Abstract
The dysregulation of the ubiquitously transcribed TPR gene on the X chromosome (UTX) has been reported to be involved in the oncogenesis of several types of cancers. However, the expression and significance of UTX in esophageal squamous cell carcinoma (ESCC) remains largely undetermined. Immunohistochemistry was performed in 106 ESCC patients, and correlated with clinicopathological features and survival. The functional role of UTX in ESCC cells was determined by UTX-mediated siRNA. Univariate analyses showed that high UTX expression was associated with superior overall survival (OS, p = 0.011) and disease-free survival (DFS, p = 0.01). UTX overexpression was an independent prognosticator in multivariate analysis for OS (p = 0.013, hazard ratio = 1.996) and DFS (p = 0.009, hazard ratio = 1.972). The 5-year OS rates were 39% and 61% in patients with low expression and high expression of UTX, respectively. Inhibition of endogenous UTX in ESCC cells increased cell viability and BrdU incorporation, and decreased the expression of epithelial marker E-cadherin. Immunohistochemically, UTX expression was also positively correlated with E-cadherin expression. High UTX expression is independently associated with a better prognosis in patients with ESCC and downregulation of UTX increases ESCC cell growth and decreases E-cadherin expression. Our results suggest that UTX may be a novel therapeutic target for patients with ESCC.
Keywords: esophageal cancer, squamous cell carcinoma, UTX, E-cadherin
1. Introduction
Esophageal cancer ranks as the ninth leading cause of cancer deaths in Taiwan, and more than 90% of esophageal cancer was esophageal squamous cell carcinoma (ESCC) [1]. Despite advances in the diagnosis and treatment of ESCC in recent decades, the prognosis of patients with ESCC still remains unsatisfactory [2,3,4]. The 5-year survival rate of patients diagnosed with ESCC is approximately 20~40% [4,5]. Therefore, the discovery of biomarkers for ESCC prognosis may improve risk-adapted treatment strategies and lead to the identification of a novel target for ESCC.
The ubiquitously transcribed TPR gene on the X chromosome (UTX), also known as KDM6A, has been identified as a histone demethylase that specifically targets di- and tri-methyl groups on lysine 27 of histone H3 (H3K27me2/3) and has been proven to be essential during cellular reprogramming [6], embryonic development, and tissue-specific differentiation [7]. In 2009, inactivated somatic mutations and deletions targeting the UTX gene were identified in a variety of human cancers including multiple myeloma, medulloblastoma, esophageal, colon, bladder, prostate, and renal cancer [8,9,10,11]. Constitutional inactivation of UTX causes a specific hereditary disorder called the Kabuki syndrome which may develop into several types of cancer such as neuroblastoma, hepatoblastoma, acute leukemia, and fibromyxoid sarcoma, suggesting that Kabuki syndrome is a cancer predisposition syndrome [12].
Kabuki individuals with mutations in UTX have been identified in both female and male patients [13]. Kabuki syndrome results from hypomorphic female heterozygous mutation and null male hemizygous mutation of UTX [14]. A recent study indicated that Kabuki causative UTX protein mutations vary from complete UTX deletion to single amino acid point substitutions. However, more precise molecular mechanisms of these UTX mutations in cells or mouse models should be further investigated.
In addition, UTX gene was identified as one of the 127 significantly mutated genes in The Cancer Genome Atlas (TCGA) study in which whole-exome sequencing was performed on 3281 tumors derived from 12 tumor types [15]. UTX was downregulated in multiple myeloma cell lines leading to an increase in cell growth [16]. Decreased UTX also induced the expression of adhesion factors, including AOC3, CDHR5, and NCAM1 that are involved in cell reattachment upon dissemination. On the other hand, UTX was identified as a prooncogenic cofactor essential for leukemia maintenance in class II basic helix–loop–helix (bHLH) protein TAL1-positive (but not TAL1-negative) T-cell acute lymphoblastic leukemia [17]. Meanwhile, Kim et al. reported that UTX contributes to breast cancer cell proliferation with high levels of UTX being associated with poor prognosis in patients with breast cancer [18]. In cervical and head and neck tumors, HPV (human papillomavirus)-positive tumors were found to express higher levels of KDM6A [19]. These results indicated the complicated role of UTX in the pathogenesis of cancer. To the best of our knowledge, although UTX defects have been reported in ESCC [11], the prognostic significance of UTX expression in patients with ESCC remains largely undefined. Therefore, we conducted the present study to investigate this issue further.
