Abstract
Lung adenocarcinoma is frequently occurring type of lung cancer with high metastatic risk. We performed a DNA methylation profiling analysis to identify possible prognostic markers involved in lung adenocarcinoma. DNA methylation profiling data (GSE66386) were downloaded from the Gene Expression Omnibus (GEO) database. Differentially methylated genes were identified using a limma package. GO enrichment analysis was performed to identify vital functions related to differential gene methylation, and pathway analysis was performed to assess the associations between different proteins with regard to regulation of cell function and metabolism. The screening results showed a total of 112,662 differentially methylated genes in lung adenocarcinoma patients compared with those of the normal controls. These CpGs were involved in 16,705 genes. The skeletal system development (P=9.46E-27) and embryonic organ morphogenesis (P=8.67E-24) were found to be involved in lung adenocarcinoma. The cancer (P=3.64E-07), Rap1 signaling (P=9.21E-05) and calcium signaling (P=9.21E-05) pathways constituted the important pathways associated with lung adenocarcinoma. In conclusion, methylated PTPRF, HOXD3, HOXD13 and CACNA1A are potential markers and may be utilized for the diagnosis and therapy of lung adenocarcinoma.
Keywords: DNA methylation, diagnostic biomarker, lung adenocarcinoma, pathway
Introduction
Lung adenocarcinoma is a common type of lung cancer with high incidence (40%) (1). It has high risk of distant metastasis, which poses a severe threat to the survival rates of patients (2). However, prognosis varies considerably at different stages of this disease (3). Therefore, an effective screening tool is imperative and of critical importance in the clinical management of lung adenocarcinoma, which provides early and accurate detection thereof.
DNA methylation markers are effective biomarkers and promising candidates in the diagnosis of disease (4,5). Aberrant methylation in cancer usually occurs at CpG islands, which results in changes in the transcription of tumor suppressor genes (6). For lung adenocarcinoma, several in vitro studies have shown the difference of the DNA methylation level between patients and normal controls. For example, F2RL3 methylation has been demonstrated to be a strong predictor for the incidence of lung adenocarcinoma (7). The p16 gene promoter methylation is also associated with smoking-induced lung adenocarcinoma (7). Tomizawa et al (8) suggested that RASSF1A methylation was significantly associated with pleural involvement, vascular invasion and decreased patient survival time, which may be a powerful marker for the prognosis of patients with lung adenocarcinoma at an early stage. Previous findings have shown the role of DNA methylation in a potential diagnosis of lung adenocarcinoma. However, the pathogenesis involved in lung adenocarcinoma has not been clearly understood.
In the present study, microarray data (GSE66386) were downloaded and a comprehensive bioinformatics approach was performed to screen differentially expressed methylation genes in lung adenocarcinoma samples compared with normal controls. The aim was to identify potential DNA methylation biomarkers and to determine the possible pathogenesis for lung adenocarcinoma.
Materials and methods
Microarray data and preprocessing
The gene expression dataset (GSE66386) was collected from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database. The dataset included 183 samples, of which 164 were lung adenocarcinoma samples, and 19 were matched normal lung tissue samples. The dataset was uploaded by Bjaanæs et al and cited in their study (9). The platform of this microarray was Illumina Infinium 450 K Human Methylation BeadChip. The differentially methylated genes were identified using the limma package available at http://www.bioconductor.org-packages/release/bioc/html/limma.html. As a result, the DNA methylation data with 456947 CpGs were used for analysis. For preprocessing of the row data, the following probes were removed: i) The distance value (from CpG to SNP) <2; ii) the minor allele frequency <0.05; iii) the cross-hybridising probes and sex chromosome-specific DNA probes.
Identification of genes with methylation differences
The percentage methylation values were expressed as β values. The differential methylation of CpGs between lung adenocarcinoma and control samples were identified if the absolute mean difference between the two groups was <0.05, and the P-value was <0.05. Consequently, the P-value was calculated by t-test.
