome.nih.gov; accessed 23rd July 2016), including 22,547 human genes and the corresponding clinical data. A total of 3 patients with missing data were excluded. Next, the 410 ovarian cancer samples were randomly divided into a training set (n=204) and a testing set (n=206). The training set was used to identify gene expression signature, and the testing set was used for validation.
Statistical analysis
A univariate Cox regression analysis was used to evaluate the association between the expression level of genes and patient OS. Next, based on the corresponding result, a risk score formula was built to calculate the risk score for each patient. Risk score (RS)=∑Ni=1(explg × coef), where N is the number of genes, explg is the expression value of genes and ‘coef’ is the estimated regression coefficient of genes in the univariate Cox regression analysis. Considering that a model with a smaller number of genes would be more practical, genes that were significantly associated with patient survival were identified using the RSFVH algorithm (P<0.001). Kaplan-Meier and Cox proportional-hazard regression analyses were performed for two genes, with the expectation of identifying an improved model for predicting survival. The cut-off values for the two genes were computed with X-tile (18). The survival differences between the low- and high-risk groups were evaluated, and the sensitivity and specificity of the model in the survival prediction was also compared using receiver operating characteristic (ROC) analysis. All analyses were performed using R program (http://www.r=project.org) including packages named survival ROC. Survival and random Forest SRC was downloaded from Bio-conductor.
Results
Patient characteristics
All 410 patients involved in the present study were clinically and pathologically diagnosed with ovarian serous cystadenocarcinoma, and the data were downloaded from TCGA database. The mean age of these 410 patients was 60 years (range, 30–87). Using the International Federation of Gynecology and Obstetrics classification (19), clinical stages of the tumor were classified into stages I–IV. In the present study, there were 0 patients with stage I, 22 patients with stage II, 326 patients with stage III, and 62 patients with stage IV disease. The 3 patients lacking clinical staging data were not included in any analysis. All other patient information is summarized in Table I. A total of 2 patients whose tissues, obtained from surgical resection, were used were clinically and were pathologically diagnosed with ovarian serous cystadenocarcinoma. Their ages were 44 and 53 respectively, the clinical stages of tumor were stage III, and they were labeled Patients 1 and 2.