2. Results
2.1. Patient Characteristics
A total of 106 patients with ESCC who had received surgery were considered in this study. The patients had a median age of 55 years (range, 29–80 years), and the characteristics of the patients are further summarized in Table 1. Among them, 101 (95%) were men and 5 (5%) were women. In terms of T classification, 42 (40%) of the patients were T1; 28 (26%) were T2; 26 (25%) were T3; and 10 (9%) were T4. Furthermore, in terms of N classification, 70 (66%) of the patients were N0; 25 (24%) were N1; 9 (8%) were N2; and 2 (2%) were N3. In terms of the 7th edition American Joint Committee on Cancer AJCC stages staging system 5 (5%) of the patients were stage IA, 17 (16%) were stage IIA; 26 (24%) were stage IIB; 11 (10%) were stage IIIA; 3 (3%) were stage IIIB; 9 (9%) were stage IIIC; and 2 (2%) were stage IV. Further analyses of histologic grades showed a grade 1 lesion in of the 10 (9%) patients, grade 2 in 70 (66%) of the patients, and grade 3 in 26 (25%) of the patients. Primary tumor location was found to be upper in 19 (18%) of the patients, middle in 36 (34%) of the patients, and lower in 51 (48%) of the patients. Among all 106 patients, resection margins were positive for residual tumor in 15 (14%) of the patients. At the time of analysis, the median periods of follow-up were 66 months (range, 61–112 months) for the 46 survivors and 53 months (range, 3.5–112 months) for all 106 patients. The 5-year survival (OS) and disease-free survival (DFS) of these 106 patients were 48% and 43%, respectively.
Table 1.
Characteristics of 106 patients with esophageal squamous cell carcinoma (ESCC) receiving esophagectomy.
| Age (years) | Parameters | No. of Cases (%) |
|---|---|---|
| median | 55 | |
| mean | 56 | |
| range | 29–80 | |
| Sex | ||
| male | 101 (95%) | |
| female | 5 (5%) | |
| Primary tumor location | ||
| Upper | 19 (18%) | |
| Middle | 36 (34%) | |
| Lower | 51 (48%) | |
| T classification | ||
| T1 | 42 (40%) | |
| T2 | 28 (26%) | |
| T3 | 26 (25%) | |
| T4 | 10 (9%) | |
| N classification | ||
| N0 | 70 (66%) | |
| N1 | 25 (24%) | |
| N2 | 9 (8%) | |
| N3 | 2 (2%) | |
| 7th AJCC Stage | ||
| IA | 5 (5%) | |
| IB | 33 (31%) | |
| IIA | 17 (16%) | |
| IIB | 26 (24%) | |
| IIIA | 11 (10%) | |
| IIIB | 3 (3%) | |
| IIIC | 9 (9%) | |
| IV | 2 (2%) | |
| Histological grading | ||
| Grade 1 | 10 (9%) | |
| Grade 2 | 70 (66%) | |
| Grade 3 | 26 (25%) | |
| Surgical margin | ||
| Negative | 91 (86%) | |
| Positive | 15 (14%) | |
| UTX expression | ||
| Low expression | 62 (58%) | |
| High expression | 44 (42%) | |
| E-cadherin | ||
| Low expression | 50 (47%) | |
| High expression | 56 (53%) |
AJCC, American Joint Committee on Cancer.
2.2. Correlation between Clinicopathologic Parameters and UTX Expression
Among the 106 patients considered, high UTX expression was identified in 44 (42%) of the patients (Figure 1). The associations between the clinicopathological parameters and UTX expression are summarized in Table 2. We did not observe any association between UTX expression and any clinicopathologic parameters including age, primary tumor location, histologic grading, T classification, N classification, and 7th edition American Joint Committee on Cancer (AJCC) Stage.
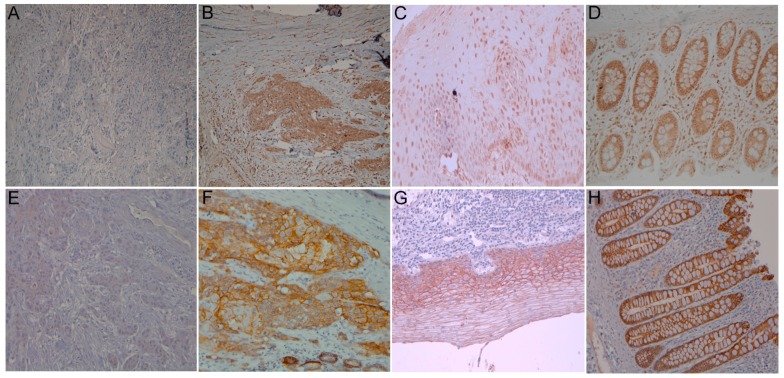
Figure 1.
