Abstract
Plants are able to discriminate and respond to structurally related chitooligosaccharide (CO) signals from pathogenic and symbiotic fungi. In model plants Arabidopsis thaliana and Oryza sativa LysM-receptor like kinases (LysM-RLK) AtCERK1 and OsCERK1 (chitin elicitor receptor kinase 1) were shown to be involved in response to CO signals. Based on phylogenetic analysis, the pea Pisum sativum L. LysM-RLK PsLYK9 was chosen as a possible candidate given its role on the CERK1-like receptor. The knockdown regulation of the PsLyk9 gene by RNA interference led to increased susceptibility to fungal pathogen Fusarium culmorum. Transcript levels of PsPAL2, PsPR10 defense-response genes were significantly reduced in PsLyk9 RNAi roots. PsLYK9’s involvement in recognizing short-chain COs as most numerous signals of arbuscular mycorrhizal (AM) fungi, was also evaluated. In transgenic roots with PsLyk9 knockdown treated with short-chain CO5, downregulation of AM symbiosis marker genes (PsDELLA3, PsNSP2, PsDWARF27) was observed. These results clearly indicate that PsLYK9 appears to be involved in the perception of COs and subsequent signal transduction in pea roots. It allows us to conclude that PsLYK9 is the most likely CERK1-like receptor in pea to be involved in the control of plant immunity and AM symbiosis formation.
Keywords: pea Pisum sativum L., Lysin-motif receptor-like kinase PsLYK9, AM symbiosis, phytopathogenic fungi, gene expression, knockdown regulation, Agrobacterium rhizogenes
1. Introduction
The ability of plants to distinguish between pathogenic and beneficial microorganisms determines their ability to successfully exist in soil. Plants possess a well-developed system to recognize certain surface components and the compounds of microorganisms released into the medium, which are called microbe-associated molecular patterns (MAMPs); these are perceived by plant pattern recognition receptors [1,2,3]. The recognition of a microorganism as a pathogen during early interactions triggers activation of the immune responses that prevent microbial penetration into plant cells. Plants are also able to develop symbiotic relationships with beneficial microorganisms like arbuscular mycorrhizal (AM) fungi and nitrogen-fixing bacteria belonging to the order Rhizobiales, known as rhizobia [4,5]. The development of symbiotic interactions is based on the ability of microorganisms to avoid the activation of the plant’s immune system or to suppress it for intracellular infection progression. To establish different types of plant–microbial relationships, microorganisms enter into a molecular dialog with plants.
Chitin and its derivatives, chitooligosaccharides (COs), with a high degree of polymerization, are typical MAMPs that are perceived by plants [6,7,8,9,10]. Long-chain COs produced by pathogenic fungi and invertebrates are detected as elicitors of defense reactions in plants that prevent microbial infection. In contrast, short-chain COs with degrees of polymerization 4 and 5 (CO4 and CO5) and lipochitooligosaccharides (LCOs), Myc factors and Nod factors, have been identified as the key signals of AM fungi and rhizobia, which enable entry of the microbes into plant root cells. Legume plants may be considered as unique objectives to study the pathogenic and mutualistic relationships of plants, as they are able to perceive all four types of chitooligosaccharidic molecular signals coming from microorganisms, but inducing various responses.
Receptors with LysM motifs in extracellular domains were shown to be required in the recognition of both bacterial and fungal LCOs and COs in plants [7,8,11,12,13,14,15,16,17,18,19,20,21,22]. In model plants Arabidopsis thaliana and Oryza sativa, Lysin-motif receptor-like kinases (LysM-RLKs) AtCERK1 and OsCERK1 (chitin elicitor receptor kinase 1) were shown to be essential for the recognition of chitin and long-chain COs with degree of polymerization 8 (CO8). It was found that in A. thaliana and O. sativa, the receptors AtCERK1 and OsCERK1 form homooligomeric AtCERK1/AtCERK1 or heterooligomeric complexes with additional receptor proteins (like AtLYK5/AtCERK1 and OsCEBiP/OsCERK1), which are involved in the regulation of plant resistance to infection [9,17,19]. In addition, studies based on phenotypic analyses of the chitin receptor mutant (cerk1) in rice O. sativa that—in contrast to Arabidopsis—was able to form symbiosis with AM fungi, showed that OsCERK1 was also involved in the recognition of short-chain COs with degree of polymerization 4 and 5 (CO4–CO5) through complex formation with an unknown co-receptor [20,21]. It is known that AtCERK1 and OsCERK1 were also important for peptidoglycan recognition; this has a structure that is similar to that of chitin, and it comprises the main component of the bacterial cell wall. To recognize peptidoglycan, receptor proteins form complexes like AtCERK1/AtLYM1/AtLYM3 and OsCERK1/OsLYP4/OsLYP6 [17,23]. This formation establishes CERK1 as playing a key role in the recognition of signal molecules of both fungal and bacterial origin.
Based on the results of a series of studies, an evolutionary relationship between LysM-RLK CERK1 and LysM-RLKs, which are involved in Nod factor perception and symbiosis regulation between plants and rhizobial bacteria like NFR1 (Nod factor receptor 1) in Lotus japonicus and LYK3 (LysM-type receptor-like kinase 3) in Medicago truncatula, was suggested [15,21,24]. Evidence in support of this assumption primarily comes from experiments on the activation of symbiotic genes, not only by Nod factors, but also by chitin and CO8 in L. japonicus wild-type plants and mutants impaired in nfr1 gene [24]. Similar results were obtained during the analysis of transcriptomes of L. japonicus roots treated with CO8 [25]. It points toward the participation of receptor to chitin/long-chain COs in induction of symbiotic genes. At the same time, the treatment of nfr1 mutant plants with chitin or CO8 showed that their recognition related to specific receptor, and recognition occurred independently of Nod factor receptor. Consequently, there are specific receptors for chitin and oligomers of chitin, which remain unknown in legume plants.
To investigate the interplay between symbiotic and defense pathways in the legume plant pea P. sativum, we were interested in searching for receptors involved in controlling plant immunity and symbiosis development. In this study, we questioned whether the PsLyk9 gene regulates plant immunity and AM symbiosis development in pea plants, and whether it may also regulate the expression of the AM symbiosis-related markers and defense-response genes in roots treated with COs differing in degree of polymerization. We found that the PsLyk9 gene was upregulated in response to pathogenic fungi infection, and its enhanced expression was observed in its roots during mycorrhization. Data on PsLyk9 RNA interference (RNAi) suggest that the PsLyk9 gene may regulate plant defense resistance to phytopathogenic fungi and AM symbiosis development. This indicates that PsLYK9 may be a suitable candidate for a CERK1-like receptor in pea.
2. Results
2.1. Searching for a Candidate among Pea Lyk Genes
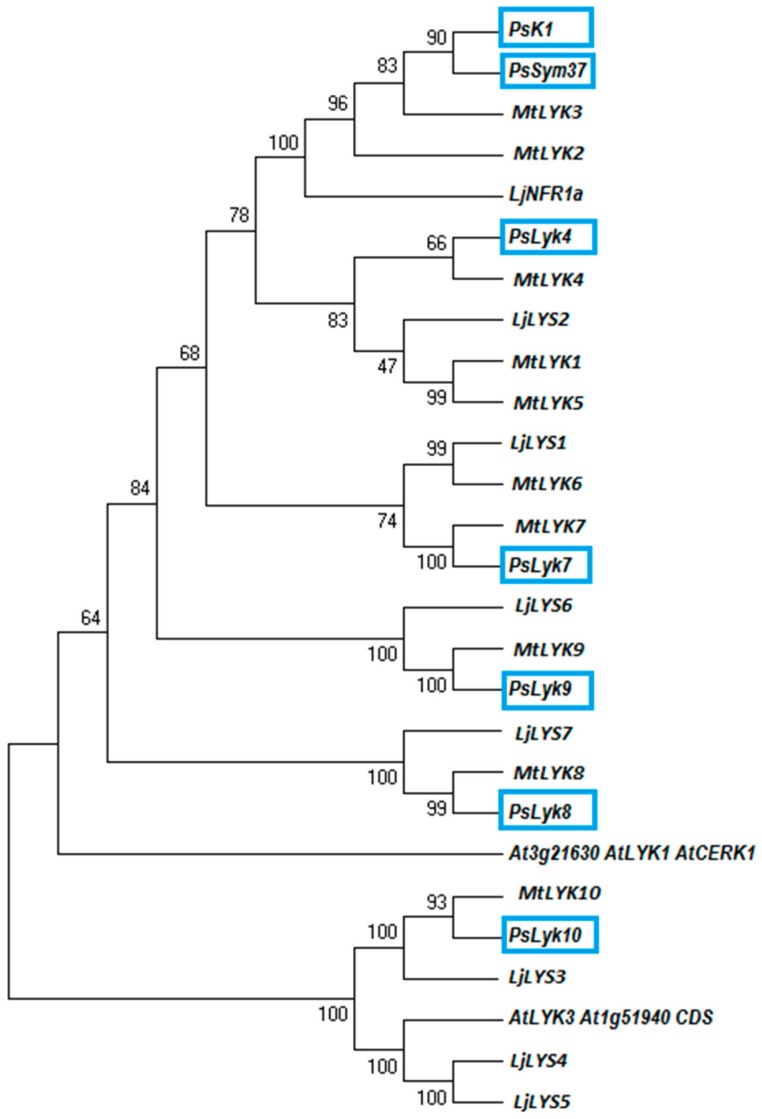
To identify CERK1-like receptor in pea, we searched for genes encoding LysM-RLKs in this legume; 26 and 19 LysM-RLKs genes have been found in model legumes like Medicago truncatula and Lotus japonicus [26,27,28], while only three P. sativum L. genes of this family have been annotated in databases to date—PsSym10 and PsSym37, PsK1 [11,29]. Previously, it was shown that the AtCERK1 in Arabidopsis has a high homology to genes in M. truncatula and L. japonicus, which belong to LysM-I phylogenetic groups including homologs of MtLYK3 (LysM-type receptor-like kinase 3) and LjNFR1 (Nod factor receptor 1) genes in these legumes [26,27,30]. OsCERK1 in rice also has a high homology with LjNFR1/MtLYK3 in M. truncatula and L. japonicus that belong to the same groups [21]. LysM-I phylogenetic groups include additional ten (MtLYK1–10) and seven (LjLYS1–7) LysM-RLK genes that demonstrate a high level of homology with MtLYK3 and LjNFR1, correspondently [26,28,30]. Representatives of LysM-I phylogenetic groups have from 10 to 12 exons and encode LysM receptors with potentially functional kinases (subfamily of LysM receptor like kinases, LYKs).