Table I.
Summary of patient demographics and clinical characteristics.
| Characteristics | Training set | Testing set | Total |
|---|---|---|---|
| Age | |||
| Median, years | 60 | 60 | 60 |
| Range, years | 30–87 | 30–87 | 30–87 |
| Clinical stage | |||
| Stage I, n | 0 | 0 | 0 |
| Stage II, n | 11 | 11 | 22 |
| Stage III, n | 163 | 163 | 326 |
| Stage IV, n | 30 | 32 | 62 |
| Patient status | |||
| Alive, n | 84 | 95 | 179 |
| Deceased, n | 120 | 111 | 231 |
Detection of genes associated with overall survival time of patients with ovarian cancer in the training set by RT-PCR
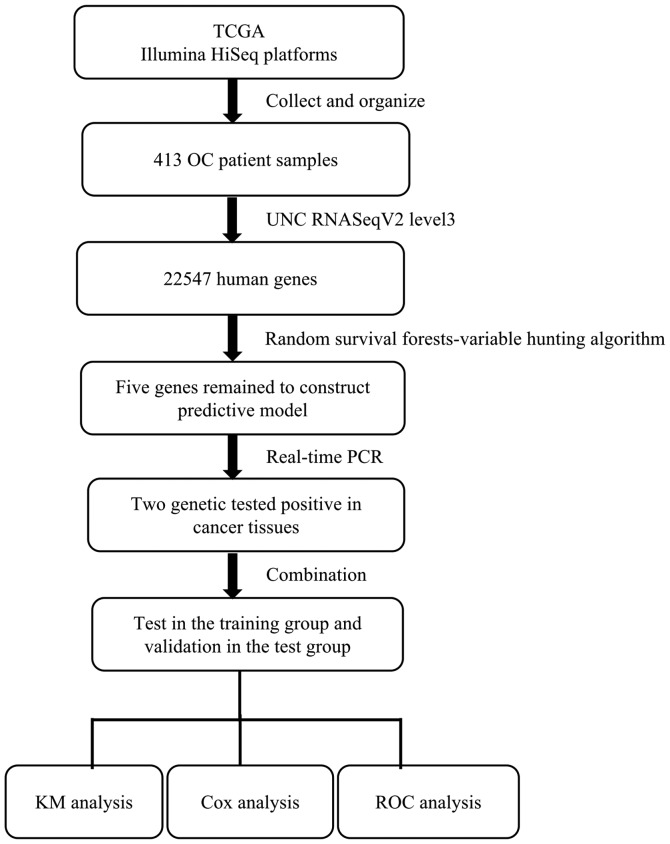
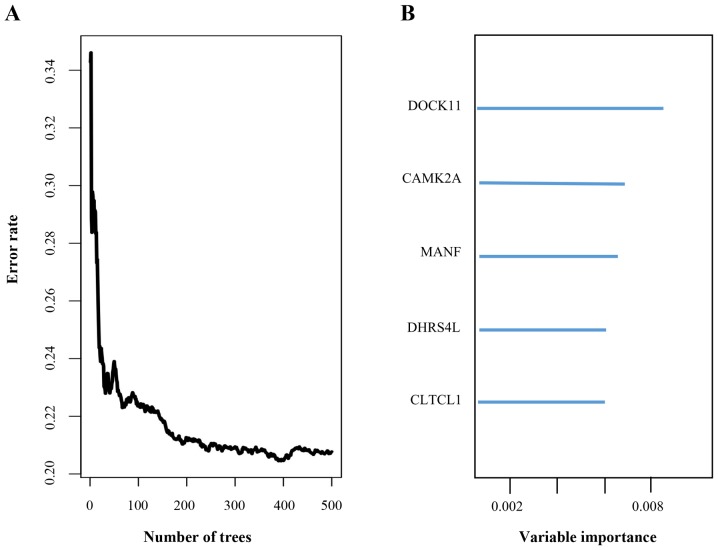
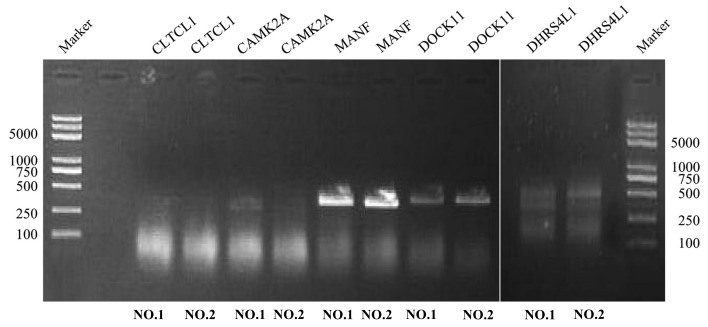
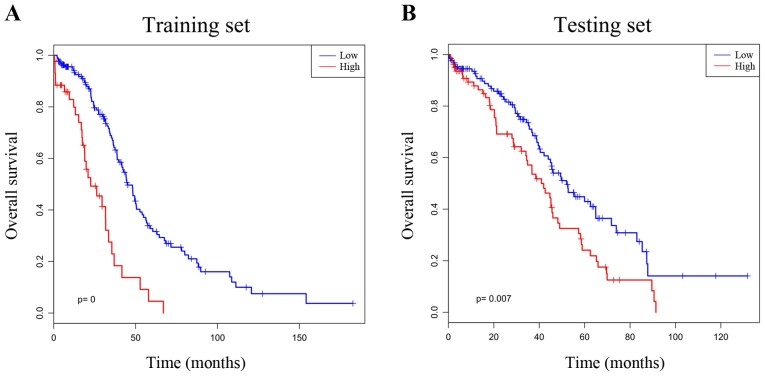
To identify the genes potentially associated with overall survival time in patients with ovarian serous cystadenocarcinoma, a total of 22,547 genes were identified by random survival forests analysis. The order of analyses to develop the risk score model and validate the efficiency of the signature to predict prognostic outcomes is demonstrated in Fig. 1. A univariate Cox proportional hazards regression analysis of the genes expression profiling data with survival time, and survival status as the dependent variable was conducted. Using a random forest supervised classification algorithm, a total of 5 genes (MANF, DOCK11, CLTCL1, CAMK2A, DHRS4L1) with the highest association with the prognostic classification were selected according to the permutation importance scores for verification with PCR in 2 fresh ovarian serous cystadenocarcinoma tissues (Fig. 2; Table II). Within the selection of the five genes and subsequent RT-PCR analysis, despite a number of changes in the amplification conditions and primers referring to the optimization of the RT-PCR step, only two genes exhibited positive expression (Fig. 3). The information concerning these two genes is summarized in Table III. Following this comparison, the optimum model including these two genes was determined. The risk score formula for this model was (−0.53179 × expression value of MANF) + (0.324759 × expression value of DOCK11). Using X-tile to determine the cut-off values, the values of the training and testing sets was −2.60 and −2.86, respectively (18). These values were included in the low group. Survival analysis was performed by using the Kaplan-Meier method with a log-rank statistical test between the high-risk group and low-risk group. As demonstrated in Fig. 4A, Kaplan-Meier curves indicated that patients in the high-risk group exhibited significantly (P<0.001) poorer prognosis than those in the low-risk group.
