Abstract
The function of a protein is of great interest in the cutting-edge research of biological mechanisms, disease development and drug/target discovery. Besides experimental explorations, a variety of computational methods have been designed to predict protein function. Among these in silico methods, the prediction of BLAST is based on protein sequence similarity, while that of machine learning is also based on the sequence, but without the consideration of their similarity. This unique characteristic of machine learning makes it a good complement to BLAST and many other approaches in predicting the function of remotely relevant proteins and the homologous proteins of distinct function. However, the identification accuracies of these in silico methods and their false discovery rate have not yet been assessed so far, which greatly limits the usage of these algorithms. Herein, a comprehensive comparison of the performances among four popular prediction algorithms (BLAST, SVM, PNN and KNN) was conducted. In particular, the performance of these methods was systematically assessed by four standard statistical indexes based on the independent test datasets of 93 functional protein families defined by UniProtKB keywords. Moreover, the false discovery rates of these algorithms were evaluated by scanning the genomes of four representative model organisms (Homo sapiens, Arabidopsis thaliana, Saccharomyces cerevisiae and Mycobacterium tuberculosis). As a result, the substantially higher sensitivity of SVM and BLAST was observed compared with that of PNN and KNN. However, the machine learning algorithms (PNN, KNN and SVM) were found capable of substantially reducing the false discovery rate (SVM < PNN < KNN). In sum, this study comprehensively assessed the performance of four popular algorithms applied to protein function prediction, which could facilitate the selection of the most appropriate method in the related biomedical research.
Keywords: false discovery rate, machine learning, protein function prediction, support vector machine, BLAST
1. Introduction
The function of a protein is of great interest in the current research of biological mechanisms [1], disease development [2] and drug/target discovery [3,4,5,6,7], and a variety of databases is available for providing functional annotations from the perspectives of the sequence [8], protein-protein interaction [9,10], the biological network [11,12,13,14,15] and many specific functional classes [16,17,18,19,20,21,22]. However, a substantial gap is still observed between the total number of protein sequences discovered and that of proteins characterized with known function [23]. To cope with this gap, thousands of high-throughput genome projects are under study [24], and over 13 million sequences have been discovered, but only 1% of these validated by experimental annotation [25]. Apart from those experimental approaches, many in silico methods have been designed and extensively used to discover protein functions [26]. These include clustering of sequences [27], gene fusion [28], sequence similarity [29,30], evolution study [31], structural comparison [32], protein-protein interaction [33,34], functional classification via the sequence-derived [35,36,37,38] and domain [39,40,41,42,43] feature, omics profiling [44,45,46,47] and integrated methods, which collectively consider multiple methods and data to promote the performance of function prediction [48,49,50,51].
Among these in silico methods [52], the basic local alignment search tool (BLAST) [53] revealing protein functions based on excess sequence similarity [54] demonstrated great capacity and attracted substantial interest from the researchers of this field [55,56]. Apart from BLAST, machine learning algorithms have been frequently applied in recent years for functional prediction [57,58,59,60,61,62], and a variety of online software tools based on machine learning was developed as predictors without considering the similarity in sequence or structure [36,63]. This unique characteristic makes machine learning a good complement to other in silico approaches in predicting the function of remotely relevant protein and the homologous proteins of distinct functions [64,65].
So far, three machine learning algorithms, including K-nearest neighbor (KNN), probabilistic neural network (PNN) and support vector machine (SVM), have been extensively explored to classify proteins into certain functional families by analyzing the sequence-based physicochemical property [64,65] and to assess protein functional classes collectively [63]. These algorithms are recognized as powerful alternative methods for predicting the function of both proteins [66,67,68,69,70] and other molecules [71]. However, over one third of the protein sequences in UniProt [26] are still labeled as “putative”, “uncharacterized”, “unknown function” or “hypothetical”, and the difficulty in discovering the function of the remaining proteins is reported to come mainly from the false discovery rate of in silico algorithms [55,56,72]. Moreover, the identification accuracies of those approaches still need to be further improved [55,56,73]. Thus, it is urgently needed to assess the identification accuracies and false discovery rates among those different in silico approaches.
In this study, the performances of four popular functional prediction algorithms (BLAST, SVM, KNN and PNN) were comprehensively evaluated from two perspectives. In particular, the identification accuracies (measured by four standard statistical indexes) of various algorithms were systematically evaluated based on the independent test data of 93 functional families. Secondly, the false discovery rates of these algorithms were compared by scanning the genomes of four representative model species (Homo sapiens, Arabidopsis thaliana, Saccharomyces cerevisiae and Mycobacterium tuberculosis). In sum, these findings provided detailed information on the performances of those algorithms that are popular for protein function prediction, which may facilitate the choice of the appropriate algorithm(s) in the related biomedical research.
2. Results and Discussion
2.1. Assessment of the Identification Accuracies Measured by Four Popular Metrics
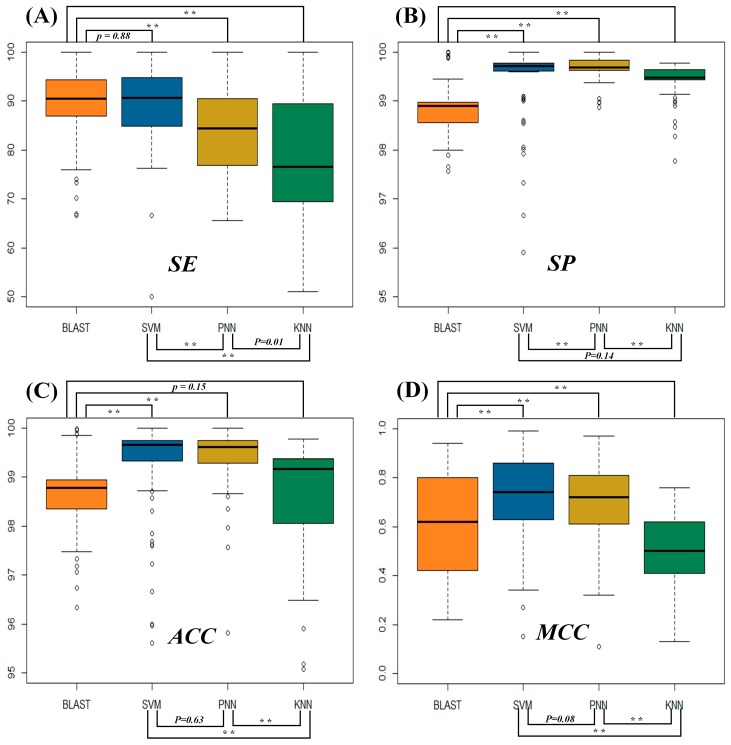
The statistical differences in sensitivity (SE) (Figure 1A), specificity (SP) (Figure 1B), accuracy (ACC) (Figure 1C) and Matthews correlation coefficient (MCC) (Figure 1D) among four popular functional prediction algorithms are illustrated. As illustrated in Figure 1A, the SE of BLAST measured by the independent test dataset of 93 families was roughly equivalent to that of SVM, but statistically higher than that of both PNN and KNN. In particular, the SE of 93 functional families was 50.00~99.99% for SVM, 43.93~99.99% for BLAST, 65.52~99.99% for PNN and 51.09~99.99% for KNN, and the SE median values of BLAST, SVM, PNN and KNN equaled 90.59%, 90.52%, 84.38% and 76.54%, respectively. As shown in Figure 1B, the majority of the SPs of all algorithms surpassed 98.00%; SPs of 93 functional families were 95.90~99.99% for SVM, 97.56~99.99% for BLAST, 98.87~99.99% for PNN and 97.77~99.43% for KNN; and the SP median value of BLAST, SVM, PNN and KNN was 98.90%, 99.72%, 99.67% and 99.44%, respectively. These results revealed a relatively low level of false discovery rates for all popular functional prediction algorithms.
Figure 1.
Statistical differences in the performance of four protein function prediction algorithms (BLAST, SVM, PNN and KNN) assessed by four metrics: (A) sensitivity (SE); (B) specificity (SP); (C) accuracy (ACC); and (D) Matthews correlation coefficient (MCC). Significant and moderately significant differences were shown by a p-value of (**), respectively.