Functional enrichment analysis of differential methylation of genes
Gene ontology (GO) analysis for screening of differential methylation of genes was conducted to search the vital functions related to the differential gene methylation in lung adenocarcinoma (10). The P-values was adjusted into false discovery rate (FDR) by Benjaminiand Hochberg study (11). FDR <0.01 was considered as the cutoff value. The proteins of annotated differential gene methylation were compared with their homologous proteins based on COG database (http://www.ncbi.nlm.nih.gov/COG). P-value <0.05 was regarded as the cut off threshold value. By this strategy, the screened genes with methylation differences were then assigned to different gene functions including cellular component (CC), molecular function (MF) and biological process (BP).
Pathway analysis
A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (12) analysis was performed to evaluate the associations between different proteins in the regulation of cell function and metabolism. Fisher's test was used to identify significant pathways that were enriched by genes with methylation differences with the threshold of FDR <0.05.
Results
Identification of methylation genes
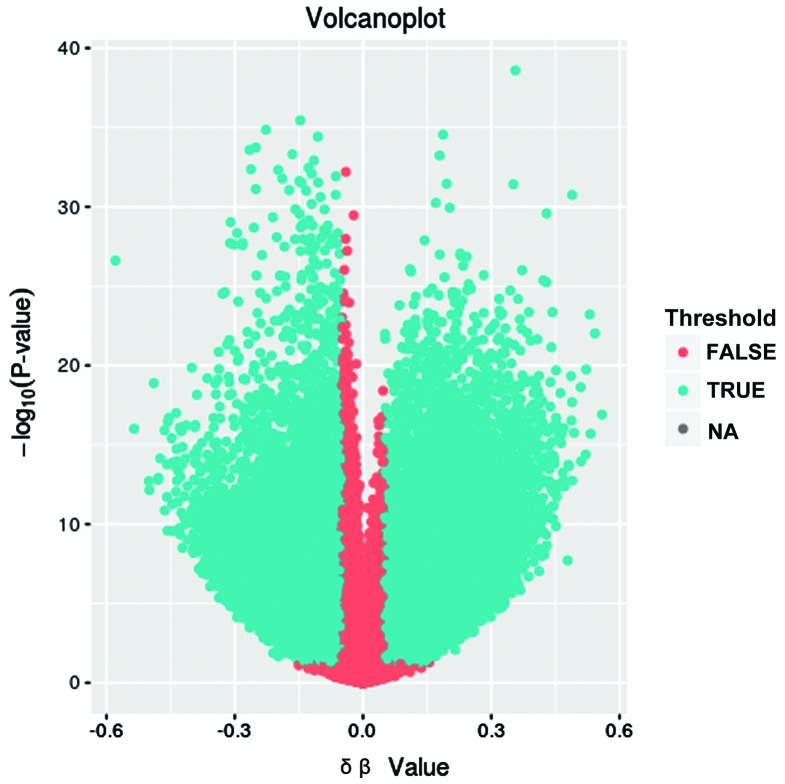
After standardized pretreatment, we collected 413043 CpGs for the subsequent analysis. After further screening, 112662 differentially methylated regions (DMRs) in lung adenocarcinoma patients compared with normal controls, including 57235 upmethylated CpGs and 55427 downmethylated ones. These CpGs were involved in 16,705 genes. The results are summarized as volcano plots (Fig. 1). The difference of the differentially methylated genes between lung adenocarcinoma and normal control is shown on the x-axis, and the negative P-value was shown on the y-axis.
Figure 1.
Volcanoplot of DNA methylation microarray results in lung adenocarcinoma versus normal controls. The difference of differentially methylated genes between lung adenocarcinoma and normal controls is shown on the x-axis, and the negative P-value is shown on the y-axis.