Figure 1.
Flow chart of the protocols performed in the present study. TCGA, The Cancer Genome Atlas; OC, ovarian cancer; PCR, polymerase chain reaction; KM, Kaplan-Meier; ROC, receiver operating characteristic.
Figure 2.
(A) Random survival forests-variable hunting analysis reveals the error rate for the data as a function of trees. (B) The importance values for the 5 predictive genes. MANF, mesencephalic astrocyte-derived neurotrophic factor; DOCK11, dedicator of cytokinesis 11; CLTCL1, clathrin heavy chain-like 1; CAMK2A, calcium/calmodulin dependent protein kinase 11α; DHRS4L1, dehydrogenase/reductase 4-like 1.
Table II.
Five genes significantly associated with the survival time of patients in the training set.
| Gene name | Coefficient | HR | P-value |
|---|---|---|---|
| MANF | −0.53179 | 0.587553 | 0.002083 |
| DOCK11 | 0.324759 | 1.383697 | 0.022425 |
| CLTCL1 | 0.550443 | 1.734021 | 0.029124 |
| CAMK2A | 4.112609 | 61.10596 | 0.000217 |
| DHRS4L1 | −0.81199 | 0.443976 | 0.040793 |
HR, hazard ratio; MANF, mesencephalic astrocyte-derived neurotrophic factor; DOCK11, dedicator of cytokinesis 11; CLTCL1, clathrin heavy chain-like 1; CAMK2A, calcium/calmodulin dependent protein kinase 11α; DHRS4L1, dehydrogenase/reductase 4-like 1.
Figure 3.
Reverse transcription PCR. The expression of five genes was assessed in two cases of ovarian serous cystadenocarcinoma. The molecular length of the five genes (CLTCL1, CAMK2A, MANF, DOCK11, DHRS4L1) was 1,146, 657, 396, 1,349 and 710 bp, respectively. Experiments were repeated in triplicate. MANF, mesencephalic astrocyte-derived neurotrophic factor; DOCK11, dedicator of cytokinesis 11; CLTCL1, clathrin heavy chain-like 1; CAMK2A, calcium/calmodulin dependent protein kinase 11α; DHRS4L1, dehydrogenase/reductase 4-like 1.
Table III.
Analysis of the function of the two-gene model.
| Gene name | Chromosomal position | Start site | End site | Function |
|---|---|---|---|---|
| MANF | chr3 | 50674969 | 51536662 | Inhibits cell proliferation and ER stress-induced cell death |
| DOCK11 | chrX | 118146063 | 118826973 | GEF that activates CDC42 by exchanging bound GDP for free GTP |
ER, endoplasmic reticulum; GEF, guanine nucleotide-exchange factor; GDP, guanosine diphosphate; GTP, guanosine triphosphate; CDC42, cell division control protein Cdc42 homolog; chr, chromosome; MANF, mesencephalic astrocyte-derived neurotrophic factor; DOCK11, dedicator of cytokinesis 11.
Figure 4.
Kaplan-Meier curves with two-sided log-rank test demonstrate association between the score resulting from the two-gene model and patient survival. Using X-tile to compute a cut-off value, patients were divided into high- and low-risk groups. (A) Kaplan-Meier curves for patients in the training set (n=206). (B) Kaplan-Meier curves for patients in the testing set (n=207). The survival differences between the high- and low-risk groups were determined by two-sided long-rank tests.