Due to the dominant number of negative samples in the independent test datasets, the statistical difference in ACC was very similar to that of SP (Figure 1C). The majority of the ACCs of all algorithms surpassed 97%. The ACCs of 93 functional families were between 95.61% and 99.99% for SVM, between 66.68% and 99.98% for BLAST, between 95.81% and 99.99% for PNN and between 81.39% and 99.77% for KNN. Moreover, median values of ACCs of BLAST, SVM, PNN and KNN equaled 98.78%, 99.66%, 99.61% and 99.16%, respectively. MCC was frequently applied to reflect the stability of the protein function predictor and was considered as one of the most comprehensive parameters because of its full consideration of TP, TN, FP and FN. As shown in Figure 1D, the MCC of both SVM and PNN was better than that of BLAST and KNN. The majority of MCCs were over 0.6 and 0.4 for SVM-PNN and BLAST-KNN, respectively. In particular, MCCs of 93 functional families were between 0.15 and 0.99 for SVM, between 0.22 and 0.94 for BLAST, between 0.11 and 0.97 for PNN and between 0.13 and 0.76 for KNN. The median values of MCCs for BLAST, SVM, PNN and KNN equaled 0.62, 0.74, 0.72 and 0.50, respectively. In sum, there were consistently low levels of the false discovery rate among all algorithms as assessed by the metric SP. However, when the positive discovery rates (SEs) and the stability of prediction (MCC) were considered, SVM, PNN and BLAST stood out as more powerful algorithms for protein function prediction.
2.2. Evaluating the Statistical Differences in SE and MCC among Four Metrics
For the machine learning algorithms (SVM, PNN and KNN), there was a significant statistical difference in their SEs and MCCs. As shown in Figure 1A, the statistical difference in SEs between SVM and PNN equaled 3.5 × 10−6, while that between SVM and KNN was 1.0 × 10−11. Moreover, there was a significant statistical difference between PNN and KNN (p-value = 0.01). In particular, the number of families with SEs of >90%, ≤90% and >80% and ≤80% for SVM equaled 49, 33 and 11, respectively; the number of families with SEs of >90%, ≤90% and >80% and ≤80% for PNN equaled 17, 25 and 20, respectively; and the number of functional families with SEs of >90%, ≤90% and >80% and ≤80% for KNN equaled 19, 13 and 45, respectively. Similar to the SE, the statistical difference in MCC between SVM and PNN was 0.08, and that between SVM and KNN was 2.2 × 10−16. Moreover, there was a clear statistical difference between PNN and KNN (p-value = 2.2 × 10−16). In particular, the number of families with MCCs of >0.85, ≤0.85 and >0.7 and ≤0.7 for SVM was 26, 26 and 41, respectively; the number of functional families with MCCs of >0.85, ≤0.85 and >0.7 and ≤0.7 for PNN equaled 6, 29 and 27, respectively; and there were no protein families with MCCs over 0.7 for KNN. In summary, there were clear ascending trends in both SE and MCC as shown in Figure 1A,D (from KNN to PNN to SVM).
Similar to SVM, BLAST also demonstrated great performances in both SE and MCC. The statistical differences (measured by p-value) in the SE and MCC between BLAST and SVM were 0.88 and 2.0 × 10−7, respectively. As demonstrated in Table 1 and Table S1, the SE of BLAST surpassed that of SVM in 51 families, but was worse than that of SVM in 40 families. Moreover, the SEs’ median values (90.52% for BLAST and 90.59% for SVM) and mean values (88.92% for BLAST and 89.08% for SVM) indicated that the SE of SVM was slightly better than that of BLAST and significantly better than that of PNN and KNN. Meanwhile, MCC of SVM was higher than that of BLAST in 68 families, but was lower than that of BLAST in 20 families. The MCCs’ median values (0.62 for BLAST, 0.74 for SVM) and mean values (0.61 for BLAST, 0.73 for SVM) indicated a slight improvement in prediction stabilities by SVM.
Table 1.
The performance of four protein function prediction algorithms assessed by four popular metrics: sensitivity (SE), specificity (SP), accuracy (ACC) and Matthews correlation coefficient (MCC).
| UniProt Keyword | Protein Functional Family | GO Category | BLAST | SVM | PNN | KNN | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SE % | SP % | AC % | MCC | SE % | SP % | AC % | MCC | SE % | SP % | AC % | MCC | SE % | SP % | AC % | MCC | |||
| KW-0020 | Allergen | - | 76.32 | 98.92 | 98.78 | 0.48 | 84.81 | 99.69 | 99.66 | 0.57 | 86.42 | 99.84 | 99.81 | 0.69 | 74.07 | 99.48 | 99.32 | 0.41 |
| KW-0049 | Antioxidant | GO:0016209 | 94.15 | 99.23 | 99.20 | 0.60 | 89.00 | 99.76 | 99.73 | 0.67 | 86.00 | 99.84 | 99.80 | 0.71 | 69.00 | 99.42 | 99.24 | 0.43 |
| KW-0117 | Actin capping | GO:0051693 | 94.55 | 99.08 | 99.07 | 0.35 | 93.98 | 99.75 | 99.74 | 0.70 | 91.18 | 99.80 | 99.78 | 0.71 | 73.53 | 99.42 | 99.22 | 0.43 |
| KW-0147 | Chitin-binding | GO:0008061 | 86.96 | 98.96 | 98.94 | 0.34 | 91.75 | 99.72 | 99.68 | 0.78 | 75.36 | 99.61 | 99.47 | 0.63 | 93.84 | 98.57 | 98.05 | 0.37 |
| KW-0157 | Chromophore | GO:0018298 | 96.70 | 98.54 | 98.51 | 0.70 | 93.83 | 99.74 | 99.68 | 0.86 | 86.91 | 99.66 | 99.52 | 0.80 | 89.38 | 99.48 | 98.53 | 0.59 |
| KW-0195 | Cyclin | GO:0061575 | 89.34 | 98.92 | 98.89 | 0.44 | 97.96 | 99.78 | 99.78 | 0.60 | 89.80 | 99.84 | 99.83 | 0.62 | 75.51 | 99.63 | 99.53 | 0.39 |
| KW-0251 | Elongation factor | GO:0003746 | 99.51 | 98.57 | 98.60 | 0.83 | 96.72 | 99.67 | 99.62 | 0.92 | 84.14 | 99.67 | 99.29 | 0.85 | 95.84 | 99.46 | 97.21 | 0.63 |
| KW-0339 | Growth factor | GO:0008083 | 94.05 | 98.99 | 98.95 | 0.65 | 84.30 | 99.69 | 99.62 | 0.76 | 86.01 | 99.81 | 99.72 | 0.80 | 76.54 | 99.66 | 99.16 | 0.61 |
| KW-0343 | GTPase activation | GO:0005096 | 76.06 | 98.57 | 98.40 | 0.47 | 92.45 | 99.67 | 99.65 | 0.66 | 86.73 | 99.82 | 99.78 | 0.72 | 61.95 | 99.44 | 99.25 | 0.46 |
| KW-0344 | Guanine-nucleotide releasing factor | GO:0005085 | 74.09 | 98.57 | 98.44 | 0.39 | 83.33 | 99.72 | 99.69 | 0.57 | 89.74 | 99.64 | 99.62 | 0.56 | 93.59 | 99.15 | 98.95 | 0.31 |