To obtain more important methylation genes, we initially removed 109 CpGs with β value ≤0.2 or ≥0.8, and then excluded 99954 CpGs with an absolute value of difference between the average β values of the two groups ≥0.2. As a result, 12599 CpGs related to 5,489 genes were obtained.
Functional enrichment analysis
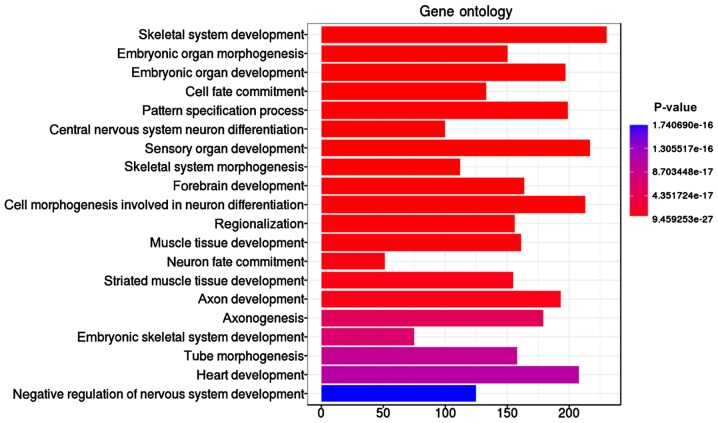
The ‘skeletal system development’ function, including 230 significant genes with methylation differences, was identified as the most significant (P=9.46E-27) GO term with 5,489 methylation genes (Table I and Fig. 2). In addition, embryonic organ morphogenesis (P=8.67E-24), embryonic organ development (P=1.20E-23), and cell fate commitment (P=5.90E-23) were found to be involved in the lung adenocarcinoma.
Table I.
The top 10 enriched GO descriptions for differentially expressed methylation genes.
| ID | Description | P-value | P-adjust | Count |
|---|---|---|---|---|
| GO:0001501 | Skeletal system development | 9.46E-27 | 5.19E-23 | 230 |
| GO:0048562 | Embryonic organ morphogenesis | 8.67E-24 | 2.20E-20 | 150 |
| GO:0048568 | Embryonic organ development | 1.20E-23 | 2.20E-20 | 197 |
| GO:0045165 | Cell fate commitment | 5.90E-23 | 8.10E-20 | 133 |
| GO:0007389 | Pattern specification process | 5.27E-21 | 5.79E-18 | 199 |
| GO:0021953 | Central nervous system neuron differentiation | 1.81E-20 | 1.66E-17 | 100 |
| GO:0007423 | Sensory organ development | 1.87E-19 | 1.47E-16 | 217 |
| GO:0048705 | Skeletal system morphogenesis | 4.66E-19 | 3.20E-16 | 112 |
| GO:0030900 | Forebrain development | 1.11E-18 | 6.75E-16 | 164 |
| GO:0048667 | Cell morphogenesis involved in neuron differentiation | 1.79E-18 | 9.50E-16 | 213 |
GO, Gene Ontology.
Figure 2.
Gene ontology biological process enrichment analysis of the screened differentially expressed methylation genes.
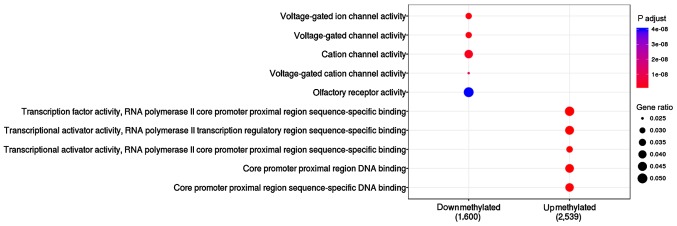
The function of upmethylated and downmethylated gene sets were compared in this study (Table II and Fig. 3). Results showed that 2,539 upmethylated genes were enriched in the following pathways: transcription factor activity (P=4.13E-19), RNA polymerase II core promoter proximal region sequence-specific binding (P=1.82E-17), and transcriptional activator activity (P=4.43E-13). A total of 1,600 differential downmethylated genes were mainly enriched in voltage-gated ion channel activity (P=1.13E-09), voltage-gated channel activity (P=1.13E-09) and cation channel activity (P=2.66E-09).