Verification of survival-associated genes in the testing set
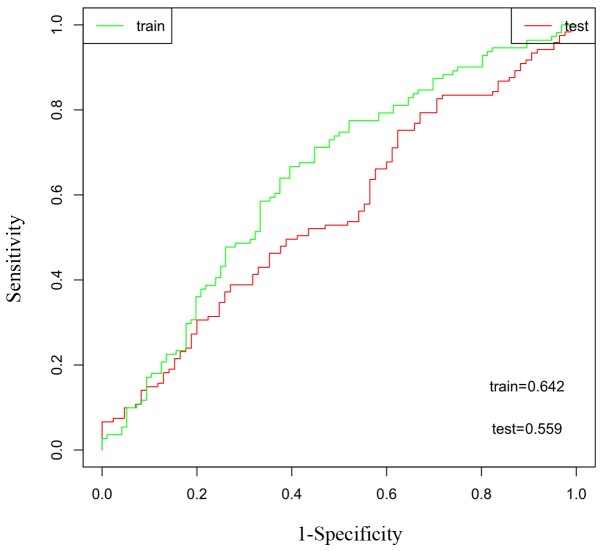
To determine the prognostic potential of the two-gene signature, Kaplan-Meier survival analysis was performed on the testing set. There was a statistically significant difference (P=0.007) between the high- and low-risk groups, which was in agreement with that in the training test, revealing that this two-gene signature may serve a role in predicting the survival of ovarian cancer patients (Fig. 4B). To confirm the clinical performance of the two-gene model as a biomarker for predicting prognosis further, ROC analysis was used to evaluate the validity of the gene signature on patient survival. The area under the ROC curve (AUC) of the training and testing sets were 0.642 and 0.559, respectively, demonstrating that the two-gene model exhibited high sensitivity and specificity, and could be used as a biomarker to predict the prognostic survival of patients (Fig. 5).
Figure 5.
Receiver operating characteristic analysis of the two-gene model. The area under the curve of the training and testing sets was 0.642 and 0.559, respectively, indicating that the two-gene model exhibited sensitivity and specificity in predicting the survival time of patients with ovarian serous cystadenocarcinoma.
To examine whether the two-gene model distinguished between the high- and low-risk patients, Cox regression analyses were performed in the training and testing sets. The results confirmed that the two-gene model was an independent prognostic factor for the prognosis of ovarian cancer in the training and the test sets (Table IV).
Table IV.
Two-gene model of Cox regression in training and testing sets.
| Sample sets | Parameter estimate | Standard error | P-value | HR | 95% CI |
|---|---|---|---|---|---|
| Training set | 0.64009 | 0.21858 | 0.0034 | 1.897 | 1.236–2.911 |
| Testing set | 0.59023 | 0.25254 | 0.0194 | 1.804 | 1.100–2.960 |
HR, hazard ratio; CI confidence interval.
Discussion
Ovarian cancer (OC) is one of the leading causes of cancer mortality in gynecological oncology, exhibiting a 5-year survival rate of 44% (20). The serous ovarian cancer high-grade subtype is one of the most aggressive and metastatic forms of ovarian cancer (21). In the present study, a five-gene signature that was significantly associated with patient survival in ovarian serous cystadenocarcinoma was predicted, based on genome-wide RNA profiling of 413 ovarian cancer patients from the TCGA database using the RSFVH algorithm and Cox analysis. Using PCR, 2 genes (MANF and DOCK11) were verified to exhibit positive expression in ovarian serous cystadenocarcinoma tissues. Subsequently, it was confirmed that the two-gene model was an independent prognostic predictor of survival using Cox regression analysis on the training and testing sets.
MANF has been discussed in previous studies as a survival-promoting factor for embryonic midbrain dopaminergic neurons in vitro (22). In HeLa cells, MANF is localized in the endoplasmic reticulum, which is expressed at particularly high levels in secretory tissues with extensive protein production (23). Notably, prior studies have indicated that MANF is important for protein homeostasis in the endoplasmic reticulum, as the knockdown of MANF in cultured cells and the knockout of MANF in mice and Drosophila resulted in the activation of the unfolded protein response, a signaling pathway induced by endoplasmic reticulum stress (23–25). Evidence indicates that the endoplasmic reticulum is involved in apoptotic signaling pathways (26), and that it participates in the occurrence and development of many types of cancer, including cervical cancer (27), hepatocellular carcinoma (28) and head and neck squamous cell carcinoma (29). In addition, MANF may reduce the inflammatory response and prevent proliferation of inflamed cells by inhibiting DNA binding of the transcription factor p65 subunit, consequently suppressing the inflammatory pathways induced by nuclear factor-κ light-chain-enhancer of activated B cells binding to its target genes (30).