Based on the sequences of identified members of the M. truncatula and L. japonicus LysM-I phylogenetic groups, homologs were identified in the genome of P. sativum by searching in all available Transcriptome Shotgun Assemblies [31,32,33,34,35]. In silico searches for such LysM-RLKs resulted in identification of the full-length and partial transcripts for five additional predicted pea genes PsLyk4, PsLyk7, PsLyk8, PsLyk9 and PsLyk10 (Table S1; the names were given based on homology with M. truncatula LYK genes). Together with two other known PsSym37 and PsK1 genes (EU564096 and EU564088) from this phylogenetic group all these coding sequences were used for phylogenetic analysis (Figure 1). In addition, two LYK genes encoding receptor proteins with potentially functional kinases like AtLYK1 (AtCERK1) and AtLYK3 (At3g21630 and At1g51940) have also been used for analysis (Figure 1).
Figure 1.
Phylogenetic tree analysis showing the evolutionary relationships of the legume representatives of the LYK group of LysM-RLKs. Maximum-likelihood branch lengths were calculated using full length encoding sequences of LYK genes. The blue boxes represent pea genes. The branches show the bootstrap values. Ps, Pisum sativum; Lj, Lotus japonicus; Gm, Glycine max; Mt, Medicago truncatula; At, Arabidopsis thaliana.
Pea Lyk genes demonstrated high level of homology with M. truncatula and L. japonicus genes (Figure 1). Among these genes, PsSym37 and PsK1 showed the highest level of homology with MtLYK3 and MtLYK2 as well as with LjNFR1 encoding putative receptors to Nod factors in legumes [14,36]. PsLyk10 demonstrated the highest level of homology with MtLYK10 and LjLYS3 that encode the legume receptor to rhizobial exopolysaccharides [37]. Based on the phylogenetic analysis, AtCERK1 in Arabidopsis seems to show an evolutionary relationship with MtLYK1/PsLyk1–MtLYK9/PsLyk9 genes, like it was previously shown for kinase domains of these receptor proteins (Figure 1) [26].
Among these genes, the AtCERK1 (At3g21630) had the highest percent of identity 63.4% with PsLyk9, 61.9% with PsSym37, 61.4% with PsK1. Other pea Lyk genes showed a lower percent of homology with AtCERK1. Moreover, PsLyk9 encoded the receptor protein with YAQ sequence in kinase domain [24] that makes it suitable candidate for the role of receptor involved in symbiosis development. We hypothesized that the PsLyk9 gene may encode a CERK1-like receptor in pea.
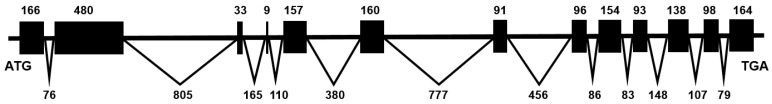
To verify the structure of the PsLyk9 gene in pea, we have cloned the full-length gene and its complementary DNA (cDNA) in cv. Finale using RACE analysis. Analysis showed that the PsLyk9 gene in pea has 11 introns (Figure 2). The full-length cDNA of PsLyk9 contained an open reading frame of 1845 nucleotides encoding a peptide of 614 amino acid residues with the predicated molecular mass of 67 kD.
Figure 2.
PsLyk9 gene structure. The exon (boxes) and intron (line) size in nucleotides are presented in cv. Finale background.
2.2. Analysis of PsLYK9 Participation in Response to Exogenously Applied Elicitors and Infection with Phytopathogenic Fungi Fusarium Culmorum
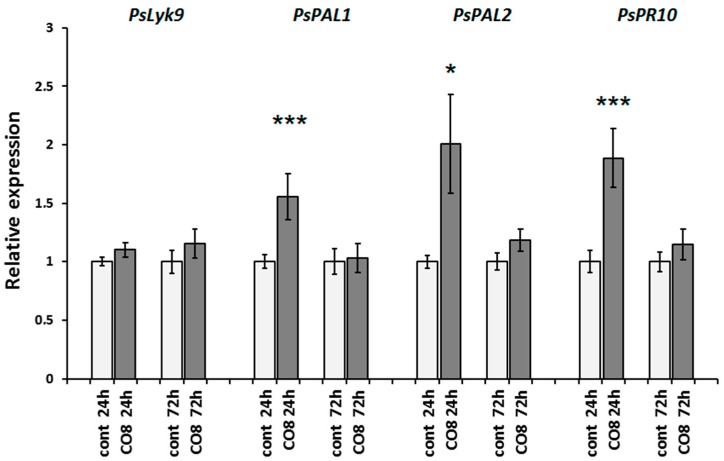
It was previously shown that pea plants can respond to chitosan oligomer treatment with a degree of polymerization of 7 or higher [38,39]. Indeed, our experiments revealed that pea plants treated with chitosan oligomers with a degree of polymerization of around 8 (CO8) respond best to the essential induction of PsPAL1 and PsPAL2 genes that encode various isoforms of the phenylalanine-ammonium-lyase (E.C.4.3.1.5) involved in phytoalexin synthesis in pea. It also induces the PsPR10 gene (pathogenesis-related 10), which is used as a defense-response marker to elicitor treatment (Figure 3). The most significant response was detected 24 h after treatment, while at 72 h, the effect was reduced. Treatment with CO8 did not result in the significant induction of the PsLyk9 gene, probably because the level of PsLyk9 expression in untreated roots was high (Figure 3).
Figure 3.
Transcript levels of the PsLyk9 gene encoding LysM-RLK and PsPAL1, PsPAL2, PsPR10 defense-response genes in pea roots treated with 10−6 M CO8 for 24 and 72 h. As a control, water-treated plants were used. The relative expression was normalized against the constitutively expressed ubiquitin and actin genes. Values are means ± SEM of three technical repeats. The graphs show the results of one biological repeat, representative for three biological independent experiments. Asterisks indicate statistically significant differences compared with the respective water-treated control: *** p < 0.001, * p < 0.05.