Immunohistochemical staining of UTX in esophageal squamous cell carcinoma (ESCC). (A) Representative example of low UTX expression. Original magnification, ×100; (B) Representative example of high UTX expression. Original magnification, ×100; (C) UTX immunoreactivity was present in adjacent normal esophageal mucosa. Original magnification, ×200; (D) UTX immunoreactivity was present in colon mucosa used as a positive control. Original magnification, ×200; (E) Representative example of low E-cadherin expression. Original magnification, ×200; (F) Representative example of high E-cadherin expression. Original magnification, ×200; (G) E-cadherin immunoreactivity was present in adjacent normal esophageal mucosa. Original magnification, ×200; (H) UTX immunoreactivity was present in colon mucosa used as a positive control. Original magnification, ×200.
Table 2.
Associations between UTX expression and clinicopathological parameters in 106 patients with ESCC receiving esophagectomy.
| Parameters | UTX Expression | E-Cadherin Expression | |||||
|---|---|---|---|---|---|---|---|
| Low | High | p Value | Low | High | p Value | ||
| Age (years) | <55 | 29 | 23 | 0.58 | 25 | 27 | 0.85 |
| ≥55 | 33 | 21 | 25 | 29 | |||
| Primary tumor location | U + M | 30 | 25 | 0.39 | 22 | 33 | 0.13 |
| L | 32 | 19 | 28 | 23 | |||
| T classification | T1 + T2 | 38 | 32 | 0.22 | 26 | 44 | 0.004 * |
| T3 + T4 | 24 | 12 | 24 | 12 | |||
| N classification | N0 | 37 | 33 | 0.10 | 25 | 45 | 0.001 * |
| N1 + 2 + 3 | 25 | 11 | 25 | 11 | |||
| 7th AJCC Stage | I | 20 | 18 | 0.36 | 12 | 26 | 0.016 * |
| II + III + IV | 42 | 26 | 38 | 30 | |||
| Histological grading | Grade 1 + 2 | 44 | 36 | 0.20 | 34 | 46 | 0.091 |
| Grade 3 | 18 | 8 | 16 | 10 | |||
| Surgical margin | Negative | 50 | 41 | 0.068 | 40 | 51 | 0.10 |
| Positive | 12 | 3 | 10 | 5 | |||
| E-cadherin expression | Low | 36 | 14 | 0.008 * | - | - | - |
| High | 26 | 30 | - | - | - | ||
AJCC, American Joint Committee on Cancer. * Statistically significant.
2.3. Survival Analyses
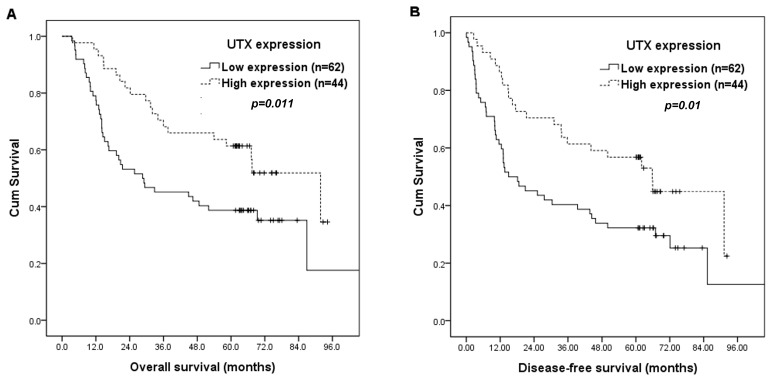
The correlations of patients’ survival with clinicopathological parameters and UTX expression are summarized in Table 3. By log-rank tests, 7th edition AJCC stage I (p = 0.008), T classification, T1/2 (p = 0.013), N classification, N0 (p < 0.001), negative surgical margin (p = 0.015), and high UTX expression (p = 0.011, Figure 2A) were associated with superior OS. Furthermore, 7th AJCC stage I (p = 0.002), T classification, T1/2 (p = 0.022), N classification, N0 (p < 0.001), and high UTX expression (p = 0.01, Figure 2B) were associated with better DFS.
Table 3.