| KW-0396 | Initiation factor | GO:0003743 | 96.88 | 98.92 | 98.86 | 0.83 | 91.36 | 99.66 | 99.50 | 0.87 | 74.21 | 99.82 | 99.32 | 0.81 | 77.64 | 99.45 | 97.98 | 0.65 |
| KW-0497 | Mitogen | GO:0051781 | 89.25 | 98.98 | 98.96 | 0.40 | 92.74 | 99.73 | 99.66 | 0.85 | 83.60 | 99.61 | 99.45 | 0.75 | 85.22 | 99.62 | 98.78 | 0.62 |
| KW-0505 | Motor protein | GO:0098840 | 93.38 | 98.96 | 98.91 | 0.63 | 89.47 | 99.75 | 99.72 | 0.69 | 80.70 | 99.86 | 99.80 | 0.72 | 64.04 | 99.45 | 99.25 | 0.46 |
| KW-0514 | Muscle protein | - | 94.22 | 98.95 | 98.92 | 0.57 | 95.38 | 99.75 | 99.73 | 0.74 | 89.23 | 99.69 | 99.65 | 0.67 | 80.00 | 99.60 | 99.32 | 0.51 |
| KW-0515 | Mutator protein | GO:1990633 | 97.65 | 98.97 | 98.97 | 0.42 | 83.82 | 99.79 | 99.76 | 0.60 | 77.94 | 99.84 | 99.80 | 0.61 | 70.59 | 99.45 | 99.32 | 0.38 |
| KW-0568 | Pathogenesis related protein | GO:0009607 | 92.86 | 98.98 | 98.97 | 0.29 | 93.36 | 99.78 | 99.74 | 0.89 | 94.87 | 99.63 | 99.58 | 0.84 | 91.20 | 99.71 | 98.72 | 0.64 |
| KW-0734 | Signal transduction inhibitor | GO:0009968 | 81.25 | 98.97 | 98.94 | 0.31 | 84.62 | 99.71 | 99.69 | 0.45 | 84.62 | 99.68 | 99.66 | 0.43 | 87.18 | 99.63 | 99.54 | 0.34 |
| KW-0786 | Thiamine pyrophosphate binding | - | 97.08 | 98.95 | 98.93 | 0.71 | 96.04 | 99.73 | 99.70 | 0.85 | 87.70 | 99.89 | 99.79 | 0.87 | 74.76 | 99.43 | 98.80 | 0.58 |
| KW-0830 | Ubiquinone binding | - | 98.37 | 98.50 | 98.49 | 0.87 | 94.07 | 99.72 | 99.56 | 0.92 | 82.58 | 99.46 | 98.98 | 0.82 | 91.47 | 99.73 | 97.20 | 0.68 |
| KW-0847 | Vitamin C binding | GO:0031418 | 94.21 | 98.96 | 98.94 | 0.46 | 91.89 | 99.79 | 99.78 | 0.53 | 97.30 | 99.69 | 99.69 | 0.48 | 81.08 | 99.64 | 99.56 | 0.35 |
The amphibian defense peptide family (KW-0878; KW, keyword) was the family with the highest SE (99.99%) for SVM, BLAST and KNN, which was known to be a rich source of antimicrobial peptides with a broad spectrum of antimicrobial activities against pathogenic microorganisms [74,75,76]. The superior SE of this family may come from its nature as a conserved element of the defense system of various species [77].
2.3. In-Depth Assessment of the False Discovery Rate by Genome Scanning
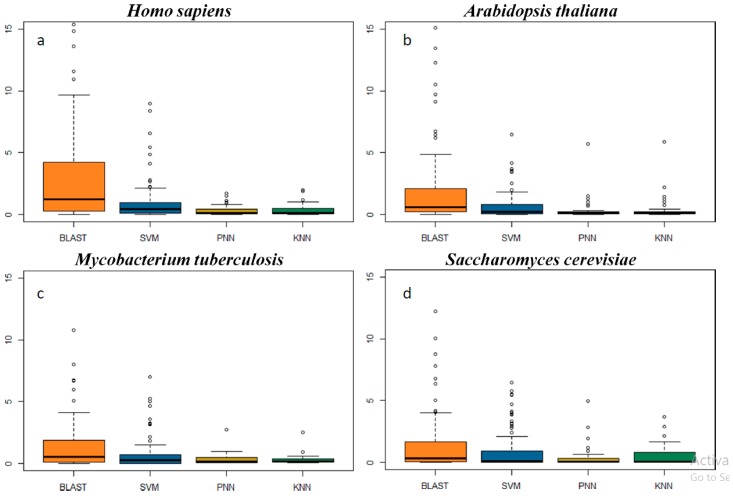
Genome scanning has been frequently used to evaluate the false discovery rate of function prediction tools [78,79]. To have a comprehensive understanding of methods’ false discovery rate, the genomes of four model organisms representing four kingdoms (Homo sapiens from Animalia, Arabidopsis thaliana from Plantae, saccharomyces cerevisiae from Fungi and Mycobacterium tuberculosis from Bacteria) were collected. As demonstrated in Table 2 and Table S2, the genome scanning revealed that the number of proteins in any of those 93 studied families predicted by SVM, PNN and KNN did not exceed 10% of the total number of proteins in the whole genome, and this was the same situation for the majority (82%) of the 93 studied families by BLAST. The higher number of proteins predicted for a certain functional family may indicate a higher false discovery rate [78,79]. For the human genome, the number of proteins identified by SVM was equivalent to or was slightly higher than that of both PNN and KNN, but was significantly lower than that of BLAST (Figure 2a). In addition, the proteins identified by PNN were lower than that of KNN in 11 families and higher in 20 families.
Table 2.
The false discovery rate assessed by the percentage of proteins identified from human and thaliana genomes by different algorithms.
| UniProt Keyword | Protein Functional Family | Homo Sapiens | Arabidopsis Thaliana | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UniProt (%) | SVM (%) | BLAST (%) | PNN (%) | KNN (%) | UniProt (%) | SVM (%) | BLAST (%) | PNN (%) | KNN (%) | ||
| KW-0117 | Actin capping | 0.09 | 0.12 | 0.72 | 0.10 | 0.10 | 0.05 | 0.07 | 0.11 | 0.05 | 0.05 |
| KW-0020 | Allergen | 0.02 | 0.18 | 3.68 | 0.11 | 0.04 | 0.01 | 0.17 | 6.22 | 0.07 | 0.09 |
| KW-0049 | Antioxidant | 0.07 | 0.09 | 0.50 | 0.08 | 0.07 | 0.09 | 0.16 | 1.11 | 0.12 | 0.13 |
| KW-0147 | Chitin-binding | 0.02 | 0.16 | 0.36 | 0.02 | 0.10 | 0.08 | 0.24 | 3.57 | 0.08 | 0.18 |
| KW-0157 | Chromophore | 0.07 | 0.15 | 2.10 | 0.07 | 0.10 | 0.28 | 0.38 | 0.88 | 0.23 | 0.30 |
| KW-0195 | Cyclin | 0.16 | 0.24 | 0.40 | 0.18 | 0.19 | 0.33 | 0.36 | 0.61 | 0.34 | 0.34 |
| KW-0251 | Elongation factor | 0.08 | 0.11 | 0.45 | 0.08 | 0.09 | 0.15 | 0.19 | 0.48 | 0.14 | 0.16 |
| KW-0339 | Growth factor | 0.65 | 0.93 | 2.50 | 0.71 | 0.73 | 0.12 | 0.18 | 0.24 | 0.13 | 0.14 |
| KW-0343 | GTPase activation | 0.97 | 1.19 | 5.47 | 0.93 | 1.02 | 0.28 | 0.24 | 1.36 | 0.21 | 0.23 |
| KW-0344 | Guanine-nucleotide releasing factor | 0.73 | 0.86 | 5.37 | 0.73 | 0.75 | 0.18 | 0.20 | 2.12 | 0.17 | 0.19 |
| KW-0396 | Initiation factor | 0.24 | 0.39 | 1.70 | 0.26 | 0.25 | 0.26 | 0.38 | 1.71 | 0.24 | 0.28 |
| KW-0497 | Mitogen | 0.20 | 0.65 | 4.37 | 0.30 | 0.35 | 0.00 | 0.07 | 0.52 | 0.01 | 0.02 |
| KW-0505 | Motor protein | 0.66 | 0.75 | 4.07 | 0.67 | 0.67 | 0.59 | 0.45 | 2.14 | 0.34 | 0.42 |
| KW-0514 | Muscle protein | 0.31 | 0.42 | 4.35 | 0.37 | 0.39 | 0.00 | 0.17 | 1.26 | 0.11 | 0.13 |
| KW-0515 | Mutator protein | 0.01 | 0.02 | 0.05 | 0.01 | 0.01 | 0.01 | 0.01 | 0.05 | 0.01 | 0.01 |
| KW-0568 | Pathogenesis-related protein | 0.00 | 0.08 | 0.09 | 0.04 | 0.05 | 0.13 | 0.20 | 0.91 | 0.15 | 0.16 |
| KW-0734 | Signal transduction inhibitor | 0.22 | 0.23 | 1.22 | 0.21 | 0.21 | 0.01 | 0.01 | 0.74 | 0.01 | 0.01 |
| KW-0786 | Thiamine pyrophosphate binding | 0.06 | 0.07 | 0.13 | 0.06 | 0.06 | 0.12 | 0.15 | 0.28 | 0.13 | 0.14 |
| KW-0830 | Ubiquinone binding | 0.08 | 0.71 | 0.12 | 0.19 | 0.60 | 0.13 | 0.25 | 0.42 | 0.17 | 0.18 |
| KW-0847 | Vitamin C binding | 0.10 | 0.12 | 0.18 | 0.10 | 0.09 | 0.07 | 0.11 | 0.53 | 0.07 | 0.08 |
Figure 2.