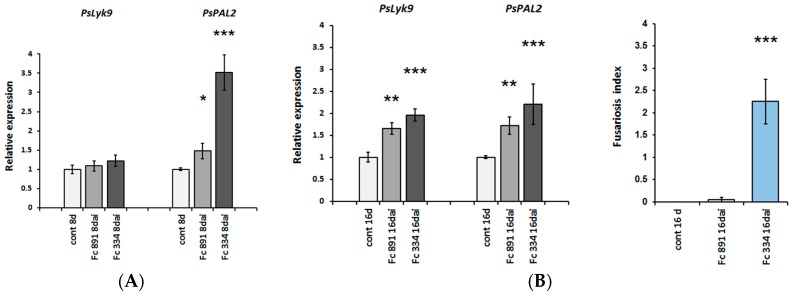
To verify the influence of PsLYK9 on plant interactions with phytopathogenic fungi, two F. culmorum (Wm. G. Sm.) Sacc. 334 and 891 strains, which differ in their ability to cause symptoms of disease in pea plants, were applied [40]. On day 16 after infection, when the visible signs of disease began to appear on pea plants, both F. culmorum strains induced the expression of PsLyk9 and PsPAL2 in plant roots (Figure 4). The effect was the most pronounced for the highly aggressive strain 334, probably due to higher pathogenic load. Since the PsLyk9 gene is induced in response to infection with both weakly and highly pathogenic fungal strains, it may indicate that the receptor kinase encoded by this gene participates in the development of non-specific immune response in plants toward infection with phytopathogenic fungi acting as a typical pattern recognition receptor. To further investigate it, the transcript level of the PsLyk9 gene can be evaluated at earlier stages of pea—F. culmorum interaction.
Figure 4.
PsLyk9, PsPAL2 gene expression in pea roots on days 8 (A) and 16 ((B), left) after infection with phytopathogenic fungi F. culmorum. Weakly pathogenic Fc 891 and highly pathogenic Fc 334 strains were used for infection. As a control, non-infected plants were used. Disease symptoms were evaluated in points on day 16 ((B), right), when the signs of disease began to appear on pea plants. No visible signs of disease were found on day 8 after infection. Asterisks indicate statistically significant differences compared with control non-infected plants: *** p < 0.001, ** p < 0.01, * p < 0.05.
2.3. Effect of PsLyk9 Gene Repression on the Transcription Level of Defense-Response Genes
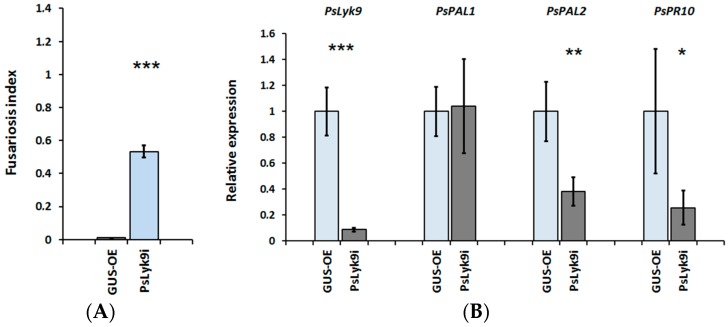
To analyze the PsLyk9 gene function in detail, an RNAi construct was generated (PsLyk9i) and introduced in pea plants, targeting PsLyk9 mRNA (Figure 5). In PsLyk9i transgenic roots, the expression of the PsLyk9 gene decreased significantly by 80% compared with control roots (GUS overexpressing control, GUS-OE). In transgenic roots with PsLyk9 knockdown, we expected to see an increase in their sensitivity to the weakly pathogenic fungus strain F. culmorum 891. Indeed, it was found that in PsLyk9i pea roots, suppression of PsLyk9 correlated with an increased sensitivity to the weakly pathogenic strain F. culmorum 891 in comparison with the control GUS-OE plants (Figure 5A). Signs of Fusarium damage were observed in all examined roots, while no evidence of disease was detected in control plants. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analyses of RNA extracted from individual transgenic roots (n = 7–10) demonstrated that roots carrying the PsLyk9-RNAi construct displayed reduced transcription levels in PsPAL2 and PsPR10 gene expression (Figure 5B).
Figure 5.
Effect of PsLyk9 RNAi on the resistance of pea plants to the weakly pathogenic fungus F. culmorum 891 and the transcript levels of defense-response genes. (A) Disease level in PsLyk9-RNAi and GUS-OE transgenic pea plants; (B) The expression levels of the PsPAL1 and PsPAL2, PsPR10 defense-response genes in transgenic pea roots with PsLyk9 knockdown; GUS-OE—transgenic pea plants transformed with pB7WG2D-GFP construct (control); PsLyk9-RNAi—transgenic pea plants transformed with the pK7GWIWG2D-PsLyk9i construct. Error bars indicate the SEM of seven-ten biological repeats. Asterisks indicate statistically significant differences compared with control water-treated roots: *** p < 0.001, ** p < 0.01, * p < 0.05.
In addition, PsLyk9 RNAi composite plants were transferred in glass jars containing CO8 and exposed for 24 h. The analysis showed decreased expression levels of PsPAL2 and PsPR10 in PsLyk9 RNAi roots in response to exogenously applied CO8 compared with GUS-control plants. Thus, we have shown that the receptor kinase PsLYK9 can have a direct effect on plants’ resistance to infection with respect to phytopathogenic fungi and the expression of defense-response genes during plant treatment with CO8.
2.4. Analysis of Expression Levels of the Genes Involved in Developing Arbuscular–Mycorrhizal Symbiosis in Wild-Type Pea Plants and Transgenic Plants with PsLyk9 Gene Repression
To test the hypothesis that the PsLYK9 receptor is involved not only in the control of plant resistance to infection with phytopathogenic fungi, but also in the regulation of symbiosis development with AM fungi, the analysis of marker gene expression levels, activated in response to AM fungi inoculation, was performed. Previously, the search for such markers was performed in pea plans. Three genes—PsAM1, PsAM3 and PsAM5 (PSU43401, AJ006305, AJ276507)—were shown to be induced at the initial stages of the plant’s interaction with AM fungi, corresponding to the presymbiotic phase and early penetration [41,42]. Subtractive hybridization and macro-array analysis enabled the identification of a set of pea mycorrhiza-regulated genes like PsMAPK (AJ308148) and PsDRR 232a (AF139018) [43]. We also found homologs of well-known M. truncatula AM symbiosis markers like MtGST (Medtr5g076900), MtPT4 (Medtr1g028600), Myb-TF (Medtr7g068600), MtRAM1 (Medtr7g027190), and MtRAM2 (Medtr1g040500) in pea [44,45,46,47,48,49,50,51]. In addition, an analysis of PsNSP1 (KT454934), PsNSP2 (EU736106), PsDWARF27 (GDTM01031662), and PsDELLA1–3 (DQ848351, DQ845340, GDTM01011926) gene expression was performed in pea plants, because it was found that they were important in AM symbiosis development in pea and other legumes [52,53,54,55,56,57,58].
Finally, the expression level of 16 genes was tested in the roots of cv. Finale inoculated with AM fungus, Rhizophagus irregularis, at 14 and 28 days after inoculation. At both terms, all analyzed pea plants were extensively colonized by the fungus. The intensity of internal colonization of the root system (M%) was on an average 17.5% and 35.0%, respectively, and the arbuscule abundance in mycorrhizal root fragments (a%) was 29.4% and 36.5%, respectively.
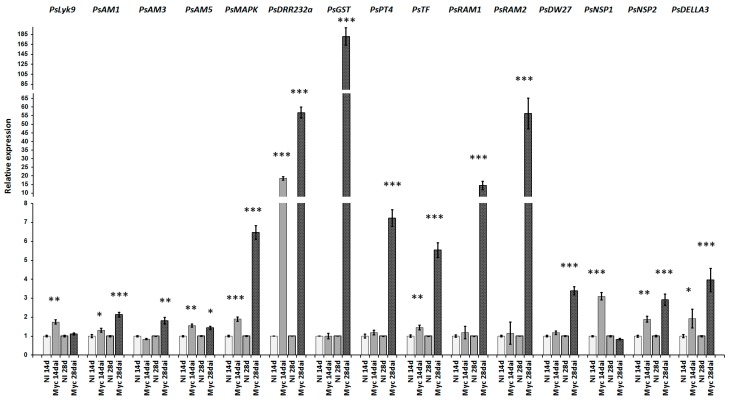
The analysis showed an increase in the expression of 13 genes (PsAM1, PsAM3, PsMAPK, PsDRR232a, PsGST, PsPT4, PsMyb-TF, PsRAM1, PsRAM2, PsNSP1, PsNSP2, PsDWARF27, and PsDELLA3), that may be considered as markers of AM symbiosis development in pea (Figure 6).
Figure 6.
Up-regulation of the AM symbiosis-related marker genes in pea roots on 14 and 28 days after inoculation with R. irregularis. The relative expression was normalized against the constitutively expressed ubiquitin and actin genes. Results are means ± SEM of three technical repeats. The graphs show the results of one biological repeat, representative for three independent experiments. Asterisks indicate statistically significant differences compared with control water-treated roots: *** p < 0.001, ** p < 0.01, * p < 0.05.