Results of univariate log-rank analysis of prognostic factors for overall survival (OS) and disease-free survival (DFS) in 106 patients with ESCC receiving esophagectomy.
| Factors | No. of Patients | Overall Survival (OS) | Disease-Free Survival (DFS) | ||
|---|---|---|---|---|---|
| 5-year OS (%) | p Value | 5-year DFS (%) | p Value | ||
| Age (years) | |||||
| <55 | 52 | 56% | 0.32 | 52% | 0.17 |
| ≥55 | 54 | 41% | 33% | ||
| Location | |||||
| U + M | 55 | 55% | 0.20 | 46% | 0.42 |
| L | 51 | 41% | 39% | ||
| T classification | |||||
| T1 + 2 | 70 | 56% | 0.013 * | 49% | 0.022 * |
| T3 + 4 | 36 | 33% | 31% | ||
| N classification | |||||
| N0 | 70 | 63% | <0.001 * | 54% | <0.001 * |
| N1 + 2 + 3 | 36 | 19% | 19% | ||
| 7th AJCC stage | |||||
| I | 38 | 63% | 0.008 * | 58% | 0.002 * |
| II + III + IV | 68 | 40% | 34% | ||
| Histological grading | |||||
| Grade 1 + 2 | 80 | 50% | 0.33 | 43% | 0.62 |
| Grade 3 | 26 | 42% | 42% | ||
| Surgical margin | |||||
| Negative | 91 | 52% | 0.015 * | 45% | 0.11 |
| Positive | 15 | 27% | 27% | ||
| UTX expression | |||||
| Low expression | 62 | 39% | 0.011 * | 32% | 0.01 * |
| High expression | 44 | 61% | 57% | ||
| E-cadherin expression | |||||
| Low expression | 50 | 38% | 0.026 * | 28% | 0.005 * |
| High expression | 56 | 57% | 55% | ||
| Low UTX/E-cadherin expression | |||||
| Presence | 36 | 31% | 0.001 * | 22% | <0.001 * |
| Absence | 70 | 57% | 53% | ||
AJCC, American Joint Committee on Cancer. * Statistically significant.
Figure 2.
Kaplan-Meier curves according to UTX status. (A) OS rate according to UTX status; (B) DFS rate according to UTX status.
In multivariate analysis, high UTX expression (p = 0.013, hazard ratio = 1.996, 95% confidence interval: 1.160~3.436) remained independently associated with superior OS, together with N classification, N0 (p < 0.001, hazard ratio = 3.819, 95% confidence interval: 2.250~6.480). For DFS, high UTX expression (p = 0.009, hazard ratio = 1.972, 95% confidence interval: 1.189~3.279) and N classification (p < 0.001, hazard ratio = 3.350, 95% confidence interval: 2.040~5.502) represented an independent adverse prognosticator. The 5-year OS and DFS rates were 61% and 57% respectively, in patients with high UTX expression, and 39% and 32% respectively, in patients with low UTX expression.
Moreover, we further tested whether the combination of UTX and E-cadherin expression can improve the prognostic value. We found that the combination of low UTX and low E-cadherin expression robustly identifies the group of patients with extremely poor prognosis. The 5-year OS (p = 0.001) and DFS (p < 0.001) rates were 31% and 22%, respectively, in the 36 patients with low UTX and E-cadherin expression, and 57% and 53%, respectively, in the 70 patients without low UTX and E-cadherin expression.
2.4. Inhibition of UTX Promoted ESCC Cells Proliferation and Epithelial-Mesenchymal Transition Process
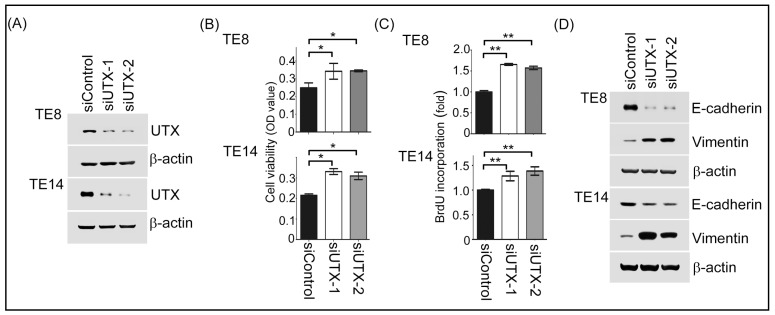
To study the functions of UTX in ESCC, we generated UTX-depleted ESCC cells in the TE8 and TE14 cell lines. The knockdown efficiency was revealed to be effective through Western blotting analysis (Figure 3A). Next, to examine if UTX modulated ESCC proliferation, we performed MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. As shown in Figure 3B, TE8 and TE14 cells transfected with UTX-mediated siRNA were grew faster than those transfected with siControl cells. Using the same cell panels, similar results were also observed with a bromodeoxyuridine BrdU incorporation assay (Figure 3C). Moreover, UTX-depleted TE8 and TE14 cells reduced the expressions of epithelial marker E-cadherin and increased those of Vimentin, a mesenchymal marker (Figure 3D). Together, these results reveal that UTX prevention may induce cell proliferation and participate in the epithelial-mesenchymal transition (EMT) process in ESCC cells. To evaluate the potential relevance of the above in vitro findings in ESCC in a clinical setting, we also analyzed the protein expressions of E-cadherin in tissues samples from the 106 human ESCC patients using immunohistochemistry. There was a significant correlation between UTX expression and E-cadherin expression (p = 0.008, Table 2). Representative staining results of E-cadherin expression are shown in Figure 1.