The false discovery rates reflected by the percentage of proteins identified from the genomes of (a) Homo sapiens, (b) Arabidopsis thaliana, (c) Saccharomyces cerevisiae and (d) Mycobacterium tuberculosis.
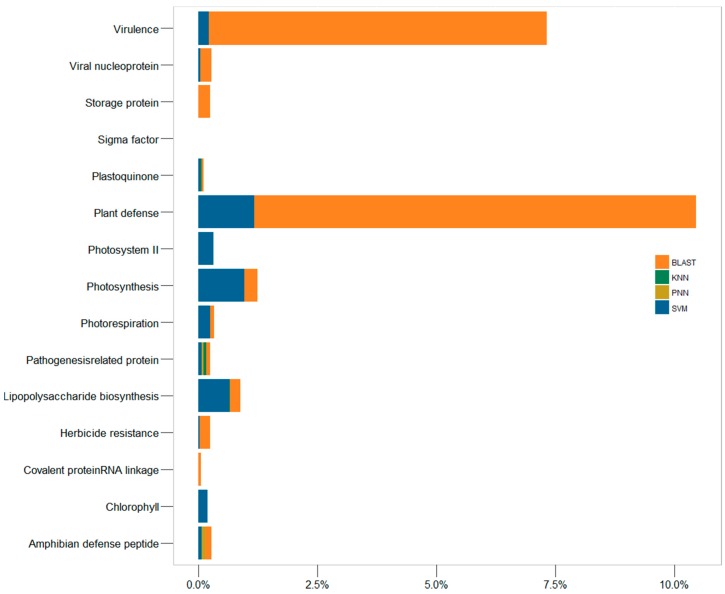
Moreover, 15 protein families only existing in plants, microbes or viruses (Table S3, not existing in the human genome) were collected for assessing the false discovery rate of each algorithm. For example, the covalent protein-RNA linkage family (KW-0191) contained proteins attaching covalently to the RNA molecules in virus [80], and the storage protein (KW-0758) included the proteins as a source of nutrients for the development or growth of the organism in plants. For these families (Table S3), SVM did not identify any proteins from the human genome, while 0.06% and 0.25% of the proteins in the human genome were falsely assigned by BLAST to the family of covalent protein-RNA linkage protein and storage protein, respectively. As illustrated in Figure 3, several other families (such as plant defense, virulence) also demonstrated a significantly higher false discovery rate by BLAST than that of SVM.
Figure 3.
The false discovery rates reflected by the percentage of proteins of 15 protein families only existing in plants, microbes or viruses, but not existing in the human genome identified from the genomes of Homo sapiens.
For the other three genomes, their situation was similar to the human genome. Take the Arabidopsis thaliana genome as an example: proteins identified by SVM were equivalent to or slightly higher than those by PNN and KNN in all protein families, but lower than that of BLAST in 77 families, and the number of protein discovered by PNN was lower than that of KNN in 26 families. In summary, the level of false discovery rate (Figure 2b–d) could be ordered as BLAST > SVM > PNN and KNN. These results revealed that BLAST was more prone to generate a false discovery rate than the other three machine learning methods (SVM > PNN ≈ KNN).
As reported [81,82,83,84,85], an open web-server is recognized as useful for constructing effective methods and tools. A variety of web-servers have increasing impacts on medical sciences [86], driving medicinal chemistry to an unprecedented revolution [87], and efforts will be further made to develop web-based services for the performance assessment discussed in this study.
3. Materials and Methods
To construct a valid statistical model for a biology problem based on protein sequences [88,89,90,91,92,93,94,95,96,97], a rule of five steps is needed [98]. Firstly, a valid construction of datasets for both training and testing the model is required. Secondly, an effective conversion of the sequence to the digital feature vector is asked to represent their targeted properties. Thirdly, a powerful statistical method should be designed for the functional prediction. Fourthly, the accuracies of the constructed statistic model should be validated correctly. Fifthly, a web-server based on the constructed model may be further developed for public access. The corresponding methods and steps adopted in this study are provided and described below.
3.1. Collecting the Protein Sequences of Different Functional Families
Table 1 provides a full list of 93 protein families collected from UniProt [43], and the performances of the popular protein function prediction methods (BLAST, KNN, PNN, SVM) were measured via independent test datasets (the way to generate an independent dataset is shown in the following Section 2.2). These 93 included 12 families of binding molecules (e.g., sodium-, potassium-, SH3- and RNA-binding), 15 ligand families (e.g., plastoquinone ligand, vitamin C ligand and ubiquinone ligand), 58 families defined by Gene Ontology (40 molecular functions and 18 biological processes) and 8 broad families defined by UniProt [43]. All families were contained in the keyword categories of UniProt, and the majority (82.7%) of these 93 families were able to be mapped to GO terms (Table 1). Protein entries that have not been manually annotated and reviewed by UniProtKB curators in a keyword category were not considered for analysis in this study. As a result, 107~49,517 protein-entries from 93 families were collected.
3.2. Construction of the Training and Testing Datasets
The independent test dataset was frequently constructed to evaluate the performances of protein function predictors in recent years [99,100,101,102,103,104]. To construct a valid set of data for building the predictor of each family, the datasets of the training, testing and independent test were generated by a strictly defined process after the data collection described in Section 2.1. Firstly, all proteins of different sequences in a specific family are assigned randomly with a number, which is within the range of the total number of proteins in that family. Secondly, these sequences in each protein function family were sequentially selected based on the number assigned and then iteratively added to the training, testing and independent test datasets. Samples in these datasets are all known as the positive samples. Thirdly, the Pfam families [16] of the proteins of a certain functional family were retrieved from the Pfam database [16] for generating negative samples. The Pfam family with protein(s) of this functional family was defined as the “positive” one, and the remaining families were grouped into the “negative” ones. Finally, 3 representatives were randomly picked out of the negative families and sequentially added to the training, testing and independent test datasets, and samples in these datasets are thus known as the negative samples. It is necessary to emphasize that there was no overlap among the datasets of the training, testing and independent test [60,61].
To assess the false discovery rate among algorithms, the genomes of four model organisms representing four kingdoms (Homo sapiens from Animalia, Arabidopsis thaliana from Plantae, Saccharomyces cerevisiae from Fungi and Mycobacterium tuberculosis from Bacteria) were collected from UniProt. The protein entries without any manual annotation and review by the UniProtKB curators were not taken into consideration. In total, 20,183, 15,169, 6721 and 2166 protein sequences in FASTA format were collected for human, Arabidopsis thaliana, Saccharomyces cerevisiae and Mycobacterium tuberculosis, respectively.
3.3. Feature Vectors Used for Representing the Protein Sequence
The conversion of the protein sequence into the digital feature vector was conducted based on properties of each residue within that protein. These properties include: (1) charge; (2) polarizability; (3) polarity; (4) surface tension; (5) amino acid (AA) composition; (6) van der Waals volume via normalizing; (7) hydrophobicity; (8) solvent accessibility; and (9) protein secondary structure [36,105,106,107]. Then, 3 features were applied to describe each property [36]. These features contained: (a) composition (No. of AAs of a particular property over the total No. of AAs; (b) transition (the percentage of AAs with a certain property was followed by AAs with a different property); and (c) distribution (the sequence lengths within which the first, one fourth, half, three-quarters and all of the AAs of specific property were localized). The detailed procedure for generating the feature vector from the sequence was described in previous publications [36,65]. These features have already been successfully applied to facilitate the prediction of enzyme functional [108] and structural classes [107].