The other positive gene identified in the present study, DOCK11, is a gene that belongs to the dedicator of cytokinesis (Dock) protein family, a class of guanine nucleotide exchange factors (GEFs) that activate the Rho GTPases, and one of the three members of the Dock-D subfamily. Dock proteins are large proteins, which constitute a major class, together with the Dbl-homology proteins, of Rho GEFs (31,32). Membrane receptors promote the reorganization of the actin cytoskeleton downstream of the Rho GTPases by their GEFs to regulate cell adhesion and migration (33). There are two classes of exchange factors that are associated with GTPases: The classical Dbl-associated exchange factors and, the more recently identified atypical Dock-family exchange factors. The Dock family of exchange factors was identified only 12 years ago as a novel class of Rho GTPase activators, particularly Ras-related C3 botulinum toxin substrates 1, 2 and 3, and Cdc42 (34,35). In mammals, there are 11 Dock genes, which are grouped into 4 subfamilies: A, B, C and D (35). The D subfamily, characterized by an N-terminal pleckstrin homology domain, is made up of 3 members, Dock9, Dock10, and Dock11 (32,36,37). Dock11 mediates a positive feedback activation of cell division control protein Cdc42 homolog (Cdc42), as active Cdc42 may in turn bind to Dock11 and enhance its GEF activity (37). Sakabe et al (38) revealed that Dock11 recruitment downstream of Fc γ-receptor III and TLR4 activated Cdc42 to promote cell migration. As TLR4 has been demonstrated to promote the epithelial-mesenchymal transition and cancer cell migration (39–41), we hypothesized that Dock11 activity is associated with cancer-induced pathological cell migration. In support of this hypothesis, Dock11 was detected among the top-20 highest expressed genes in testicular carcinoma (42).
Expression of MANF and DOCK11 in human tissues has not been extensively studied, including in ovarian tissues. The Human Protein Atlas (www.proteinatlas.org) database (43) was used to identify that MANF was not expressed in the follicle cells and was expressed at low levels in the stroma cells of normal ovarian tissues; DOCK11 was not detected in the stromal cells and detected at low levels in the follicle cells of the normal ovarian tissue. MANF exhibited high, medium and low expression, and DOCK11 presented a medium, low, no expression in different clinical staging tissue of ovarian serous cystadenocarcinoma. In the present study, five genes associated with ovarian cancer survival were predicted through analysis of TCGA database, and the positive expression of two genes (MANF and DOCK11) was validated in ovarian cancer. In subsequent experiments, the expression of MANF and DOCK11 at the protein level in ovarian serous cystadenocarcinoma and adjacent normal tissues should be verified, and the clinical relevance should be identified to determine the use of these genes as novel biomarkers to predict the treatment outcomes of patients with ovarian cancer.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Holschneider CH, Berek JS. Ovarian cancer: Epidemiology, biology, and prognostic factors. Semin Surg Oncol. 2000;19:3–10. doi: 10.1002/1098-2388(200007/08)19:1<3::AID-SSU2>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 3.Klint A, Tryggvadóttir L, Bray F, Gislum M, Hakulinen T, Storm HH, Engholm G. Trends in the survival of patients diagnosed with cancer in female genital organs in the Nordic countries 1964–2003 followed up to the end of 2006. Acta Oncol. 2010;49:632–643. doi: 10.3109/02841861003739330. [DOI] [PubMed] [Google Scholar]