Table II.
The top 5 enriched down- and upmethylated pathways involved in the development of lung adenocarcinoma.
| Cluster | ID | P-value | P-adjust | Count |
|---|---|---|---|---|
| Downmethylated | GO:0005244 | 2.52E-12 | 1.13E-09 | 52 |
| Downmethylated | GO:0022832 | 2.52E-12 | 1.13E-09 | 52 |
| Downmethylated | GO:0005261 | 8.86E-12 | 2.66E-09 | 69 |
| Downmethylated | GO:0022843 | 5.10E-11 | 1.15E-08 | 40 |
| Downmethylated | GO:0004984 | 2.64E-10 | 4.00E-08 | 84 |
| Upmethylated | GO:0000982 | 4.19E-22 | 4.13E-19 | 124 |
| Upmethylated | GO:0001228 | 3.7E-20 | 1.82E-17 | 116 |
| Upmethylated | GO:0001077 | 1.35E-15 | 4.43E-13 | 85 |
| Upmethylated | GO:0001159 | 1.92E-13 | 4.74E-11 | 113 |
| Upmethylated | GO:0000987 | 1.55E-12 | 3.05E-10 | 110 |
GO, Gene Ontology.
Figure 3.
The function of upmethylated and downmethylated genes.
Pathway analysis
Pathway analysis was used to evaluate the possible pathway associated with lung adenocarcinoma induced by DNA methylation (Table III). The results showed that significant genes with methylation differences were involved in 74 pathways, among which, the cancer pathways (P=3.64E-07), Rap1 signaling pathway (P=9.21E-05) and calcium signaling pathway (P=9.21E-05) were the important pathways associated with lung adenocarcinoma.
Table III.
The top 10 enriched pathways involved in development of lung adenocarcinoma.
| ID | Pathway name | P-value | P-adjust | Count |
|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 1.28E-09 | 3.64E-07 | 152 |
| hsa04015 | Rap1 signaling pathway | 9.70E-07 | 9.21E-05 | 85 |
| hsa04020 | Calcium signaling pathway | 7.74E-07 | 9.21E-05 | 75 |
| hsa04921 | Oxytocin signaling pathway | 1.78E-06 | 0.000127156 | 67 |
| hsa04024 | cAMP signaling pathway | 2.75E-06 | 0.000156673 | 80 |
| hsa03008 | Ribosome biogenesis in eukaryotes | 6.25E-06 | 0.000296837 | 5 |
| hsa04010 | MAPK signaling pathway | 9.25E-06 | 0.000376788 | 96 |
| hsa00190 | Oxidative phosphorylation | 1.08E-05 | 0.000384468 | 13 |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 1.34E-05 | 0.000423855 | 59 |
| hsa04022 | cGMP-PKG signaling pathway | 1.73E-05 | 0.000448535 | 54 |
Discussion
Alterations of epigenetics such as DNA methylation are of great importance in carcinogenesis. Increased knowledge has shown the great importance of DNA methylation in lung cancer (13). Thus it is of great importance to evaluate the connection between the level of methylation and the increased expression in epigenetic diagnosis and therapy. In the present study, we identified a great amount of genomic regions that showed different methylations in tumor and normal lung adenocarcinoma tissues by microarray based on the methylation screening approach. A total of 12599 DMRs associated with 5,489 genes were eventually screened, including PTPRF, HOXD3, HOXD13 and CACNA1A. These genes are promising in the development of biomarkers for diagnostic or therapy purposes.