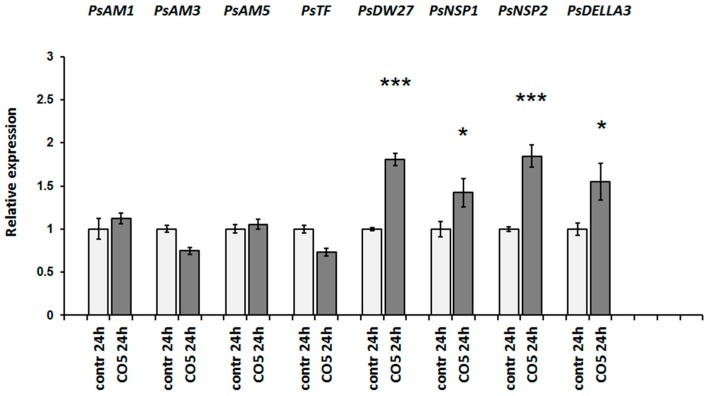
Since the changes in gene expression during mycorrhiza inoculation occur in response to the combined effect of both short COs and Myc factors, we also tested the activation of these markers in response to exogenously applied chitin oligosaccharide CO5. In the case of CO5, concentrations of 10−6 and 10−5 M were used to estimate the effect of these signal molecules on plants. All CO5 treatments were carried out for 24 h. Both CO5 concentrations were active in the induction of responses, but the effect of 10−5 M was more pronounced. The activation of at least four genes (PsDELLA3, PsNSP1, PsNSP2, and PsDWARF27) after treatment with CO5 was found in our experiments (Figure 7).
Figure 7.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of transcription levels of the AM symbiosis-related marker genes in pea roots treated with 10−5 M CO5 for 24 h. As a control mock-treated plants were used. The relative expression was normalized against the constitutively expressed ubiquitin and actin genes. Values are means ± SEM of three technical repeats. The graphs show the results of one biological repeat, representative for three biological independent experiments. Asterisks indicate statistically significant differences compared with the respective mock-treated control: *** p < 0.001, * p < 0.05.
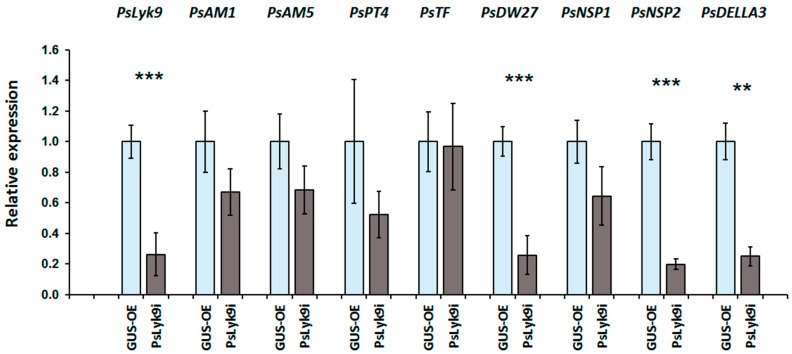
At the next stage of our study, we analyzed the expression levels of AM symbiosis development marker genes in the PsLyk9 RNAi transgenic roots in response to CO5 treatment (Figure 8). It was shown that in response to treating the PsLyk9 RNAi roots with CO5, there was no activation of the expression of four markers (Figure 8), although the decrease in PsNSP1 transcription was not statistically significant due to its high variability. Thus, this finding suggests that the receptor kinase PsLYK9 participates in the development of AM symbiosis in pea and controls plants’ response to recognizing COs with a low degree of polymerization.
Figure 8.
Reduction in gene expression level of PsDWARF27, PsNSP1, PsNSP2, and PsDELLA3 in the transgenic pea roots with PsLyk9 knockdown treated with 10−5 M CO5 for 24 h. GUS-OE—transgenic pea plants transformed with pB7WG2D-GUS construct (control); PsLyk9i—transgenic pea plants transformed with the pK7GWIWG2D-PsLyk9i construct. Error bars indicate the SEM of seven-ten biological repeats. Asterisks indicate statistically significant differences compared with control (GUS-overexpressing plants (GUS-OE)): *** p < 0.001, ** p < 0.01.
Activation of PsNSP1, PsNSP2, and PsDWARF27 may be related to strigolactone (SL) biosynthesis in plants during AM symbiosis development [59]. Therefore, the regulation of SL biosynthesis by NSP1 and NSP2 may be an ancestral function conserved in higher plants.
2.5. Analysis of Transcription Levels of the Cytokinin Response Genes in Pea Roots
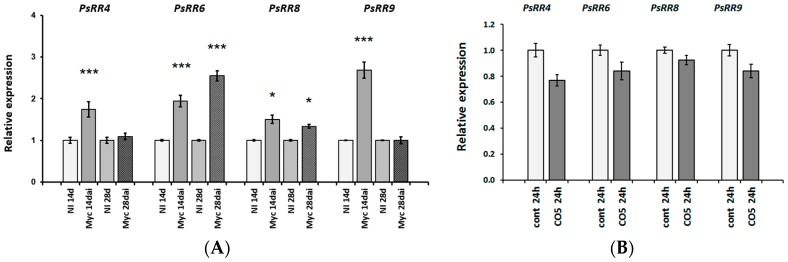
It is known that hormones, particularly cytokinins, may participate in the formation of AM symbiosis in plants. Increased levels of trans-zeatin riboside were found in the mycorrhizal roots when compared with non-mycorrhizal roots in some legume plants [60]. The activation of type-A and type-B cytokinin response regulators (type-A RR and type-B RR) was shown to be related to cytokinin receptor activation in legume plants [61,62,63]. Indeed, our experiments showed that the expression of type-A cytokinin response genes RR4, RR6, RR8, and RR9 increased in wild-type roots within 14 days of infection with AM fungi (Figure 9A).
Figure 9.
Relative expression of type-A cytokinin response regulator genes PsRR4, PsRR6, PsRR8, PsRR9 was determined using qRT-PCR. (A) Analysis of pea roots on 14 and 28 days after inoculation with Rhizophagus irregularis (dai). NI, not inoculated; (B) Effect was estimated in pea roots treated with 10−5 M CO5 for 24 h. As a control mock-treated roots were used. The relative expression was normalized against the constitutively expressed ubiquitin and actin genes. Asterisks indicate statistically significant differences compared with control (not inoculated or not treated plants): *** p < 0.001, * p < 0.05.
To verify whether these cytokinin response genes can be activated by CO5 during the development of AM symbiosis, we estimated the level of A-type RR4, RR6, RR8, and RR9 expression after treatment with signal molecules. However, we were not able to see any differences in the expression of these genes in treated and control pea roots (Figure 9B). Therefore, during mycorrhization, the induction of cytokinin response genes was probably related to Myc factors, not to CO4–CO5 molecules.
3. Discussion
Here, we report the characterization of the pea LysM-RLK PsLYK9 involved in recognizing COs with various degrees of polymerization, and which is implicated in the regulation of both plant resistance to phytopathogenic fungi and the control of AM symbiosis development.
In plants, LysM-RLKs have been involved in the recognition of chitin and COs from fungi, bacterial peptidoglycans, and various LCOs like Myc factors and Nod factors, which are known to be symbiotic signals produced by rhizobia and AM fungi [8,9,12,13,14,15,17,23]. Among them, the LysM-RLKs AtCERK1 and OsCERK1 can recognize fungal chitin/COs and bacterial peptidoglycans [8,9]. They act as typical pattern-recognition receptors that activate pathogen-triggered immunity pathways and confer resistance through the activation of defense responses to limit pathogen growth. Recent studies have also shown that OsCERK1 may fulfill dual functions and involved in recognizing structurally similar signals not only from phytopathogenic fungi (like long-chain CO8), but also from symbiotic AM fungi (like short-chain CO4 and CO5) [20,21]. We suggested that in pea plants that can form symbiotic relationships with AM fungi, a CERK1-like receptor may also fulfill a dual function.
Based on the phylogenetic analysis, we determined that the pea LysM-RLK LYK9 may be a possible candidate as a CERK1-like receptor. The knockdown of the PsLyk9 gene by RNA interference (PsLyk9-RNAi) resulted in the down-regulation of PsPAL2 and PsPR10 defense-response genes in roots, and this led to greater susceptibility to fungal pathogens F. culmorum producing a mixture of COs with various degrees of polymerization. In addition, the responses to exogenously applied CO8 elicitors were reduced in PsLyk9 RNAi plants. These results clearly indicate that PsLYK9 appears to be involved in the perception of CO elicitors and subsequent signal transduction in pea roots.
Since legume plants can form symbiotic relationships with AM fungi, we also evaluated PsLYK9 involvement in the recognition of short CO5 as primary signals of AM fungi. The knockdown of PsLyk9 greatly affected the gene responses induced by short COs (i.e., CO5), the most numerous signals of AM fungi. The results of this study further indicate that PsLYK9 may also be involved in the recognition of CO5 in pea roots.