3.4. Functional Prediction of Protein Constructed by Machine Learning
To construct the prediction model, the parameters of machine learning methods were optimized using the testing dataset for each training process. Once suitable parameters were discovered, a new training set was constructed by combining the original training and testing datasets, and the corresponding parameters were directly accepted for training a new model. To assess the performance of the constructed models and detect possible over-fitting, the independent test set was further applied. It is necessary to emphasize that all duplicates in the protein sequence were removed during datasets’ construction.
3.5. Construction of Protein Functional Prediction Model Based on Sequence Similarity
Sequence similarity was assessed by the NCBI Protein-Protein BLAST (Version 2.6.0+) [53,54]. Firstly, the combined training and testing dataset was adopted to form the BLAST database, and the sequences in the independent test dataset were used as queries. The BLAST E-value and percentage sequence identity were usually applied to represent the level of similarity between sequences [109]. The functional variation between proteins was reported to be rare when their sequence identity was more than 40% [110,111]. Thus, an E-value of 0.001 and a sequence identity of 40% were adopted as the cutoffs in this study to assess the functional conservation of BLAST hits.
3.6. Assessing the Identification Accuracies of the Studied Methods
The performance of protein function prediction algorithms was systematically assessed by four popular metrics, sensitivity (SE), specificity (SP), accuracy (ACC) and Matthews correlation coefficient (MCC), based on the independent test datasets generated from the 93 studied families (Supplementary Materials Table S1). All 4 metrics were widely used in assessing the performance of protein function predictors [112,113,114,115,116,117]. In particular, SE is defined by the percentage of true positive samples correctly identified as “positive” [118,119] (shown in Equation (1)):
| (1) |
SP indicates the proportion of true negative samples that were correctly predicted as “negative” [118,119] (in Equation (2)):
| (2) |
ACC refers to the number of true samples (positive plus negative) divided by the number of all samples studied (shown in Equation (3)):
| (3) |
The MCC was an important metric reflecting the stability of a protein function predictor, which described the correlation between a predictive value and an actual value [118,119]. It has been considered as one of the most comprehensive parameters in any category of predictors due to its full consideration of all four results. In particular, the MCC could be calculated by Equation 4:
| (4) |
In particular, those four results were TP (No. of true positive samples), TN (No. of true negative samples), FP (No. of false positive samples) and FN (No. of false negative samples) [118,119]. It is very important to emphasize that these four metrics are applicable to the single-class situations (each protein is grouped into just one family). For the multi-class situations frequently observed in complicated biological networks [81,82,83,84] and biomedical researches [84,89,117], different metrics should be defined [120].
3.7. The Rates of False Discovery of the In Silico Methods Studied Here
As reported, genome scanning was a comprehensive method to evaluate the capacity of protein functional prediction tools in identifying and classifying protein families [78,79]. In this paper, an evaluation of the false discovery rate of the studied protein function predictors was performed by scanning the genomes of 4 model organisms representing 4 kingdoms (Homo sapiens from Animalia, Arabidopsis thaliana from Plantae, Saccharomyces cerevisiae from Fungi and Mycobacterium tuberculosis from Bacteria). The false discovery rates were assessed by reconstructing the prediction models of those in silico algorithms. In particular, the sequences of proteins in a certain functional family were all put into the reference database for BLAST scanning and were also used to reconstruct the machine learning models using the optimized parameters obtained in Section 3.4. In reality, the total amount of proteins not belonging to a certain family should be much larger than that of proteins in that family. Therefore, a tiny reduction in the value of SP may lead to a significant discovery of false positive hits, which reminded us to use SP as an effective indicator when evaluating the model’s false discovery rates.
4. Conclusions
This study discovered substantially higher sensitivity (SP) and stability (MCC) of BLAST and SVM than that of PNN and KNN. However, the machine learning algorithms (PNN, KNN and SVM) were found capable of significantly reducing the false discovery rate (with PNN and KNN performed the best). In conclusion, this study comprehensively assessed the performances of popular algorithms applied to protein function prediction, which could facilitate the selection of the appropriate method in the related biomedical research.
Acknowledgments
This work was funded by the Precision Medicine Project of the National Key Research and Development Plan of China (2016YFC0902200); the Innovation Project on Industrial Generic Key Technologies of Chongqing (cstc2015zdcy-ztzx120003); the Fundamental Research Funds for Central Universities (10611CDJXZ238826, CDJZR14468801, CDJKXB14011); and the National Natural Science Foundation of China (21505009).
Supplementary Materials
The Supplementary Materials are available online at www.mdpi.com/1422-0067/19/1/183/s1.
Author Contributions
Feng Zhu and Lin Tao conceived of and designed the experiments. Chun Yan Yu and Xiao Xu Li carried out most of the experiments in this paper. Chun Yan Yu, Ying Hong Li, Hong Yang, Wei Wei Xue and Yu Zong Chen performed the bioinformatics analysis. Feng Zhu wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Baldi P., Brunak S., Chauvin Y., Andersen C.A., Nielsen H. Assessing the accuracy of prediction algorithms for classification: An overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 2.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg S.E., Chandel N.S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant M.A. Integrating computational protein function prediction into drug discovery initiatives. Drug Dev. Res. 2011;72:4–16. doi: 10.1002/ddr.20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B., Tang J., Yang Q., Li S., Cui X., Li Y., Chen Y., Xue W., Li X., Zhu F. Noreva: Normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017;45:162–170. doi: 10.1093/nar/gkx449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B., Tang J., Yang Q., Cui X., Li S., Chen S., Cao Q., Xue W., Chen N., Zhu F. Performance evaluation and online realization of data-driven normalization methods used in lc/ms based untargeted metabolomics analysis. Sci. Rep. 2016;6:38881. doi: 10.1038/srep38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J., Wang P., Yang H., Zhou J., Li Y., Li X., Xue W., Yu C., Tian Y., Zhu F. Comparison of FDA approved kinase targets to clinical trial ones: Insights from their system profiles and drug-target interaction networks. BioMed Res. Int. 2016;2016:2509385. doi: 10.1155/2016/2509385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M., et al. Eggnog 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:286–293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D., Jensen L.J. Protein-protein interaction databases. Methods Mol. Biol. 2015;1278:39–56. doi: 10.1007/978-1-4939-2425-7_3. [DOI] [PubMed] [Google Scholar]