Figure 3.
UTX expression regulated cell growth and epithelial-mesenchymal transition (EMT) expression in TE8 and TE14 cells. (A) The protein expression level of UTX was demonstrated in TE8 cells transfected with siControl and siUTX using Western blotting analysis; (B,C) The cell growth abilities of siControl and siUTX in TE8 and TE14 cells were measured by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and BrdU assays; (D) The E-cadherin, and Vimentin protein expression levels were investigated with Western blotting analysis in TE8 and TE14 cells transfected with siControl and siUTX. *, p < 0.05; **, p < 0.01.
3. Discussion
As is well known, epigenetic modifications, such as histone methylation or demethylation are involved in gene activation. UTX, a histone demethylase, removes di- and tri-methyl groups on histone H3 lysine 27 (H3K27) which is essential for tissue differentiation, cell maintenance, cell reprogramming and cancer development [20]. Using exome- or genome-wide sequencing strategies and TCGA databases, various somatic mutations and deletions of UTX have been found in several human cancers, with a prevalence of 24.24% in bladder cancer, 10% in prostate cancer, and 1.85% in esophageal cancer [20]. Interestingly, induction of UTX expression in UTX-depleted cells results in a K3K27me3 decrease and attenuates cell proliferation [11]. In addition, in normal fibroblast cells, UTX expression activates the Rb (Retinoblastoma) pathway to suppress cell growth, indicating that UTX may play the role of tumor suppressor in human cancers [21]. Conversely, in breast tumors, UTX is rarely mutated and its upregulation is correlated with poor prognosis [22]. UTX knockdown in leukemia cell lines exhibits an anti-growth effect [23]. These conflicting results imply that the function of UTX in human cancers may depend on cell-context manner. In the present study, we found that positive UTX expression was associated with better clinical outcomes in 106 patients with ESCC receiving esophagectomy and blockage of endogenous UTX by the siRNA approach led to an increase in cell proliferation and elevation of the BrdU incorporation ability in ESCC cells, suggesting that UTX acts a tumor suppressor in ESCC.
EMT plays a critical role in cancer progression. EMT allows cells to prevent death and promotes migratory and invasive abilities. An abundance of evidence indicates that an epigenetic regulator plays a vital role in controlling EMT and cancer progression. Several previous studies [24] have shown that reduced E-cadherin expression is a poor prognosticator in patients with ESCC. In our study, we found that decreased E-cadherin expression was associated with higher T classification, higher N classification, advanced 7th edition AJCC stages, and inferior OS rates in 106 ESCC patients receiving esophagectomy. With regard to colon cancer cells, Zha et al. [25] and Zhou et al. [8] reported the inactivation of UTX down-regulated E-cadherin gene expression. For breast cancer cells, Choi et al. [26] found that UTX loss induced EMT. However, the role of UTX in the regulation of EMT expression is still unclear in ESCC cells. Here, we demonstrated that loss-of function of UTX decreased E-cadherin expression and increased vimentin expression in ESCC cells, indicating that UTX might be involved in the regulation of the EMT process in ESCC.
Our study has one critical limitation in that it is a retrospective analysis and is based on a relatively small number of patients.
In conclusion, high UTX expression is independently associated with better prognosis in patients with ESCC. In ESCC cell lines, downregulation of UTX increases cell growth and decreases E-cadherin expression. Our results may further elucidate the role of UTX in ESCC and provide a potential therapeutic target for patients with ESCC.
4. Materials and Methods
4.1. Patient Population
We retrospectively reviewed ESCC patients receiving esophagectomy at Kaohsiung Chang Gung Memorial Hospital. Approval to analyze and publish the aggregated anonymous data was given by the Institutional Review Board committee of Kaohsiung Chang Gung Memorial Hospital at Kaohsiung, Taiwan. The approval number of this project was 104-7233B. The approval date was on 17 November 2015. We excluded patients with synchronous cancers in other organs and patients receiving preoperative chemoradiotherapy, preoperative chemotherapy, or preoperative radiotherapy. Finally, 106 patients were identified. Patients undergoing surgery had a radical esophagectomy with cervical esophagogastric anastomosis (McKeown procedure) or an Ivor Lewis esophagectomy with intrathoracic anastomosis, reconstruction of the digestive tract with a gastric tube, and pylorus drainage procedures. All patients received two-field lymph node dissection. The pathological tumor node metastasis (TNM) staging system was determined according to the 7th edition AJCC staging system [27]. The OS rate was determined from the time of surgery to death as a result of all causes. The DFS rate was computed from the time of surgery to the recurrence of disease or death from any cause without evidence of recurrence.