While preparing this manuscript, a paper by Bozsoki et al. [64] was published that described similar results; the team’s investigation also showed the critical role played by LysM-RLK LjLYS6/MtLYK9 when recognizing chitin-based signal molecules in L. japonicus and M. truncatula. The screening of mutant collections enabled the identification of those mutants impaired in lys6 and lyk9 genes, which were defective in triggering chitin/COs-based responses in these legume plants. Therefore, our results were consistent with findings of Bozsoki et al. [64] in L. japonicus and M. truncatula. It seems like the pea PsLyk9 gene is a likely ortholog of the LjLYS6 and MtLYK9 genes in two model legumes.
One of the aims of our research was to identify those genes that are specifically involved in signal transduction in response to CO5 recognition. Studies of model legumes showed that components of a common signal pathway, like MtDMI2/LjSYMRK, MtDMI1/LjPOLLUX, MtDMI3/LjCCaMK, MtIPD3/LjCYCLOPS, MtNSP1/LjNSP1, and MtNSP2/LjNSP2 were involved in triggering signal transduction via signal molecules, the Myc factors from AM fungi [53,55,65,66]. Myc factors trigger calcium spiking, lateral root formation and a definite set of genes in legume plants [53,67]. An analysis of mutants showed that the MtDMI2/LjSYMRK and MtDMI1/LjPOLLUX regulators were also necessary for triggering signal transduction by short CO4 and CO5, which can induce calcium spiking and a specific gene expression [25,68]. Here, we demonstrated that exogenously applied CO5 upregulated PsDELLA3, PsNSP1, and PsNSP2 in pea roots. This may suggest that DELLA3, NSP1, and NSP2 are also involved in signal transduction activated by short COs. Although the transcriptome profiling of M. truncatula and L. japonicus roots revealed upregulation of the NSP1 and NSP2 genes encoding TFs by Myc factors [67], but not CO5 10−8 М [25], we suppose it may be connected to higher concentrations of applied CO5 in our experiments. To further investigate whether the PsDELLA3, PsNSP1, and PsNSP2 are involved in the CO4–CO5-activated signaling pathway, the effect of exogenously applied CO5 on della, nsp1 and nsp2 mutants should be estimated.
Since DELLA proteins are able to stimulate NSP1/NSP2 complex formation [58], a model may be proposed for a CO5-activated signal pathway in which DELLA3 and NSP1/NSP2 function as symbiotic regulators to directly activate the expression of DWARF27. DWARF27 is required for strigolactone biosynthesis; thus, cross-talk between gibberellin and strigolactone signaling during AM symbiosis may be mediated through DELLA3, NSP1, and NSP2. Conversely, no changes in the transcription level of the RAM1 and RAM2 genes, which are involved in the regulation of cutin synthesis during AM symbiosis [49], were found in our experiments. This may suggest that Myc factors, and not CO4–CO5, are involved in the activation of this pathway.
Recent studies showed that ERN and RAD1 transcription factors may play an important role in AM symbiosis development [45,69,70]. Therefore, whether CO5 can stimulate the expression of other transcription factors like ERN1, ERN2, and RAD1, which are involved in the regulation of various stages of AM symbiosis development, remains to be elucidated.
It was previously shown that trans-zeatin riboside accumulates to higher levels in mycorrhizal when compared with non-mycorrhizal roots [60]. This suggests that the cross-talk between plant hormones, cytokinins, and LCO or CO signaling may take place in AM symbiosis. It was interesting for us to determine that CO5 may induce the expression of cytokinin response genes in the roots of pea plants. However, the absence of cytokinin response gene activation in our experiment suggests that the function of CO signals is probably restricted to the presymbiotic AM stages, and that they do not activate genes related to cytokinin activation.
4. Materials and Methods
4.1. Plant Material and Germination Conditions
Pisum sativum L. cv. Finale seeds were surface sterilized for 5 min in concentrated sulfuric acid, then washed 3 times with sterile water, transferred to Petri dishes with 1% water agar and germinated in the dark at room temperature for 4–5 days. After germination (for 4–5 days), plants were transferred into pots with vermiculite or mineral substrate and grown in a growth chamber at 21 °C in a 16 h/8 h light/dark cycle at 60% humidity.
Seeds of chives Allium schoenoprasum L. were surface-disinfected as follows: 1 min in 96% ethanol, a rinse with sterile water, 5 min in 0.15% potassium permanganate aqueous solution, and a thorough rinse with sterile water. They were germinated on sterile humid filter paper in Petri dishes for 3–5 days at 27 °C in the dark and for 1–2 days at room temperature in the light. The seedlings were planted into pots (15 per pot) containing substrate with Rhizophagus irregularis inoculum. After 4–6 weeks, chive plants with mycorrhized roots were transferred to individual pots (1 per pot) and grown for 2–4 weeks.
4.2. Treatment of Pea Seedlings
Firstly, 4–5 days old pea seedlings were transferred in glass jars with 10−6–10−5 M water solutions of CO5 or CO8 and incubated for 24 h. Fully acetylated COs with degree of polymerization five (CO5) (Megazyme) and deacetylated COs with main degree of polymerization around eight (CO8) Mn = 1089, Mw = 1514, Ip = 1.39, CDS = 93% in Cl-form (The Center of Bioengineering Russian Academy of Science) were used for treatment. After treatment, the pea root fragments corresponding to responsive zones were harvested for RNA extraction and gene expression analysis.
4.3. Fusarium Culmorum Infection
Two strains of phytopathogenic fungi Fusarium culmorum (Wm. G. Sm.) Sacc. (weakly pathogenic 891 and highly pathogenic 334 strains) were used to infect pea plants. To obtain inoculum, F. culmorum 891 or 334 strains were grown on Chapek’s agar for 10–14 days and then washed from the plates with sterile water. 4–5 day old pea seedlings cv. Finale or PsLyk9i transgenic pea plants were transferred in pots with vermiculite saturated with Jensen medium [71], containing 3 × 105 conidia of the phytopathogenic fungus F. culmorum 891 or 334 strains. The control plants were grown in sterile vermiculite saturated with Jensen medium. Infected root tissue was harvested at different stages after inoculation together with the uninoculated control roots.
4.4. Rhizophagus Irregularis Infection
The isolate of the symbiotic fungus Rhizophagus irregularis BEG144 was provided by the International Bank of Glomeromycota (Dijon, France) as a soil-root based inoculum from onion (Allium cepa L.) pot cultures. The isolate was used to produce the chives A. schoenoprasum L. nurse culture to infect wild type pea plants with mycorrhizal fungus (see Section 4.1).
Three pea seedlings were planted into each nurse pot around a mycorrhizal chive plant (3 pots per variant). All plants were grown in 300 mL ceramic pots, with a mineral substrate, the silica rich marl, supplemented with 1 g/L calcium orthophosphate. Pots with substrate were sterilized by autoclaving for 60 min at 134 °C and 0.22 MPa. Plants were grown in a growth room under the following conditions: 16/8 h day/night, 21 °C, relative humidity 60% and were fed once a week with modified Hoagland’s solution without phosphate (50 mL per pot) [57,72], and watered as needed. The expression of AM symbiosis specific genes was assessed on 14 and 28 days after infection. AM development was quantitatively assessed as described earlier [57].
4.5. Molecular Cloning
To obtain a genetic construct for downregulation of the PsLyk9 gene, a fragment of 281 b.p. was amplified using cDNA cv. Finale as a matrix with primers flanked by the attB sites (Table S2) and cloned in the pDONR221 vector using BP clonase (Thermo Fisher Scientific, Waltham, MA, USA). Finally, the fragment was cloned in the pK7GWIWG2D vector using LR clonase enzyme. pK7GWIWG2D vector used in this work contained the green fluorescent protein (GFP) marker gene for the selection of transgenic roots. The resulting vector (PsLyk9-RNAi) contained a hairpin construct flanked by the 35S promoter and terminator sequences. The primers used for cloning are listed in Table S2. The verified construct was transferred into Agrobacterium rhizogenes strain Arqua 1 using electroporation method.