- 10.Jeanquartier F., Jean-Quartier C., Holzinger A. Integrated web visualizations for protein-protein interaction databases. BMC Bioinform. 2015;16:195. doi: 10.1186/s12859-015-0615-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M. Stitch 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:380–384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., et al. String v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The string database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The string database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:362–368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y.H., Yu C.Y., Li X.X., Zhang P., Tang J., Yang Q., Fu T., Zhang X., Cui X., Tu G., et al. Therapeutic target database update 2018: Enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H., Qin C., Li Y.H., Tao L., Zhou J., Yu C.Y., Xu F., Chen Z., Zhu F., Chen Y.Z. Therapeutic target database update 2016: Enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:1069–1074. doi: 10.1093/nar/gkv1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu F., Shi Z., Qin C., Tao L., Liu X., Xu F., Zhang L., Song Y., Liu X., Zhang J., et al. Therapeutic target database update 2012: A resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40:1128–1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu F., Han B., Kumar P., Liu X., Ma X., Wei X., Huang L., Guo Y., Han L., Zheng C., et al. Update of ttd: Therapeutic target database. Nucleic Acids Res. 2010;38:787–791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y.H., Wang P.P., Li X.X., Yu C.Y., Yang H., Zhou J., Xue W.W., Tan J., Zhu F. The human kinome targeted by FDA approved multi-target drugs and combination products: A comparative study from the drug-target interaction network perspective. PLoS ONE. 2016;11:e0165737. doi: 10.1371/journal.pone.0165737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu F., Ma X.H., Qin C., Tao L., Liu X., Shi Z., Zhang C.L., Tan C.Y., Chen Y.Z., Jiang Y.Y. Drug discovery prospect from untapped species: Indications from approved natural product drugs. PLoS ONE. 2012;7:e39782. doi: 10.1371/journal.pone.0039782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erdin S., Lisewski A.M., Lichtarge O. Protein function prediction: Towards integration of similarity metrics. Curr. Opin. Struct. Biol. 2011;21:180–188. doi: 10.1016/j.sbi.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K., Chetvernin V., Church D.M., Dicuccio M., Federhen S., et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2012;40:13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrell D., Dimmer E., Huntley R.P., Binns D., O’Donovan C., Apweiler R. The goa database in 2009—An integrated gene ontology annotation resource. Nucleic Acids Res. 2009;37:396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The UniProt Consortium Activities at the universal protein resource (UniProt) Nucleic Acids Res. 2014;42:191–198. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bork P., Koonin E.V. Predicting functions from protein sequences—where are the bottlenecks? Nat. Genet. 1998;18:313–318. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- 28.Chitale M., Hawkins T., Park C., Kihara D. ESG: Extended similarity group method for automated protein function prediction. Bioinformatics. 2009;25:1739–1745. doi: 10.1093/bioinformatics/btp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahraeian S.M., Luo K.R., Brenner S.E. Sifter search: A web server for accurate phylogeny-based protein function prediction. Nucleic Acids Res. 2015;43:141–147. doi: 10.1093/nar/gkv461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teichmann S.A., Murzin A.G., Chothia C. Determination of protein function, evolution and interactions by structural genomics. Curr. Opin. Struct. Biol. 2001;11:354–363. doi: 10.1016/S0959-440X(00)00215-3. [DOI] [PubMed] [Google Scholar]
- 32.Enright A.J., Iliopoulos I., Kyrpides N.C., Ouzounis C.A. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 33.Aravind L. Guilt by association: Contextual information in genome analysis. Genome Res. 2000;10:1074–1077. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- 34.Kotlyar M., Pastrello C., Pivetta F., Lo Sardo A., Cumbaa C., Li H., Naranian T., Niu Y., Ding Z., Vafaee F., et al. In silico prediction of physical protein interactions and characterization of interactome orphans. Nat. Methods. 2015;12:79–84. doi: 10.1038/nmeth.3178. [DOI] [PubMed] [Google Scholar]
- 35.Jensen L.J., Gupta R., Staerfeldt H.H., Brunak S. Prediction of human protein function according to gene ontology categories. Bioinformatics. 2003;19:635–642. doi: 10.1093/bioinformatics/btg036. [DOI] [PubMed] [Google Scholar]
- 36.Cai C.Z., Han L.Y., Ji Z.L., Chen X., Chen Y.Z. SVM-prot: Web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003;31:3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lobley A.E., Nugent T., Orengo C.A., Jones D.T. Ffpred: An integrated feature-based function prediction server for vertebrate proteomes. Nucleic Acids Res. 2008;36:297–302. doi: 10.1093/nar/gkn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu F., Qin C., Tao L., Liu X., Shi Z., Ma X., Jia J., Tan Y., Cui C., Lin J., et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc. Natl. Acad. Sci. USA. 2011;108:12943–12948. doi: 10.1073/pnas.1107336108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das S., Sillitoe I., Lee D., Lees J.G., Dawson N.L., Ward J., Orengo C.A. Cath funfhmmer web server: Protein functional annotations using functional family assignments. Nucleic Acids Res. 2015;43:148–153. doi: 10.1093/nar/gkv488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P., Zhang X., Fu T., Li S., Li B., Xue W., Yao X., Chen Y., Zhu F. Differentiating physicochemical properties between addictive and nonaddictive adhd drugs revealed by molecular dynamics simulation studies. ACS Chem. Neurosci. 2017;8:1416–1428. doi: 10.1021/acschemneuro.7b00173. [DOI] [PubMed] [Google Scholar]
- 41.Xue W., Wang P., Li B., Li Y., Xu X., Yang F., Yao X., Chen Y.Z., Xu F., Zhu F. Identification of the inhibitory mechanism of fda approved selective serotonin reuptake inhibitors: An insight from molecular dynamics simulation study. Phys. Chem. Chem. Phys. 2016;18:3260–3271. doi: 10.1039/C5CP05771J. [DOI] [PubMed] [Google Scholar]
- 42.Zheng G., Xue W., Wang P., Yang F., Li B., Li X., Li Y., Yao X., Zhu F. Exploring the inhibitory mechanism of approved selective norepinephrine reuptake inhibitors and reboxetine enantiomers by molecular dynamics study. Sci. Rep. 2016;6:26883. doi: 10.1038/srep26883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P., Yang F., Yang H., Xu X., Liu D., Xue W., Zhu F. Identification of dual active agents targeting 5-ht1a and sert by combinatorial virtual screening methods. Biomed. Mater. Eng. 2015;26(Suppl. 1):2233–2239. doi: 10.3233/BME-151529. [DOI] [PubMed] [Google Scholar]
- 44.Li D., Ju Y., Zou Q. Protein folds prediction with hierarchical structured SVM. Curr. Proteom. 2016;13:79–85. doi: 10.2174/157016461302160514000940. [DOI] [Google Scholar]
- 45.Wei L., Tang J., Zou Q. Skipcpp-pred: An improved and promising sequence-based predictor for predicting cell-penetrating peptides. BMC Genom. 2017;18(Suppl. 7):742. doi: 10.1186/s12864-017-4128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan S., Duan Y., Zou Q. Hpslpred: An ensemble multi-label classifier for human protein subcellular location prediction with imbalanced source. Proteomics. 2017;17 doi: 10.1002/pmic.201700262. [DOI] [PubMed] [Google Scholar]
- 47.Wei L., Xing P., Su R., Shi G., Ma Z.S., Zou Q. Cppred-rf: A sequence-based predictor for identifying cell-penetrating peptides and their uptake efficiency. J. Proteome Res. 2017;16:2044–2053. doi: 10.1021/acs.jproteome.7b00019. [DOI] [PubMed] [Google Scholar]
- 48.Friedberg I., Harder T., Godzik A. JAFA: A protein function annotation meta-server. Nucleic Acids Res. 2006;34:379–381. doi: 10.1093/nar/gkl045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wass M.N., Barton G., Sternberg M.J. Combfunc: Predicting protein function using heterogeneous data sources. Nucleic Acids Res. 2012;40:466–470. doi: 10.1093/nar/gks489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. Interproscan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piovesan D., Giollo M., Leonardi E., Ferrari C., Tosatto S.C. Inga: Protein function prediction combining interaction networks, domain assignments and sequence similarity. Nucleic Acids Res. 2015;43:134–140. doi: 10.1093/nar/gkv523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bandyopadhyay S., Ray S., Mukhopadhyay A., Maulik U. A review of in silico approaches for analysis and prediction of hiv-1-human protein-protein interactions. Brief. Bioinform. 2015;16:830–851. doi: 10.1093/bib/bbu041. [DOI] [PubMed] [Google Scholar]