4.2. Immunohistochemistry
Immunohistochemistry staining was performed using an immunoperoxidase technique. Staining was performed on slides (4 mm) of formalin-fixed, paraffin-embedded tissue sections with primary antibodies against UTX (Clone D3Q1l, 1:400, Cell Signaling Technology, Boston, MA, USA) and E-cadherin (BD610182, 1:2000, BD Biosciences, Sparks, MD,). Briefly, after deparaffinization and rehydration, the retrieval of the antigen was performed by treating the slides in 10 mmol/L citrate buffer (pH 6.0) in a hot water bath (95 °C) for 20 min. Endogenous peroxidase activity was blocked for 15 min in 0.3% hydrogen peroxide. After blocking with 1% goat serum for 1 h at room temperature, the sections were incubated with primary antibodies for at least 18 h at 4 °C overnight. Immunodetection was performed using the LSAB2 kit (Dako, Carpinteria, CA, USA) followed by 3-3′-diaminobenzidine for color development and hematoxylin for counterstaining. Incubation without the primary antibody was used as a negative control, and normal colon mucosa was used as a positive control for UTX and E-cadherin. The staining was assessed by 2 pathologists (Sheng-Lan Wang and Wan-Ting Huang without any information on clinicopathologic features or treatment outcome. A semi-quantitative immunoreactive score (IRS) was used to evaluate the immunohistochemistry staining. [28] The IRS was calculated by multiplying the staining intensity (graded as: 0 = no staining, 1 = weak staining, 2 = moderate staining, and 3 = strong staining) with the percentage of positively stained cells (0 = no stained cell, 1 = <10% of stained cells, 2 = 10–50% of stained cells, 3 = 51–80% of stained cells, and 4 = >80% of stained cells). The criterion for positive staining was a specimen with a IRS > 4.
4.3. Cell Lines and siRNA Transfection
The human esophageal cell lines TE8 were obtained from the American Type Culture Collection, and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% fetal bovine serum at 37 °C and 5% CO2. For siRNA transfection experiments, two synthetic UTX targeting siRNAs and one sicontrol were applied. Cells were transfected with UTX-mediated siRNA (50 nM) in serum-free DMEM using Plus/Lipofetamin Transfection Reagent according to the manufacturer’s instructions. Two double-stranded synthetic RNA oligomers (5′-GAACAGCUCCGCGCAAAUA-3′ and 5′-GAGAGUAAUUCACGAAAGA-3′) deduced from human UTX, and one siRNA control (#4611G; Ambion, Austin, TX, USA) were used in the siRNA experiments.
4.4. Western Blotting Assay
TE8 and TE14 cells transfected with a negative control or UTX-siRNA were harvested and homogenized in RIPA (Radio-Immunoprecipitation Assay) lysis buffer, containing protease inhibitor cocktail on ice for 30 min. The homogenate was centrifuged at 4 °C at 15,000 rpm for 30 min, and lysates were collected. Protein concentration was determined using the BCA Protein Assay kit (Thermo Fisher, Waltham, MA, USA). The cell lysates were subjected to the 10% SDS-PAGE gels for following western blotting. The primary antibodies (anti-UTX, anti-E-cadherin, anti-Vimentin, and anti-β-actin) were obtained from Cell Signaling Technology (Danvers, MA, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from Sigma (St. Louis, MO, USA).
4.5. MTT and BrdU Assay
Cells were seeded at a density of 1 × 104 cells per well in 96-well plates and maintained in DMEM supplemented with 10% FBS (fatal bovine serum). After overnight incubation, the culture medium was removed and the cells were washed with PBS (phosphate buffer saline). The freshly completed DMEM (Dulbecco’s Modified Eagle Medium) medium was added and cultured for 48 h. Following this that the culture medium was removed and the cells were washed with PBS. The cells were then incubated with 0.5 mg/mL MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), in a culture medium without FBS, for 4 h at 37 °C in a 5% CO2 atmosphere. The medium was removed and 100 μL DMSO (Dimethyl sulfoxide) buffer was added and incubated in the dark for 10 min. Absorbance was measured on a microplate reader at 540 nm. For bromodeoxyuridine (BrdU) incorporation assay, cells were seeded at a density of 5 × 103 cells/well in 96-well culture plates for 48 h. BrdU incorporation analysis was performed using a cell proliferation ELISA kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions.