4.6. Agrobacterium Rhizogenes-Mediated Plant Transformation
For pea transformation, the seeds were sterilized as described above. Seeds were germinated on 1% water agar in the darkness. Then, 4–5 days old seedlings were transferred in sterile dark plastic bags with Jensen medium to the light and incubated for 2–3 days. The pea roots were cut off in the area of the hypocotyl and transformed with freshly grown A. rhizogenes strain Arqua 1. Plants were put in plastic jars (Duchefa Biochemie, Haarlem, The Netherlands) on Jensen agar, and the area of cut-off was covered with wet wool and foil. The seedlings were co-cultivated with A. rhizogenes for 10–14 days at 21 °C (16 h/8 h light/darkness) and subsequently transferred to Emergence medium [73] containing 150 mg/mL of cefotaxime. Plants were incubated on Emergence medium with antibiotic for 5 days and then transferred to fresh Emergence medium without antibiotic and additionally incubated for 3–5 days until the new roots were formed. Emerging roots were analyzed using an epifluorescent stereomicroscope (Stereo Discovery V8; Carl Zeiss, Göttingen, Germany). To analyze the effect of COs treatment, the plants were placed into glass jars and incubated with CO5 for 24 h.
4.7. RNA Isolation and cDNA Synthesis
Pea roots were harvested and frozen in liquid nitrogen. Total RNA was isolated using about 50–100 mg tissue per sample. Samples were thoroughly ground in a mortar to a fine powder in liquid nitrogen, at least three biological replicate per each condition. RNA was extracted using TriZol reagent (Thermo Fisher Scientific). After a DNase (Thermo Fisher Scientific) treatment, the samples were extracted with an equal volume of chloroform, and RNA was precipitated from the aqueous phase with 3 M sodium acetate and ethanol and subsequently quantified with a spectrophotometer UV-1280 (Shimadsu, Kyoto, Japan). RNA purity was checked by measuring spectrophotometric ratios of A260/A280 nm about 2. The efficacy of the DNase treatment was checked by using controls without reverse transcriptase for subsequent quantitative reverse transcription PCR (qPCR) analysis. RNA (from 1 to 2.5 μg) was used for cDNA synthesis with RevertAid Reverse Transcriptase (Thermo Fisher Scientific) for 1 h at 42 °C followed by heating to 70 °C, 10 min. Aliquots of the cDNA were diluted 1:10 for qPCR analysis.
4.8. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
The qRT-PCR analysis was performed on a CFX-96 real-time PCR detection system with a C1000 thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA), and SYBR Green intercalating dye were used for detection (Bio-Rad Laboratories). Each PCR reaction was carried out in a total volume of 10 µL. The following PCR program was used: 95 °C for 30 s, 40 cycles of 95 °C for 30 s, 54 °C for 30 s, 72 °C for 30 s. All reactions were done in triplicate and averaged. Cycle threshold (Ct) values were obtained with the accompanying software and data were analyzed with the 2–ΔΔCt method [74]. The relative expression was normalized against the constitutively expressed ubiquitin and actin genes in pea. All the primer sets used in the expression analysis are listed in Table S2. All primer pairs were designed using Vector NTI program and produced by Evrogen Company (Moscow, Russia) (www.evrogen.com). Each experiment was repeated at least three times with independent biological samples.
4.9. Statistical Methods and Computer Software
The expression levels of the gene of interest (GOI) relative to the reference genes Ubiquitin and Actin were calculated for each cDNA sample using the CFX Manager™ software version 2.1 (BioRad Laboratories, Richmond, CA, USA). The expression levels of GOI were calculated as ratio of treated samples to control samples. Statistical analysis was conducted by SIGMAPLOT 13.
Multiple alignment of nucleotide sequences was performed using Clustal W [75] using Vector NTI Advance 10 (InforMax, http://www.informaxinc.com). MEGA6 was used to generate graphic output of phylogenetic tree [76]. One-way ANOVA and Student’s t-test were used to compare gene expression levels in transgenic roots.
5. Conclusions
We identified a LysM receptor-like kinase (LysM-RLK) PsLYK9 required for CO recognition and implicated in the regulation of both plant immune responses and the control of AM symbiosis development.
Acknowledgments
This work was supported by Russian Science Foundation (RSF project No. 16-16-10043). The research was performed using the equipment of the Core Center “Genomic technologies, proteomics and cellular biology”. English-language editing of this manuscript was provided by Journal Prep.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/19/1/8/s1.
Author Contributions
Irina V. Leppyanen and Elena A. Dolgikh conceived and designed the experiments; Irina V. Leppyanen and Vlada Y. Shakhnazarova prepared the figures; Irina V. Leppyanen and Elena A. Dolgikh drafted the manuscript; Oksana Y. Shtark and Nadezhda A. Vishnevskaya contributed materials for experiments; Igor A. Tikhonovich and Elena A. Dolgikh edited the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Boller T., Felix G.A. Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 2.Dodds P.N., Rathjen J.P. Plant Immunity: Towards an Integrated View of Plant-Pathogen Interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 3.Schwessinger B., Ronald P.C. Plant Innate Immunity: Perception of Conserved Microbial Signatures. Annu. Rev. Plant Biol. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- 4.Beringer J.E., Brewin N., Johnston A.W.B., Schulman H.M., Hopwood D.A. The Rhizobium-Legume Symbiosis. Proc. R. Soc. Lond. B Biol. Sci. 1979;204:219–233. doi: 10.1098/rspb.1979.0024. [DOI] [PubMed] [Google Scholar]
- 5.Parniske M. Arbuscular Mycorrhiza: The Mother of Plant Root Endosymbioses. Nat. Rev. Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 6.Felix G., Regenass M., Boller T. Specific Perception of Subnanomolar Concentrations of Chitin Fragments by Tomato Cells—Induction of Extracellular Alkalinization, Changes in Protein Phosphorylation, and Establishment of a Refractory State. Plant J. 1993;4:307–316. doi: 10.1046/j.1365-313X.1993.04020307.x. [DOI] [Google Scholar]
- 7.Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. Plant Cells Recognize Chitin Fragments for Defense Signaling Through a Plasma Membrane Receptor. Proc. Natl. Acad. Sci. USA. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., et al. Two LysM Receptor Molecules, CEBiP And OsCERK1, Cooperatively Regulate Chitin Elicitor Signaling in Rice. Plant J. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman M.-A., Sundelin T., Nielsen J.T., Erbs G. MAMP (Microbe-Associated Molecular Pattern) Triggered Immunity in Plants. Front. Plant Sci. 2013;4:139. doi: 10.3389/fpls.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 12.Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. LysM Domain Receptor Kinases Regulating Rhizobial Nod Factor-Induced Infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 13.Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal L., et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 14.Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. A LysM Receptor-Like Kinase Plays a Critical Role in Chitin Signaling and Fungal Resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iizasa E., Mitsutomi M., Nagano Y. Direct Binding of a Plant LysM Receptor-like Kinase, LysM RLK1/CERK1, to Chitin in Vitro. J. Biol. Chem. 2010;285:2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B., Li J.-F., Ao Y., Qu J., Li Z., Su J., Zhang Y., Liu J., Feng D., Qi K., et al. Lysin Motif-Containing Proteins LYP4 and LYP6 Play Dual Roles in Peptidoglycan and Chitin Perception in Rice Innate Immunity. Plant Cell. 2012;24:3406–3419. doi: 10.1105/tpc.112.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan J., Tanaka K., Zhang X.C., Son G.H., Brechenmacher L., Nguyen T.H., Stacey G. LYK4, a Lysin Motif Receptor-Like Kinase, is Important for Chitin Signaling and Plant Innate Immunity in Arabidopsis. Plant Physiol. 2012;160:396–406. doi: 10.1104/pp.112.201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y., Liang Y., Tanaka K., Nguyen C.T., Jedrzejczak R.P., Joachimiak A., Stacey G. The Kinase LYK5 is a Major Chitin Receptor in Arabidopsis and Forms a Chitin-Induced Complex with Related Kinase CERK1. Elife. 2014;3:e03766. doi: 10.7554/eLife.03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyata K., Kozaki T., Kouzai Y., Ozawa K., Ishii K., Asamizu E., Okabe Y., Umehara Y., Miyamoto A., Kobae Y., et al. The Bifunctional Plant Receptor, OsCERK1, Regulates Both Chitin-Triggered Immunity and Arbuscular Mycorrhizal Symbiosis in Rice. Plant Cell Physiol. 2014;55:1864–1872. doi: 10.1093/pcp/pcu129. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Dong W., Sun J., Feng F., Deng Y., He Z., Oldroyd G.E., Wang E. The Receptor Kinase CERK1 has Dual Functions in Symbiosis and Immunity Signalling. Plant J. 2015;81:258–267. doi: 10.1111/tpj.12723. [DOI] [PubMed] [Google Scholar]