- 4.Markman M, Bundy BN, Alberts DS, Fowler JM, Clark-Pearson DL, Carson LF, Wadler S, Sickel J. Phase III trial of standard-dose intravenous cisplatin plus paclitaxel versus moderately high-dose carboplatin followed by intravenous paclitaxel and intraperitoneal cisplatin in small-volume stage III ovarian carcinoma: An intergroup study of the Gynecologic Oncology Group, Southwestern Oncology Group, and Eastern Cooperative Oncology Group. J Clin Oncol. 2001;19:1001–1007. doi: 10.1200/JCO.2001.19.4.1001. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, Copeland LJ, Walker JL, Burger RA. Gynecologic Oncology Group: Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 6.Coleman MP, Forman D, Bryant H, Butler J, Rachet B, Maringe C, Nur U, Tracey E, Coory M, Hatcher J, et al. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the UK, 1995–2007 (the International Cancer Benchmarking Partnership): An analysis of population-based cancer registry data. Lancet. 2011;377:127–138. doi: 10.1016/S0140-6736(10)62231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker J. Genomic Health, Inc. Pharmacoqenomics. 2007;8:397–399. doi: 10.2217/14622416.8.4.397. [DOI] [PubMed] [Google Scholar]
- 8.Zhan Y, Guo W, Zhang Y, Wang Q, Xu XJ, Zhu L. A five-gene signature predicts prognosis in patients with kidney renal clear cell carcinoma. Comput Math Methods Med. 2015;2015:842784. doi: 10.1155/2015/842784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Melo J, Tang D. Elevation of SIPL1 (SHARPIN) increases breast cancer risk. PLoS One. 2015;10:e0127546. doi: 10.1371/journal.pone.0127546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, Misek DE, Wu R, Zhai Y, Darrah DM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–4729. [PubMed] [Google Scholar]
- 12.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonome T, Lee JY, Park DC, Radonovich M, Pise-Masison C, Brady J, Gardner GJ, Hao K, Wong WH, Barrett JC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 14.Gilks CB, Vanderhyden BC, Zhu S, van de Rijn M, Longacre TA. Distinction between serous tumors of low malignant potential and serous carcinomas based on global mRNA expression profiling. Gynecol Oncol. 2005;96:684–694. doi: 10.1016/j.ygyno.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 15.Mok SC, Chao J, Skates S, Wong K, Yiu GK, Muto MG, Berkowitz RS, Cramer DW. Prostasin, a potential serum marker for ovarian cancer: Indentification through microarray technology. J Natl Cancer Inst. 2001;93:1458–1464. doi: 10.1093/jnci/93.19.1458. [DOI] [PubMed] [Google Scholar]
- 16.Spentzos D, Levine DA, Kolia S, Otu H, Boyd J, Libermann TA, Cannistra SA. Unique gene expression profile based on pathologic response in epithelial ovarian cancer. J Clin Oncol. 2005;23:7911–7918. doi: 10.1200/JCO.2005.02.9363. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen HN, Averette HE, Hoskins W, Sevin BU, Penalver M, Steren A. National survey of ovarian carcinoma. VI. Critical assessment of current International Federation of Gynecology and Obstetrics staging system. Cancer. 1993;72:3007–3011. doi: 10.1002/1097-0142(19931115)72:10<3007::AID-CNCR2820721024>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Baldwin LA, Huang B, Miller RW, Tucker T, Goodrich ST, Podzielinski I, DeSimone CP, Ueland FR, van Nagell JR, Seamon LG. Ten-year relative survival for epithelial ovarian cancer. Obstet Gynecol. 2012;120:612–618. doi: 10.1097/AOG.0b013e318264f794. [DOI] [PubMed] [Google Scholar]
- 21.McCluggage WG. Morphological subtypes of ovarian carcinoma: A review with emphasis on new developments and pathogenesis. Pathology. 2011;43:420–432. doi: 10.1097/PAT.0b013e328348a6e7. [DOI] [PubMed] [Google Scholar]
- 22.Petrova P, Raibekas A, Pevsner J, Vigo N, Anafi M, Moore MK, Peaire AE, Shridhar V, Smith DI, Kelly J, et al. MANF: A new mesencephalic, astrocyte-derived neurotrophic factor with selectivity for dopaminergic neurons. J Mol Neurosci. 2003;20:173–188. doi: 10.1385/JMN:20:2:173. [DOI] [PubMed] [Google Scholar]
- 23.Apostolou A, Shen Y, Liang Y, Luo J, Fang S. Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Exp Cell Res. 2008;314:2454–2467. doi: 10.1016/j.yexcr.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindahl M, Danilova T, Palm E, Lindholm P, Võikar V, Hakonen E, Ustinov J, Andressoo JO, Harvey BK, Otonkoski T, et al. MANF is indispensable for the proliferation and survival of pancreatic β cells. Cell Rep. 2014;7:366–375. doi: 10.1016/j.celrep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palgi M, Greco D, Lindström R, Auvinen P, Heino TI. Gene expression analysis of Drosophilaa Manf mutants reveals perturbations in membrane traffic and major metabolic changes. BMC Genomics. 2012;13:134. doi: 10.1186/1471-2164-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang YM, Yang Y, Dai WW, Li XM, Ma JQ, Tang LP. Genistein-induced apoptosis is mediated by endoplasmic reticulum stress in cervical cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:3292–3296. [PubMed] [Google Scholar]