PTPRF gene is a member of the protein tyrosine phosphatase (PTP) family, which is known to be a signaling molecule involved in the regulation of cell processes (14). It was previously reported to be expressed as a potential predictive marker for treatment with erlotinib in non-small-cell lung cancer (15), and has been found to promote cell migration in several types of cancer (16,17). The present study showed that the methylated PTPRF was focused on the role of regulation of protein binding. In lung adenocarcinomas, CRBP-1 high expression was suggested to be an aggressive phenotype (18). CPBP-1 is known as a 15 kDa cytosolic binding protein that is crucial in retinol uptake and esterification (19,20). CPBP-1 was also highly expressed in lung adenocarcinoma (18), and played an important role in protein binding. Therefore, we consider that the function of methylated PTPRF associated with protein binding is of great importance in lung adenocarcinomas.
Homeobox (HOX) genes, belonging to the homeobox family, are important in the regulation of transcription which controls the expression of morphogenesis-related genes (21) In the present study, HOXD3 was upmethylated in lung adenocarcinoma patients compared to the normal controls. In addition, HOXD3 was involved in two possible functions associated with lung adenocarcinoma, positive regulation of cell development and cell-substrate adhesion. In an in vitro experiment, Hamada et al (22) have shown that the overexpression of HOXD3 gene significantly promoted migration and invasion in human lung cancer A549 cells. Authors of that study suggested that HOXD3 overexpression prominently induced a high expression of integrin β3, which promotes pulmonary metastasis via increased endothelium and adhesion (22). Another study identified that in A549 cells, overexpressing HOXD3 gene accelerated the expression of N-cadherin and integrins (23). These results suggest that the aberrant methylation of HOXD3 plays an important role in the regulation of genes involved in the metastasis and invasion of lung adenocarcinoma via the function of cell-substrate adhesion.
Another HOX gene, HOXD13, was screened with differential hypermethylation enriched in the transcriptional activator activity in lung adenocarcinoma in the present study. Hypermethylated HOXD13 has been reported in malignant melanomas (24), breast cancer (25) and extrahepatic cholangiocarcinoma (26), with methylation rates of 30.8, 94.38 and 57.7%, respectively. Thus, the upmethylation of HOXD13 is a potential diagnostic biomarker and therapeutic target.
There is insufficient information regarding the relationship between CACNA1A and cancer. However, we found that CACNA1A was downmethylated in patients with lung adenocarcinoma compared to normal controls. As a novel possible tumor-methylated candidate involved in lung cancer, the CACNA1A methylation has been identified from microarray experiment (27,28). However, the methylated level as well as its possible carcinogenic mechanism have not been clearly understood. Further analysis in the present study showed that downmethylated CACNA1A was involved in several functions such as the calcium channel and was also voltage-dependent. The intracellular calcium overload is regarded as an important factor inducing mitochondrial-dependent apoptosis directly and indirectly (29). The function of mitochondrial transmembrane potential may be interrupted by calcium phosphate in the matrix (30). In a previous study, it was shown that in TCM, curcumin exerted an anti-proliferative effect by calcium overload via activating the calcium channel in lung cancer cells (31). Therefore, downmethylated CACNA1A may be involved in the development of lung adenocarcinoma via the calcium channel signaling pathway.
In conclusion, from the DNA methylation profiling, we achieved two purposes: i) identification of DNA methylation changes that may be associated with lung adenocarcinoma. ii) Identification of function and pathway of methylated genes that may be involved in lung adenocarcinomas. From these data, we concluded that screened genes with methylation differences, including PTPRF, HOXD3, HOXD13 and CACNA1A serve as potential markers and may be used in the diagnosis and therapy of patients with lung adenocarcinoma.