4.6. Statistical Analysis
For patient data, statistical analysis was performed using the SPSS 17 software package (SPSS Inc., Chicago, IL, USA). The chi-square test and Fisher’s exact test were used to compare data between the two groups. For the survival analysis, the Kaplan-Meier method was used for univariate analysis, and the difference between the survival curves was tested by a log-rank test. In a stepwise forward fashion, significant parameters at a univariate level were entered into a Cox regression model to analyze their relative prognostic value. For all analyses, two-sided tests of significance were used with p < 0.05 being considered significant.
Acknowledgments
This work was supported in part by grants from the National Science Council, Taiwan (MOST 106-2314-B-182A-159-MY3 and MOST 106-2320-B-182A-015 to SH Li; MOST 105-2320-B-182A-016 to CH Chen); Chang Gung Memorial Hospital (CMRPG8E1533 and CMRPG8G0891 to SH Li; CMRPG8E1471-2 to CH Chen); and Kaohsiung Medical University (Aim for the Top Universities Grant, grant Nos. KMU-TP104E27 and KMU-TP105E23). We also thank the Tissue Bank Core Lab at Kaohsiung Chang Gung Memorial Hospital (CLRPG8F1701 and CLRPG8F1702) for its excellent technical support.
Abbreviations
| UTX | ubiquitouslytranscribed TPR gene on the X chromosome |
| ESCC | Directory of open access journals |
| H3K27me2/3 | lysine 27 of histone H3 |
| TCGA | The Cancer Genome Atlas |
| bHLH | basic helix–loop–helix |
| DFS | Disease-free survival |
Author Contributions
Shau-Hsuan Li and Chang-Han Chen: Experimental design and manuscript draft. Hung-I Lu, Wan-Ting Huang, and Wei-Che Lin: Clinical sample collection, interpretation and analysis. Wan-Yu Tien, Ya-Chun Lan, and Hsin-Ting Tsai: Experimental manipulation and analysis of the results of the assays. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chen Y.H., Lu H.I., Wang Y.M., Lo C.M., Chou S.Y., Huang C.H., Shih L.H., Chen S.W., Li S.H. The prognostic significance of celiac lymph node metastasis in patients with locally advanced esophageal squamous cell carcinoma receiving curative concurrent chemoradiotherapy. Oncotarget. 2017;8:96190–96202. doi: 10.18632/oncotarget.21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J., Xie X., Zhou C., Peng S., Rao D., Fu J. Which factors are associated with actual 5-year survival of oesophageal squamous cell carcinoma? Eur. J. Cardiothorac. Surg. 2012;41:e7–e11. doi: 10.1093/ejcts/ezr240. [DOI] [PubMed] [Google Scholar]
- 3.Li S.H., Lu H.I., Chang A.Y., Huang W.T., Lin W.C., Lee C.C., Tien W.Y., Lan Y.C., Tsai H.T., Chen C.H. Angiotensin II type I receptor (AT1R) is an independent prognosticator of esophageal squamous cell carcinoma and promotes cells proliferation via mTOR activation. Oncotarget. 2016;7:67150–67165. doi: 10.18632/oncotarget.11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S.H., Chen C.H., Lu H.I., Huang W.T., Tien W.Y., Lan Y.C., Lee C.C., Chen Y.H., Huang H.Y., Chang A.Y., et al. Phosphorylated p70S6K expression is an independent prognosticator for patients with esophageal squamous cell carcinoma. Surgery. 2015;157:570–580. doi: 10.1016/j.surg.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Estes N.C., Stauffer J., Romberg M., Thomas J.H., Jewell W.R., Hermreck A. Squamous cell carcinoma of the esophagus. Am. Surg. 1996;62:573–576. [PubMed] [Google Scholar]
- 6.Rada-Iglesias A., Wysocka J. Epigenomics of human embryonic stem cells and induced pluripotent stem cells: Insights into pluripotency and implications for disease. Genome Med. 2011;3:36. doi: 10.1186/gm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shpargel K.B., Sengoku T., Yokoyama S., Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Z., Zhang H.S., Liu Y., Zhang Z.G., Du G.Y., Li H., Yu X.Y., Huang Y.H. Loss of TET1 facilitates DLD1 colon cancer cell migration via H3K27me3-mediated down-regulation of E-cadherin. J. Cell. Physiol. 2018;233:1359–1369. doi: 10.1002/jcp.26012. [DOI] [PubMed] [Google Scholar]