- 53.Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y., et al. Blast: A more efficient report with usability improvements. Nucleic Acids Res. 2013;41:29–33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearson W.R. Blast and fasta similarity searching for multiple sequence alignment. Methods Mol. Biol. 2014;1079:75–101. doi: 10.1007/978-1-62703-646-7_5. [DOI] [PubMed] [Google Scholar]
- 55.Radivojac P., Clark W.T., Oron T.R., Schnoes A.M., Wittkop T., Sokolov A., Graim K., Funk C., Verspoor K., Ben-Hur A., et al. A large-scale evaluation of computational protein function prediction. Nat. Methods. 2013;10:221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang Y., Oron T.R., Clark W.T., Bankapur A.R., D’Andrea D., Lepore R., Funk C.S., Kahanda I., Verspoor K.M., Ben-Hur A., et al. An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Biol. 2016;17:184. doi: 10.1186/s13059-016-1037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang Y., Zhang S. Predict protein structural class by incorporating two different modes of evolutionary information into chou’s general pseudo amino acid composition. J. Mol. Graph. Model. 2017;78:110–117. doi: 10.1016/j.jmgm.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Pradhan D., Padhy S., Sahoo B. Enzyme classification using multiclass support vector machine and feature subset selection. Comput. Biol. Chem. 2017;70:211–219. doi: 10.1016/j.compbiolchem.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Meher P.K., Sahu T.K., Banchariya A., Rao A.R. Dirprot: A computational approach for discriminating insecticide resistant proteins from non-resistant proteins. BMC Bioinform. 2017;18:190. doi: 10.1186/s12859-017-1587-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu F., Han L., Zheng C., Xie B., Tammi M.T., Yang S., Wei Y., Chen Y. What are next generation innovative therapeutic targets? Clues from genetic, structural, physicochemical, and systems profiles of successful targets. J. Pharmacol. Exp. Ther. 2009;330:304–315. doi: 10.1124/jpet.108.149955. [DOI] [PubMed] [Google Scholar]
- 61.Zhu F., Han L.Y., Chen X., Lin H.H., Ong S., Xie B., Zhang H.L., Chen Y.Z. Homology-free prediction of functional class of proteins and peptides by support vector machines. Curr. Protein Pept. Sci. 2008;9:70–95. doi: 10.2174/138920308783565697. [DOI] [PubMed] [Google Scholar]
- 62.Zhu F., Zheng C.J., Han L.Y., Xie B., Jia J., Liu X., Tammi M.T., Yang S.Y., Wei Y.Q., Chen Y.Z. Trends in the exploration of anticancer targets and strategies in enhancing the efficacy of drug targeting. Curr. Mol. Pharmacol. 2008;1:213–232. doi: 10.2174/1874467210801030213. [DOI] [PubMed] [Google Scholar]
- 63.Li Y.H., Xu J.Y., Tao L., Li X.F., Li S., Zeng X., Chen S.Y., Zhang P., Qin C., Zhang C., et al. SVM-prot 2016: A web-server for machine learning prediction of protein functional families from sequence irrespective of similarity. PLoS ONE. 2016;11:e0155290. doi: 10.1371/journal.pone.0155290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai C.Z., Han L.Y., Ji Z.L., Chen Y.Z. Enzyme family classification by support vector machines. Proteins. 2004;55:66–76. doi: 10.1002/prot.20045. [DOI] [PubMed] [Google Scholar]
- 65.Han L.Y., Cai C.Z., Ji Z.L., Cao Z.W., Cui J., Chen Y.Z. Predicting functional family of novel enzymes irrespective of sequence similarity: A statistical learning approach. Nucleic Acids Res. 2004;32:6437–6444. doi: 10.1093/nar/gkh984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen H.B., Yang J., Chou K.C. Fuzzy KNN for predicting membrane protein types from pseudo-amino acid composition. J. Theor. Biol. 2006;240:9–13. doi: 10.1016/j.jtbi.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 67.Nath N., Mitchell J.B. Is EC class predictable from reaction mechanism? BMC Bioinform. 2012;13:60. doi: 10.1186/1471-2105-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naveed M., Khan A. Gpcr-mpredictor: Multi-level prediction of g protein-coupled receptors using genetic ensemble. Amino Acids. 2012;42:1809–1823. doi: 10.1007/s00726-011-0902-6. [DOI] [PubMed] [Google Scholar]
- 69.Hayat M., Khan A. Predicting membrane protein types by fusing composite protein sequence features into pseudo amino acid composition. J. Theor. Biol. 2011;271:10–17. doi: 10.1016/j.jtbi.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 70.Khan Z.U., Hayat M., Khan M.A. Discrimination of acidic and alkaline enzyme using chou’s pseudo amino acid composition in conjunction with probabilistic neural network model. J. Theor. Biol. 2015;365:197–203. doi: 10.1016/j.jtbi.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 71.Li H., Yap C.W., Ung C.Y., Xue Y., Li Z.R., Han L.Y., Lin H.H., Chen Y.Z. Machine learning approaches for predicting compounds that interact with therapeutic and ADMET related proteins. J. Pharm. Sci. 2007;96:2838–2860. doi: 10.1002/jps.20985. [DOI] [PubMed] [Google Scholar]
- 72.Fujimoto M.S., Suvorov A., Jensen N.O., Clement M.J., Bybee S.M. Detecting false positive sequence homology: A machine learning approach. BMC Bioinform. 2016;17:101. doi: 10.1186/s12859-016-0955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pearson W.R. Protein function prediction: Problems and pitfalls. Curr. Protoc. Bioinform. 2015;51:1–18. doi: 10.1002/0471250953.bi0412s51. [DOI] [PubMed] [Google Scholar]
- 74.Boman H.G. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 75.Hancock R.E., Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8:402–410. doi: 10.1016/S0966-842X(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 76.Radek K., Gallo R. Antimicrobial peptides: Natural effectors of the innate immune system. Semin. Immunopathol. 2007;29:27–43. doi: 10.1007/s00281-007-0064-5. [DOI] [PubMed] [Google Scholar]
- 77.Iwamuro S., Kobayashi T. An efficient protocol for DNA amplification of multiple amphibian skin antimicrobial peptide cDNAs. Methods Mol. Biol. 2010;615:159–176. doi: 10.1007/978-1-60761-535-4_13. [DOI] [PubMed] [Google Scholar]
- 78.Brown J.B., Akutsu T. Identification of novel DNA repair proteins via primary sequence, secondary structure, and homology. BMC Bioinform. 2009;10:25. doi: 10.1186/1471-2105-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crappe J., Van Criekinge W., Trooskens G., Hayakawa E., Luyten W., Baggerman G., Menschaert G. Combining in silico prediction and ribosome profiling in a genome-wide search for novel putatively coding sorfs. BMC Genom. 2013;14:648. doi: 10.1186/1471-2164-14-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Virgen-Slane R., Rozovics J.M., Fitzgerald K.D., Ngo T., Chou W., van der Heden van Noort G.J., Filippov D.V., Gershon P.D., Semler B.L. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc. Natl. Acad. Sci. USA. 2012;109:14634–14639. doi: 10.1073/pnas.1208096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheng X., Xiao X., Chou K.C. pLoc-mPlant: Predict subcellular localization of multi-location plant proteins by incorporating the optimal go information into general PseAAC. Mol. Biosyst. 2017;13:1722–1727. doi: 10.1039/C7MB00267J. [DOI] [PubMed] [Google Scholar]
- 82.Cheng X., Xiao X., Chou K.C. pLoc-mEuk: Predict subcellular localization of multi-label eukaryotic proteins by extracting the key go information into general PseAAC. Genomics. 2018;110:50–58. doi: 10.1016/j.ygeno.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 83.Cheng X., Xiao X., Chou K.C. pLoc-mVirus: Predict subcellular localization of multi-location virus proteins via incorporating the optimal go information into general PseAAC. Gene. 2017;628:315–321. doi: 10.1016/j.gene.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 84.Cheng X., Zhao S.G., Lin W.Z., Xiao X., Chou K.C. Ploc-manimal: Predict subcellular localization of animal proteins with both single and multiple sites. Bioinformatics. 2017;33:3524–3531. doi: 10.1093/bioinformatics/btx476. [DOI] [PubMed] [Google Scholar]
- 85.Qiu W.R., Sun B.Q., Xiao X., Xu Z.C., Jia J.H., Chou K.C. iKCR-PseENs: Identify lysine crotonylation sites in histone proteins with pseudo components and ensemble classifier. Genomics. 2017 doi: 10.1016/j.ygeno.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 86.Chou K.C. Impacts of bioinformatics to medicinal chemistry. Med. Chem. 2015;11:218–234. doi: 10.2174/1573406411666141229162834. [DOI] [PubMed] [Google Scholar]