- 22.Liu S., Wang J., Han Z., Gong X., Zhang H., Chai J. Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP. Structure. 2016;24:1192–2000. doi: 10.1016/j.str.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Willmann R., Lajunen H.M., Erbs G., Newman M.-A., Kolb D., Tsuda K., Katagiri F., Fliegmann J., Bono J.-J., Cullimore J.V., et al. Arabidopsis Lysin-Motif Proteins LYM1 LYM3 CERK1 Mediate Bacterial Peptidoglycan Sensing and Immunity to Bacterial Infection. Proc. Natl. Acad. Sci. USA. 2011;108:19824–19829. doi: 10.1073/pnas.1112862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa T., Kaku H., Shimoda Y., Sugiyama A., Shimamura M., Takanashi K., Yazaki K., Aoki T., Shibuya N., Kouchi H. From Defense to Symbiosis: Limited Alterations in the Kinase Domain of LysM Receptor-Like Kinases are Crucial for Evolution of Legume-Rhizobium Symbiosis. Plant J. 2011;65:169–180. doi: 10.1111/j.1365-313X.2010.04411.x. [DOI] [PubMed] [Google Scholar]
- 25.Giovannetti M., Mari A., Novero M., Bonfante P. Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Front. Plant Sci. 2015;6:480. doi: 10.3389/fpls.2015.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arrighi J.F., Barre A., Ben Amor B., Bersoult A., Soriano L.C., Mirabella R., de Carvalho-Niebel F., Journet E.-P., Ghérardi M., Huguet T., et al. The Medicago truncatula Lysin Motif-Receptor-Like Kinase Gene Family Includes NFP and New Nodule-Expressed Genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X.C., Wu X., Findley S., Wan J., Libault M., Nguyen H.T., Cannon S.B., Stacey G. Molecular Evolution of Lysin Motif-Type Receptor-Like Kinases in Plants. Plant Physiol. 2007;144:623–636. doi: 10.1104/pp.107.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann G.V., Shimoda Y., Nielsen M.W., Jørgensen F.G., Grossmann C., Sandal N., Jørensen K., Thirup S., Madsen L.H., Tabata S., et al. Evolution and Regulation of the Lotus japonicus LysM Receptor Gene Family. Mol. Plant Microbe Interact. 2010;23:510–521. doi: 10.1094/MPMI-23-4-0510. [DOI] [PubMed] [Google Scholar]
- 29.Zhukov V., Radutoiu S., Madsen L.H., Rychagova T., Ovchinnikova E., Borisov A., Tikhonovich I., Stougaard J. The Pea Sym37 Receptor Kinase Gene Controls Infection Thread Initiation and Nodule Development. Mol. Plant Microbe Interact. 2008;21:1600–1608. doi: 10.1094/MPMI-21-12-1600. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H., Riely B.K., Burns N.J., Ané J.M. Tracing Nonlegume Orthologs of Legume Genes Required for Nodulation and Arbuscular Mycorrhizal Symbioses. Genetics. 2006;172:2491–2499. doi: 10.1534/genetics.105.051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duarte J., Rivière N., Baranger A., Aubert G., Burstin J., Cornet L., Lavaud C., Lejeune-Hénaut I., Martinant J.-P., Pichon J.-P., et al. Transcriptome Sequencing for High Throughput SNP Development and Genetic Mapping in Pea. Genomics. 2014;15:126. doi: 10.1186/1471-2164-15-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alves-Carvalho S., Aubert G., Carrère S., Cruaud C., Brochot A.L., Jacquin F., Klein A., Martin C., Boucherot K., Kreplak J., et al. Full-Length De Novo Assembly of RNA-Seq Data in Pea (Pisum sativum L.) Provides a Gene Expression Atlas and Gives Insights into Root Nodulation in this Species. Plant J. 2015;84:1–19. doi: 10.1111/tpj.12967. [DOI] [PubMed] [Google Scholar]
- 33.Zhukov V.A., Zhernakov A.I., Kulaeva O.A., Ershov N.I., Borisov A.Y., Tikhonovich I.A. De Novo Assembly of the Pea (Pisum sativum L.) Nodule Transcriptome. Int. J. Genom. 2015;2015:1–11. doi: 10.1155/2015/695947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudheesh S., Rodda M.S., Davidson J., Javid M., Stephens A., Slater A.T., Cogan N.O.I., Forster J.W., Kaur S. SNP-Based Linkage Mapping for Validation of QTLs for Resistance to Ascochyta Blight in Lentil. Front. Plant Sci. 2016;7:1604. doi: 10.3389/fpls.2016.01604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerr S.C., Gaiti F., Beveridge C.A., Tanurdzic M. De Novo Transcriptome Assembly Reveals High Transcriptional Complexity in Pisum sativum axillary Buds and Shows Rapid Changes in Expression of Diurnally Regulated Genes BMC. Genomics. 2017;18:221. doi: 10.1186/s12864-017-3577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broghammer A., Krusell L., Blaise M., Sauer J., Sullivan J.T., Maolanon N., Vinther M., Lorentzen A., Madsen E.B., Jensen K.J., et al. Legume Receptors Perceive the Rhizobial Lipochitin Oligosaccharide Signal Molecules by Direct Binding. Proc. Natl. Acad. Sci. USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawaharada Y., Kelly S., Nielsen M.W., Hjuler C.T., Gysel K., Muszyński A., Carlson R.W., Thygesen M.B., Sandal N., Asmussen M.H., et al. Receptor-Mediated Exopolysaccharide Perception Controls Bacterial Infection. Nature. 2015;523:308–312. doi: 10.1038/nature14611. [DOI] [PubMed] [Google Scholar]
- 38.Kendra D.F., Hadwiger L.A. Characterization of the Smallest Chitosan Oligomer that is Maximally Antifungal to Fusarium solani and Elicits Pisatin Formation in Pisum sativum. Exp. Mycol. 1984;8:276–281. doi: 10.1016/0147-5975(84)90013-6. [DOI] [Google Scholar]
- 39.Hadwiger L.A., Ogawa T., Kuyama H. Chitosan Polymer Sizes Effective in Inducing Phytoalexin Accumulation and Fungal Suppression are Verified with Synthesized Oligomers. Mol. Plant Microbe Interact. 1994;7:531–533. doi: 10.1094/MPMI-7-0531. [DOI] [PubMed] [Google Scholar]
- 40.Leppyanen I.V., Shahnazarova V.Y., Vishnevskaya N.A., Dolgih E.A., Strunnikova O.K. Study of Mechanisms of Interactions between Pisum sativum and Two Strains of Fusarium. Med. Mycol. 2017 in press. [Google Scholar]
- 41.Martin-Laurent F., van Tuinen D., Dumas-Gaudot E., Gianinazzi-Pearson V., Gianinazzi S., Franken P. Differential Display Analysis of RNA Accumulation in Arbuscular Mycorrhiza of Pea and Isolation of a Novel Symbiosis-Regulated Plant Gene. Mol. Gen. Genet. 1997;256:37–44. doi: 10.1007/s004380050543. [DOI] [PubMed] [Google Scholar]
- 42.Roussel H., van-Tuinen D., Franken P., Gianinazzi S., Gianinazzi-Pearson V. Signaling between Arbuscular Mycorrhizal Fungi and Plants: Identification of a Gene Expressed during Early Interactions by Differential RNA Display Analysis. Plant Soil. 2001;232:13–19. doi: 10.1023/A:1010321700347. [DOI] [Google Scholar]
- 43.Grunwald U., Nyamsuren O., Tamasloukht M., Lapopin L., Becker A., Mann P., Gianinazzi-Pearson V., Krajinski F., Franken P. Identification of mycorrhiza-regulated genes with arbuscule development-related expression profile. Plant Mol. Biol. 2004;55:553–566. doi: 10.1007/s11103-004-1303-y. [DOI] [PubMed] [Google Scholar]
- 44.Harrison M.J., Dewbre G.R., Liu J. A Phosphate Transporter from Medicago truncatula Involved in the Acquisition of Phosphate Released by Arbuscular Mycorrhizal Fungi. Plant Cell. 2002;14:2413–2429. doi: 10.1105/tpc.004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wulf A., Manthey K., Doll J., Perlick A.M., Linke B., Bekel T., Meyer F., Franken P., Küster H., Krajinski F. Transcriptional Changes in Response to Arbuscular Mycorrhiza Development in the Model Plant Medicago truncatula. Mol. Plant Microbe Interact. 2003;16:306–314. doi: 10.1094/MPMI.2003.16.4.306. [DOI] [PubMed] [Google Scholar]