Acknowledgements
The present study was funded by the Science and Technology Research projects of Guizhou province [SY: (2012)3101] and Guizhou Province Science and Technology Research ProjectsGuizhou Branch SY Word [2012] no. 3101: Induced sputum Runx3 gene promoter region methylation in value for early diagnosis of lung cancer.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shi J, Hua X, Zhu B, Ravichandran S, Wang M, Nguyen C, Brodie SA, Palleschi A, Alloisio M, Pariscenti G, et al. Somatic genomics and clinical features of lung adenocarcinoma: A retrospective study. PLoS Med. 2016;13:e1002162. doi: 10.1371/journal.pmed.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consonni D, Pierobon M, Gail MH, Rubagotti M, Rotunno M, Goldstein A, Goldin L, Lubin J, Wacholder S, Caporaso NE, et al. Lung cancer prognosis before and after recurrence in a population-based setting. J Natl Cancer Inst. 2015;107:djv059. doi: 10.1093/jnci/djv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 4.Balgkouranidou I, Liloglou T, Lianidou ES. Lung cancer epigenetics: Emerging biomarkers. Biomarkers Med. 2013;7:49–58. doi: 10.2217/bmm.12.111. [DOI] [PubMed] [Google Scholar]
- 5.Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015;34:229–241. doi: 10.1007/s10555-015-9563-3. [DOI] [PubMed] [Google Scholar]
- 6.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Schöttker B, Ordóñez-Mena J, Holleczek B, Yang R, Burwinkel B, Butterbach K, Brenner H. F2RL3 methylation, lung cancer incidence and mortality. Int J Cancer. 2015;137:1739–1748. doi: 10.1002/ijc.29537. [DOI] [PubMed] [Google Scholar]
- 8.Tomizawa Y, Kohno T, Kondo H, Otsuka A, Nishioka M, Niki T, Yamada T, Maeshima A, Yoshimura K, Saito R, et al. Clinicopathological significance of epigenetic inactivation of RASSF1A at 3p21.3 in stage I lung adenocarcinoma. Clin Cancer Res. 2002;8:2362–2368. [PubMed] [Google Scholar]
- 9.Bjaanæs MM, Fleischer T, Halvorsen AR, Daunay A, Busato F, Solberg S, Jørgensen L, Kure E, Edvardsen H, Børresen-Dale AL, et al. Genome-wide DNA methylation analyses in lung adenocarcinomas: Association with EGFR, KRAS and TP53 mutation status, gene expression and prognosis. Mol Oncol. 2016;10:330–343. doi: 10.1016/j.molonc.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium: Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the false discovery rate - A practical and powerful approach to multiple testing. JR Stat Soc Ser A Stat Soc. 1995;57:289–300. [Google Scholar]
- 12.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 13.Raungrut P, Petjaroen P, Geater SL, Keeratichananont W, Phukaoloun M, Suwiwat S, Thongsuksai P. Methylation of 14-3-3σ gene and prognostic significance of 14-3-3σ expression in non-small cell lung cancer. Oncol Lett. 2017;14:5257–5264. doi: 10.3892/ol.2017.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chagnon MJ, Uetani N, Tremblay ML. Functional significance of the LAR receptor protein tyrosine phosphatase family in development and diseases. Biochem Cell Biol. 2004;82:664–675. doi: 10.1139/o04-120. [DOI] [PubMed] [Google Scholar]
- 15.Soulières D, Hirsch FR, Shepherd FA, Bordogna W, Delmar P, Shames DS, Klughammer B. PTPRF Expression as a potential prognostic/predictive marker for treatment with erlotinib in non-small-cell lung cancer. J Thorac Oncol. 2015;10:1364–1369. doi: 10.1097/JTO.0000000000000624. [DOI] [PubMed] [Google Scholar]
- 16.Wu R, Levin AM, Kardia SLR, Cho KR. Identification of novel amplicons in ovarian cancer using a model-based scan statistic for identifying increased chromosomal regions of gene expression. Cancer Res. 2005;65:847–847. [Google Scholar]