- 28.Yeh TC, Chiang PC, Li TK, Hsu JL, Lin CJ, Wang SW, Peng CY, Guh JH. Genistein induces apoptosis in human hepatocellular carcinomas via interaction of endoplasmic reticulum stress and mitochondrial insult. Biochem Pharmacol. 2007;73:782–792. doi: 10.1016/j.bcp.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 29.El Jamal SM, Taylor EB, Elmageed Abd ZY, Alamodi AA, Selimovic D, Alkhateeb A, Hannig M, Hassan SY, Santourlidis S, Friedlander PL, et al. Interferon gamma-induced apoptosis of head and neck squamous cell carcinoma is connected to indoleamine-2,3-dioxygenase via mitochondrial and ER stress-associated pathways. Cell Division. 2016;11:11. doi: 10.1186/s13008-016-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Feng L, Wang X, Du J, Chen Y, Yang W, Zhou C, Cheng L, Shen Y, Fang S, et al. Mesencephalic astrocyte-derived neurotrophic factor is involved in inflammation by negatively regulating the NF-κB pathway. Sci Rep. 2015;5:8133. doi: 10.1038/srep08133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossman KL, Der CJ, Sondek J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 32.Meller N, Irani-Tehrani M, Kiosses WB, Del Pozo MA, Schwartz MA. Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat Cell Biol. 2002;4:639–647. doi: 10.1038/ncb835. [DOI] [PubMed] [Google Scholar]
- 33.Gadea G, Blangy A. Dock-family exchange factors in cell migration and disease. Eur J Cell Biol. 2014;93:466–477. doi: 10.1016/j.ejcb.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: Regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Côté JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J Cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- 36.Nishikimi A, Meller N, Uekawa N, Isobe K, Schwartz MA, Maruyama M. Zizimin2: A novel, DOCK180-related Cdc42 guanine nucleotide exchange factor expressed predominantly in lymphocytes. FEBS Lett. 2005;579:1039–1046. doi: 10.1016/j.febslet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Lin Q, Yang W, Baird D, Feng Q, Cerione RA. Identification of a DOCK180-related guanine nucleotide exchange factor that is capable of mediating a positive feedback activation of Cdc42. J Biol Chem. 2006;281:35253–35262. doi: 10.1074/jbc.M606248200. [DOI] [PubMed] [Google Scholar]
- 38.Sakabe I, Asai A, Iijima J, Maruyama M. Age-related guanine nucleotide exchange factor, mouse Zizimin2, induces filopodia in bone marrow-derived dendritic cells. Immun Ageing. 2012;9:2. doi: 10.1186/1742-4933-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jing YY, Han ZP, Sun K, Zhang SS, Hou J, Liu Y, Li R, Gao L, Zhao X, Zhao QD, et al. Toll-like receptor 4 signaling promotes epithelial-mesenchymal transition in human hepatocellular carcinoma induced by lipopolysaccharide. BMC Med. 2012;10:98. doi: 10.1186/1741-7015-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao SJ, Zhou YH, Yuan Y, Li D, Wu FH, Wang Q, Zhu JH, Yan B, Wei JJ, Zhang GM, Feng ZH. Triggering of Toll-like receptor 4 on metastatic breast cancer cells promotes αvβ3-mediated adhesion and invasive migration. Breast Cancer Res Treat. 2012;133:853–863. doi: 10.1007/s10549-011-1844-0. [DOI] [PubMed] [Google Scholar]
- 41.Rakhesh M, Cate M, Vijay R, Shrikant A, Shanjana A. A TLR4-interacting peptide inhibits lipopolysaccharide-stimulated inflammatory responses, migration and invasion of colon cancer SW480 cells. Oncoimmunology. 2012;1:1495–1506. doi: 10.4161/onci.22089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almstrup K, Leffers H, Lothe RA, Skakkebaek NE, Sonne SB, Nielsen JE, Rajpert-De Meyts E, Skotheim RI. Improved gene expression signature of testicular carcinoma in situ. Int J Androl. 2007;30:292–302. doi: 10.1111/j.1365-2605.2007.00758.x. [DOI] [PubMed] [Google Scholar]
- 43.Pontén F, Jirström K, Uhlen M. The human protein atlas-a tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]