- 87.Chou K.C. An unprecedented revolution in medicinal chemistry driven by the progress of biological science. Curr. Top. Med. Chem. 2017;17:2337–2358. doi: 10.2174/1568026617666170414145508. [DOI] [PubMed] [Google Scholar]
- 88.Chen W., Feng P., Yang H., Ding H., Lin H., Chou K.C. iRNA-AI: Identifying the adenosine to inosine editing sites in RNA sequences. Oncotarget. 2017;8:4208–4217. doi: 10.18632/oncotarget.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng X., Zhao S.G., Xiao X., Chou K.C. iATC-mISF: A multi-label classifier for predicting the classes of anatomical therapeutic chemicals. Bioinformatics. 2017;33:341–346. doi: 10.1093/bioinformatics/btx387. [DOI] [PubMed] [Google Scholar]
- 90.Feng P., Ding H., Yang H., Chen W., Lin H., Chou K.C. iRNA-PseCOLL: Identifying the occurrence sites of different rna modifications by incorporating collective effects of nucleotides into PseKNC. Mol. Ther. Nucleic Acids. 2017;7:155–163. doi: 10.1016/j.omtn.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu B., Wang S., Long R., Chou K.C. iRSpot-EL: Identify recombination spots with an ensemble learning approach. Bioinformatics. 2017;33:35–41. doi: 10.1093/bioinformatics/btw539. [DOI] [PubMed] [Google Scholar]
- 92.Liu B., Yang F., Chou K.C. 2l-pirna: A two-layer ensemble classifier for identifying piwi-interacting RNAS and their function. Mol. Ther. Nucleic Acids. 2017;7:267–277. doi: 10.1016/j.omtn.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu L.M., Xu Y., Chou K.C. iPGK-PseAAC: Identify lysine phosphoglycerylation sites in proteins by incorporating four different tiers of amino acid pairwise coupling information into the general PseAAC. Med. Chem. 2017;13:552–559. doi: 10.2174/1573406413666170515120507. [DOI] [PubMed] [Google Scholar]
- 94.Qiu W.R., Jiang S.Y., Xu Z.C., Xiao X., Chou K.C. iRNAm5C-PseDNC: Identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget. 2017;8:41178–41188. doi: 10.18632/oncotarget.17104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qiu W.R., Sun B.Q., Xiao X., Xu D., Chou K.C. iPhos-PseEVO: Identifying human phosphorylated proteins by incorporating evolutionary information into general PseAAC via grey system theory. Mol. Inform. 2017;36 doi: 10.1002/minf.201600010. [DOI] [PubMed] [Google Scholar]
- 96.Su Q., Lu W., Du D., Chen F., Niu B., Chou K.C. Prediction of the aquatic toxicity of aromatic compounds to tetrahymena pyriformis through support vector regression. Oncotarget. 2017;8:49359–49369. doi: 10.18632/oncotarget.17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu Y., Wang Z., Li C., Chou K.C. iPreny-PseAAC: Identify c-terminal cysteine prenylation sites in proteins by incorporating two tiers of sequence couplings into PseAAC. Med. Chem. 2017;13:544–551. doi: 10.2174/1573406413666170419150052. [DOI] [PubMed] [Google Scholar]
- 98.Chou K.C. Some remarks on protein attribute prediction and pseudo amino acid composition. J. Theor. Biol. 2011;273:236–247. doi: 10.1016/j.jtbi.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chowdhury S.Y., Shatabda S., Dehzangi A. iDNAProt-ES: Identification of DNA-binding proteins using evolutionary and structural features. Sci. Rep. 2017;7:14938. doi: 10.1038/s41598-017-14945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Filos D., Chouvarda I., Tachmatzidis D., Vassilikos V., Maglaveras N. Beat-to-beat p-wave morphology as a predictor of paroxysmal atrial fibrillation. Comput. Methods Progr. Biomed. 2017;151:111–121. doi: 10.1016/j.cmpb.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 101.Rahimi M., Bakhtiarizadeh M.R., Mohammadi-Sangcheshmeh A. Oogenesis_pred: A sequence-based method for predicting oogenesis proteins by six different modes of chou’s pseudo amino acid composition. J. Theor. Biol. 2017;414:128–136. doi: 10.1016/j.jtbi.2016.11.028. [DOI] [PubMed] [Google Scholar]
- 102.Sun M.A., Zhang Q., Wang Y., Ge W., Guo D. Prediction of redox-sensitive cysteines using sequential distance and other sequence-based features. BMC Bioinform. 2016;17:316. doi: 10.1186/s12859-016-1185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y., Li X., Tao B. Improving classification of mature microrna by solving class imbalance problem. Sci. Rep. 2016;6:25941. doi: 10.1038/srep25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meher P.K., Sahu T.K., Rao A.R. Prediction of donor splice sites using random forest with a new sequence encoding approach. BioData Min. 2016;9:4. doi: 10.1186/s13040-016-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bock J.R., Gough D.A. Predicting protein—Protein interactions from primary structure. Bioinformatics. 2001;17:455–460. doi: 10.1093/bioinformatics/17.5.455. [DOI] [PubMed] [Google Scholar]
- 106.Karchin R., Karplus K., Haussler D. Classifying g-protein coupled receptors with support vector machines. Bioinformatics. 2002;18:147–159. doi: 10.1093/bioinformatics/18.1.147. [DOI] [PubMed] [Google Scholar]
- 107.Dobson P.D., Doig A.J. Distinguishing enzyme structures from non-enzymes without alignments. J. Mol. Biol. 2003;330:771–783. doi: 10.1016/S0022-2836(03)00628-4. [DOI] [PubMed] [Google Scholar]
- 108.Des Jardins M., Karp P.D., Krummenacker M., Lee T.J., Ouzounis C.A. Prediction of enzyme classification from protein sequence without the use of sequence similarity. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:92–99. [PubMed] [Google Scholar]
- 109.Du R., Mercante D., Fang Z. An artificial functional family filter in homolog searching in next-generation sequencing metagenomics. PLoS ONE. 2013;8:e58669. doi: 10.1371/journal.pone.0058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tian W., Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 111.Wommack K.E., Bhavsar J., Ravel J. Metagenomics: Read length matters. Appl. Environ. Microbiol. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ju Z., He J.J. Prediction of lysine propionylation sites using biased svm and incorporating four different sequence features into chou’s pseaac. J. Mol. Graph. Model. 2017;76:356–363. doi: 10.1016/j.jmgm.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 113.Jia J., Liu Z., Xiao X., Liu B., Chou K.C. iPPI-Esml: An ensemble classifier for identifying the interactions of proteins by incorporating their physicochemical properties and wavelet transforms into PseAAC. J. Theor. Biol. 2015;377:47–56. doi: 10.1016/j.jtbi.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 114.Jia J., Liu Z., Xiao X., Liu B., Chou K.C. iCAR-PseCp: Identify carbonylation sites in proteins by monte carlo sampling and incorporating sequence coupled effects into general PseAAC. Oncotarget. 2016;7:34558–34570. doi: 10.18632/oncotarget.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu B., Long R., Chou K.C. iDHS-EL: Identifying DNASE I hypersensitive sites by fusing three different modes of pseudo nucleotide composition into an ensemble learning framework. Bioinformatics. 2016;32:2411–2418. doi: 10.1093/bioinformatics/btw186. [DOI] [PubMed] [Google Scholar]
- 116.Liu Z., Xiao X., Yu D.J., Jia J., Qiu W.R., Chou K.C. pRNAm-PC: Predicting n(6)-methyladenosine sites in rna sequences via physical-chemical properties. Anal. Biochem. 2016;497:60–67. doi: 10.1016/j.ab.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 117.Qiu W.R., Sun B.Q., Xiao X., Xu Z.C., Chou K.C. iPTM-mLys: Identifying multiple lysine ptm sites and their different types. Bioinformatics. 2016;32:3116–3123. doi: 10.1093/bioinformatics/btw380. [DOI] [PubMed] [Google Scholar]
- 118.Xu Y., Shao X.J., Wu L.Y., Deng N.Y., Chou K.C. iSNO-AAPair: Incorporating amino acid pairwise coupling into pseaac for predicting cysteine s-nitrosylation sites in proteins. PeerJ. 2013;1:e171. doi: 10.7717/peerj.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen W., Feng P.M., Lin H., Chou K.C. iRSpot-PseDNC: Identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res. 2013;41:e68. doi: 10.1093/nar/gks1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chou K.C. Some remarks on predicting multi-label attributes in molecular biosystems. Mol. Biosyst. 2013;9:1092–1100. doi: 10.1039/c3mb25555g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.