- 46.Javot H., Penmetsa R.V., Terzaghi N., Cook D.R., Harrison M.J. A Medicago truncatula Phosphate Transporter Indispensable for the Arbuscular Mycorrhizal Symbiosis. Proc. Natl. Acad. Sci. USA. 2007;104:1720–1725. doi: 10.1073/pnas.0608136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogekamp C., Arndt D., Pereira P.A., Becker J.D., Hohnjec N., Küster H. Laser Microdissection Unravels Cell-Type-Specific Transcription in Arbuscular Mycorrhizal Roots, Including CAAT-Box Transcription Factor Gene Expression Correlating with Fungal Contact and Spread. Plant Physiol. 2011;157:2023–2043. doi: 10.1104/pp.111.186635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogekamp C., Küster H. A Roadmap of Cell-Type Specific Gene Expression during Sequential Stages of the Arbuscular Mycorrhiza Symbiosis. Genomics. 2013;14:306. doi: 10.1186/1471-2164-14-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gobbato E., Marsh J.F., Vernié T., Wang E., Maillet F., Kim J., Miller J.B., Sun J., Bano S.A., Ratet P., et al. A GRAS-Type Transcription Factor with a Specific Function in Mycorrhizal Signaling. Curr. Biol. 2012;22:2236–2241. doi: 10.1016/j.cub.2012.09.044. [DOI] [PubMed] [Google Scholar]
- 50.Gobbato E., Wang E., Higgins G., Bano S.A., Henry C., Schultze M., Oldroyd G.E. RAM1 and RAM2 Function and Expression during Arbuscular Mycorrhizal Symbiosis and Aphanomyces euteiches Colonization. Plant Signal. Behav. 2013;8:e26049. doi: 10.4161/psb.26049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang E., Schornack S., Marsh J., Gobbato E., Schwessinger B., Eastmond P., Schultze M., Kamoun S., Oldroyd G.E. A Common Signaling Process that Promotes Mycorrhizal and Oomycete Colonization of Plants. Curr. Biol. 2012;22:2242–2246. doi: 10.1016/j.cub.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 52.Liu W., Kohlen W., Lillo A., den Camp R.O., Ivanov S., Hartog M., Limpens E., Jamil M., Smaczniak C., Kaufmann K., et al. Strigolactone Biosynthesis in Medicago truncatula and Rice Requires the Symbiotic GRAS-Type Transcription Factors NSP1 and NSP2. Plant Cell. 2011;23:3853–3865. doi: 10.1105/tpc.111.089771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maillet F., Poinsot V., André O., Puech-Pagès V., Haouy A., Gueunier M., Cromer L., Giraudet D., Formey D., Niebel A., et al. Fungal Lipochitooligosaccharide Symbiotic Signals in Arbuscular Mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 54.Takeda N., Tsuzuki S., Suzaki T., Parniske M., Kawaguchi M. CERBERUS and NSP1 of Lotus japonicus are Common Symbiosis Genes that Modulate Arbuscular Mycorrhiza Development. Plant Cell Physiol. 2013;54:1711–1723. doi: 10.1093/pcp/pct114. [DOI] [PubMed] [Google Scholar]
- 55.Floss D.S., Levy J.G., Lévesque-Tremblay V., Pumplin N., Harrison M.J. DELLA Proteins Regulate Arbuscule Formation in Arbuscular Mycorrhizal Symbiosis. Proc. Natl. Acad. Sci. USA. 2013;110:E5025–E5034. doi: 10.1073/pnas.1308973110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu N., Luo D., Zhang X., Liu J., Wang W., Jin Y., Dong W., Liu J., Liu H., Yang W., et al. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 2014;24:130–133. doi: 10.1038/cr.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shtark O.Y., Sulima A.S., Zhernakov A.I., Kliukova M.S., Fedorina J.V., Pinaev A.G., Kryukov A.A., Akhtemova G.A., Tikhonovich I.A., Zhukov V.A. Arbuscular Mycorrhiza Development in Pea (Pisum sativum L.) Mutants Impaired in Five Early Nodulation Genes Including Putative Orthologs of NSP1 and NSP2. Symbiosis. 2016;68:129–144. doi: 10.1007/s13199-016-0382-2. [DOI] [Google Scholar]
- 58.Jin Y., Liu H., Luo D., Yu N., Dong W., Wang C., Zhang X., Dai H., Yang J., Wang E. DELLA Proteins are Common Components of Symbiotic Rhizobial and Mycorrhizal Signalling Pathways. Nat. Commun. 2016;12:12433. doi: 10.1038/ncomms12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Zeijl A., Liu W., Xiao T.T., Kohlen W., Yang W.-C., Bisseling T., Geurts R. The Strigolactone Biosynthesis Gene DWARF27 is Co-Opted in Rhizobium Symbiosis. Plant Biol. 2015;15:260. doi: 10.1186/s12870-015-0651-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Rhijn P., Fang Y., Galili S., Shaul O., Atzmon N., Wininger S., Eshed Y., Lum M., Li Y., To V., et al. Expression of Early Nodulin Genes in Alfalfa Mycorrhizae Indicates that Signal Transduction Pathways Used in Forming Arbuscular Mycorrhizae and Rhizobium-Induced Nodules may be Conserved. Proc. Natl. Acad. Sci. USA. 1997;94:5467–5472. doi: 10.1073/pnas.94.10.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez-Rizzo S., Crespi M., Frugier F. The Medicago truncatula CRE1 Cytokinin Receptor Regulates Lateral Root Development and Early Symbiotic Interaction with Sinorhizobium meliloti. Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Azarakhsh M., Kirienko A.N., Zhukov V.A., Lebedeva M.A., Dolgikh E.A., Lutova L.A. KNOTTED1-LIKE HOMEOBOX 3: A New Regulator of Symbiotic Nodule Development. J. Exp. Bot. 2015;66:7181–7195. doi: 10.1093/jxb/erv414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dolgikh E.A., Shaposhnikov A.I., Dolgikh A.V., Gribchenko E.S., Bodyagina K.B., Yuzhikhin O.S., Tikhonovich I.A. Identification of Pisum sativum L. Cytokinin and Auxin Metabolic and Signaling Genes, and an Analysis of Their Role in Symbiotic Nodule Development. Int. J. Plant Physiol. Biochem. 2017;9:22–35. doi: 10.5897/IJPPB2017.0266. [DOI] [Google Scholar]
- 64.Bozsoki Z., Cheng J., Feng F., Gysel K., Vinther M., Andersen K.R., Oldroyd G., Blaise M., Radutoiu S., Stougaard J. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. USA. 2017;114:E8118–E8127. doi: 10.1073/pnas.1706795114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delaux P.M., Bécard G., Combier J.P. NSP1 is a Component of the Myc Signaling Pathway. New Phytol. 2013;199:59–65. doi: 10.1111/nph.12340. [DOI] [PubMed] [Google Scholar]
- 66.Genre A., Russo G. Does a Common Pathway Transduce Symbiotic Signals in Plant–Microbe Interactions? Front. Plant Sci. 2016;7:96. doi: 10.3389/fpls.2016.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Czaja L.F., Hogekamp C., Lamm P., Maillet F., Martinez E.A., Samain E., Dénarié J., Küster H., Hohnjec N. Transcriptional Responses Toward Diffusible Signals from Symbiotic Microbes Reveal MtNFP- and MtDMI3-Dependent Reprogramming of Host Gene Expression by Arbuscular Mycorrhizal Fungal Lipochitooligosaccharides. Plant Physiol. 2012;159:1671–1685. doi: 10.1104/pp.112.195990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Genre A., Chabaud M., Balzergue B., Puech-Pages V., Novero M., Rey T., Fournier J., Rochange S., Becard G., Bonfante P., et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 69.Young N.D., Debellé F., Oldroyd G.E., Geurts R., Cannon S.B., Udvardi M.K., Benedito V.A., Mayer K.F., Gouzy J., Schoof H., et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xue L., Cui H., Buer B., Vijayakumar V., Delaux P.M., Junkermann S., Bucher M. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 2015;167:854–871. doi: 10.1104/pp.114.255430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Brussel A.A.N., Tak T., Wetselaar A., Pees E., Wijffelman C.A. Small Leguminosae as Test Plants for Nodulation of Rhizobium leguminosarum and Other Rhizobia and Agrobacteria Harbouring a Leguminosarum Sym-Plasmid. Plant Sci. Lett. 1982;27:317–325. doi: 10.1016/0304-4211(82)90134-1. [DOI] [Google Scholar]
- 72.Hoagland D.R., Arnon D.T. The Water-Culture Method for Growing Plants without Soil. Volume 347 Agriculture Experiment Station, University of California; Berkeley, CA, USA: 1938. [Google Scholar]
- 73.Limpens E., Ramos J., Franken C., Raz V., Compaan B., Franssen H., Bisseling T., Geurts R. RNA Interference in Agrobacterium rhizogenes-Transformed Roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 2004;55:983–992. doi: 10.1093/jxb/erh122. [DOI] [PubMed] [Google Scholar]
- 74.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;4:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 75.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hall B.G. Building Phylogenetic Trees of Molecular Data with MEGA. Mol. Biol. Evol. 2013;30:1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.