- 9.Nickerson M.L., Dancik G.M., Im K.M., Edwards M.G., Turan S., Brown J., Ruiz-Rodriguez C., Owens C., Costello J.C., Guo G., et al. Concurrent alterations in TERT, KDM6A, and the BRCA pathway in bladder cancer. Clin. Cancer Res. 2014;20:4935–4948. doi: 10.1158/1078-0432.CCR-14-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suva M.L., Riggi N., Bernstein B.E. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Haaften G., Dalgliesh G.L., Davies H., Chen L., Bignell G., Greenman C., Edkins S., Hardy C., O’Meara S., Teague J., et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyake N., Mizuno S., Okamoto N., Ohashi H., Shiina M., Ogata K., Tsurusaki Y., Nakashima M., Saitsu H., Niikawa N., et al. KDM6A point mutations cause Kabuki syndrome. Hum. Mutat. 2013;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- 13.Banka S., Lederer D., Benoit V., Jenkins E., Howard E., Bunstone S., Kerr B., McKee S., Lloyd I.C., Shears D., et al. Novel KDM6A (UTX) mutations and a clinical and molecular review of the X-linked Kabuki syndrome (KS2) Clin. Genet. 2015;87:252–258. doi: 10.1111/cge.12363. [DOI] [PubMed] [Google Scholar]
- 14.Shpargel K.B., Starmer J., Wang C., Ge K., Magnuson T. UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proc. Natl. Acad. Sci. USA. 2017;114:E9046–E9055. doi: 10.1073/pnas.1705011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A., et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezponda T., Dupere-Richer D., Will C.M., Small E.C., Varghese N., Patel T., Nabet B., Popovic R., Oyer J., Bulic M., et al. UTX/KDM6A Loss Enhances the Malignant Phenotype of Multiple Myeloma and Sensitizes Cells to EZH2 inhibition. Cell Rep. 2017;21:628–640. doi: 10.1016/j.celrep.2017.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benyoucef A., Palii C.G., Wang C., Porter C.J., Chu A., Dai F., Tremblay V., Rakopoulos P., Singh K., Huang S., et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30:508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.H., Sharma A., Dhar S.S., Lee S.H., Gu B., Chan C.H., Lin H.K., Lee M.G. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gameiro S.F., Kolendowski B., Zhang A., Barrett J.W., Nichols A.C., Torchia J., Mymryk J.S. Human papillomavirus dysregulates the cellular apparatus controlling the methylation status of H3K27 in different human cancers to consistently alter gene expression regardless of tissue of origin. Oncotarget. 2017;8:72564–72576. doi: 10.18632/oncotarget.19885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van der Meulen J., Speleman F., Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658–668. doi: 10.4161/epi.28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J.K., Tsai M.C., Poulin G., Adler A.S., Chen S., Liu H., Shi Y., Chang H.Y. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev. 2010;24:327–332. doi: 10.1101/gad.1882610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Mercher T., Scholl C., Brumme K., Gilliland D.G., Zhu N. A functional role for the histone demethylase UTX in normal and malignant hematopoietic cells. Exp. Hematol. 2012;40:487–498. doi: 10.1016/j.exphem.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 24.Xu X.L., Ling Z.Q., Chen S.Z., Li B., Ji W.H., Mao W.M. The impact of E-cadherin expression on the prognosis of esophageal cancer: A meta-analysis. Dis. Esophagus. 2014;27:79–86. doi: 10.1111/dote.12024. [DOI] [PubMed] [Google Scholar]
- 25.Zha L., Cao Q., Cui X., Li F., Liang H., Xue B., Shi H. Epigenetic regulation of E-cadherin expression by the histone demethylase UTX in colon cancer cells. Med. Oncol. 2016;33:21. doi: 10.1007/s12032-016-0734-z. [DOI] [PubMed] [Google Scholar]
- 26.Choi H.J., Park J.H., Park M., Won H.Y., Joo H.S., Lee C.H., Lee J.Y., Kong G. UTX inhibits EMT-induced breast CSC properties by epigenetic repression of EMT genes in cooperation with LSD1 and HDAC1. EMBO Rep. 2015;16:1288–1298. doi: 10.15252/embr.201540244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edge S.B., Compton C.C., Fritz A.G., Greene F.L., Trotti A., editors. AJCC Cancer Satging Manual. 7th ed. Springer; New York, NY, USA: 2010. Esophagus and esophagogastric junction; pp. 103–111. [Google Scholar]
- 28.Bouvier C., Macagno N., Nguyen Q., Loundou A., Jiguet-Jiglaire C., Gentet J.C., Jouve J.L., Rochwerger A., Mattei J.C., Bouvard D., et al. Prognostic value of the Hippo pathway transcriptional coactivators YAP/TAZ and beta1-integrin in conventional osteosarcoma. Oncotarget. 2016;7:64702–64710. doi: 10.18632/oncotarget.11876. [DOI] [PMC free article] [PubMed] [Google Scholar]