- 17.Park J, Lee J, Choi C. Evaluation of drug-targetable genes by defining modes of abnormality in gene expression. Sci Rep. 2015;5:13576. doi: 10.1038/srep13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doldo E, Costanza G, Ferlosio A, Pompeo E, Agostinelli S, Bellezza G, Mazzaglia D, Giunta A, Sidoni A, Orlandi A. High expression of cellular retinol binding protein-1 in lung adenocarcinoma is associated with poor prognosis. Genes Cancer. 2015;6:490–502. doi: 10.18632/genesandcancer.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Napoli JL. Biosynthesis and metabolism of retinoic acid: roles of CRBP and CRABP in retinoic acid: roles of CRBP and CRABP in retinoic acid homeostasis. J Nutr. 1993;123(Suppl 2):362–366. doi: 10.1093/jn/123.suppl_2.362. [DOI] [PubMed] [Google Scholar]
- 20.Doldo E, Costanza G, Ferlosio A, Passeri D, Bernardini S, Scioli MG, Mazzaglia D, Agostinelli S, Del Bufalo D, Czernobilsky B, et al. CRBP-1 expression in ovarian cancer: A potential therapeutic target. Anticancer Res. 2014;34:3303–3312. [PubMed] [Google Scholar]
- 21.Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 22.Hamada Ji, Omatsu T, Okada F, Furuuchi K, Okubo Y, Takahashi Y, Tada M, Miyazaki YJ, Taniguchi Y, Shirato H, et al. Overexpression of homeobox gene HOXD3 induces coordinate expression of metastasis-related genes in human lung cancer cells. Int J Cancer. 2001;93:516–525. doi: 10.1002/ijc.1357. [DOI] [PubMed] [Google Scholar]
- 23.Ohta H, Hamada J, Tada M, Aoyama T, Furuuchi K, Takahashi Y, Totsuka Y, Moriuchi T. HOXD3-overexpression increases integrin alpha v beta 3 expression and deprives E-cadherin while it enhances cell motility in A549 cells. Clin Exp Metastasis. 2006;23:381–390. doi: 10.1007/s10585-006-9047-5. [DOI] [PubMed] [Google Scholar]
- 24.Furuta J, Nobeyama Y, Umebayashi Y, Otsuka F, Kikuchi K, Ushijima T. Silencing of peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66:6080–6086. doi: 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- 25.Zhong Z, Shan M, Wang J, Liu T, Xia B, Niu M, Ren Y, Pang D. HOXD13 methylation status is a prognostic indicator in breast cancer. Int J Clin Exp Pathol. 2015;8:10716–10724. [PMC free article] [PubMed] [Google Scholar]
- 26.Shu Y, Wang B, Wang J, Wang JM, Zou SQ. Identification of methylation profile of HOX genes in extrahepatic cholangiocarcinoma. World J Gastroenterol. 2011;17:3407–3419. doi: 10.3748/wjg.v17.i29.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez-Peña ML, Isaza CE, Pérez-Morales J, Rodríguez-Padilla C, Castro JM, Cabrera-Ríos M. Identification of potential biomarkers from microarray experiments using multiple criteria optimization. Cancer Med. 2013;2:253–265. doi: 10.1002/cam4.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S, Sánchez-Carbayo M. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86. doi: 10.1186/1479-5876-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinton P, Giorgi C, Siviero R, Zecchini E, Rizzuto R. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene. 2008;27:6407–6418. doi: 10.1038/onc.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurskii MD, Tugai VA, Fedoriv AN. Effect of serotonin and calcium on separate components of respiratory chain of mitochondria in some rabbit tissues) Ukr Biokhim Zh. 1970;42:584–588. (In Ukrainian) [PubMed] [Google Scholar]
- 31.Xu X, Chen D, Ye B, Zhong F, Chen G. Curcumin induces the apoptosis of non-small cell lung cancer cells through a calcium signaling pathway. Int J Mol Med. 2015;35:1610–1616. doi: 10.3892/ijmm.2015.2167. [DOI] [PubMed] [Google Scholar]