Abstract
Glioblastoma multiforme (GBM) is one of the most aggressive types of brain tumor worldwide. Despite the advances made in treatment and research, the median survival time for GBM patients remains <1.5 years, providing impetus for the identification of potential novel therapeutic target genes to improve GBM treatment. The deubiquitinating enzyme ubiquitin specific peptidase 15 (USP15) has emerged as a pro-oncogenic factor; however, its function in GBM has yet to be fully elucidated. The present study sought to determine whether or not USP15 is implicated in GBM cell invasion and proliferation. Following the depletion of USP15 in U87-MG and U251-MG cells by lentivirus-mediated USP15 short hairpin RNA (shRNA), the invasiveness of glioma cells was investigated. The results of the present study demonstrated that glioma cells expressing USP15 shRNA exhibited significantly lower invasiveness than cells that did not express USP15 shRNA. Additionally, USP15 depletion led to the upregulation of E-cadherin and downregulation of the mesenchymal markers, N-cadherin and vimentin. Furthermore, the influence of USP15 on glioma cell proliferation was investigated and depletion of USP15 resulted in a marked reduction in cell proliferation. Taken together, the findings of the present study clearly support the hypothesis that USP15 renders GBM cells capable of invasion and proliferation.
Keywords: ubiquitin-specific protease 15, U251-MG, U87-MG
Introduction
Glioblastoma multiforme (GBM), classified as a grade IV astrocytoma by the World Health Organization (1), is the most malignant form of intracranial glioma tumor. The standard treatment for GBM is surgery in combination with chemotherapy and/or radiotherapy. Despite advances in therapeutic research, GBM patients have a survival time of ~1 year post-diagnosis and <5% survive for 5 years (2,3). Highly invasive GBM cells may proliferate extensively and infiltrate into the brain parenchyma, leading to mortality (4,5).
The heterogeneity of GBM at the cellular level hinders the improved understanding of the disease, making it difficult to design and develop effective therapeutic strategies (1,6). To address this challenge, several studies (7,8) have investigated the global gene expression profiles and genetic alterations that drive tumor aggressiveness in GBM, and have stratified GBM into four subtypes: Pro-neural, neural, classical and mesenchymal. The majority of the established GBM cell lines belong to the fourth subtype, which has a mesenchymal gene signature (7).
In general, cadherin switching occurs during cancer cell invasion: It is a process in which expression of epithelial E-cadherin is switched to that of mesenchymal-associated cadherins, including N-cadherin and vimentin, which are typical mesenchymal markers that contribute toward the regulation of cell invasion (9–13). It should be emphasized that, although GBM cells are non-epithelial, these cells necessarily acquire their invasive capability through parts of the epithelial-mesenchymal transition (EMT) program, as exemplified by the upregulation of N-cadherin in invasive glioma cells (14). Therefore, the use of several epithelial or mesenchymal markers to determine the extent of glioma cell invasion is a valid approach.
Elucidating the molecular basis of GBM invasion and proliferation may contribute toward improving GBM therapy; therefore, a number of studies (7,15) have focused on using a variety of relevant genes in order to do so. However, the knowledge obtained from studies into these known genes is insufficient to elucidate the mechanism of GBM progression. As such, the identification of novel molecular targets involved in regulating invasion and proliferation in GBM is necessary.
Deubiquitinases (DUBs) have fundamental functions in the ubiquitin system through their ability to specifically remove ubiquitin from targeted proteins. The human genome encodes ~100 DUBs, which are stratified into 6 families (16). DUBs regulate the ubiquitinization status and the degradation of proteins, affecting the activation and localization of proteins, which in turn is essential for numerous cellular processes and signaling pathways. Therefore, a number of DUBs have been identified as pro- or anticancer factors owing to their regulatory effects on key oncogenes or other proteins involved in tumor progression (17–20). Of these DUBs, the present study focused on ubiquitin-specific protease 15 (USP15), a member of the USP family. A previous study suggested that USP15 functions as a tumor suppressor and that it is likely to be a key target for the inhibition of cancer cell progression (21). For instance, USP15 is downregulated in paclitaxel-resistant ovarian cancer (22); however, other studies have highlighted its pro-oncogenic functions. In metastatic breast cancer MDA-MB-231 cells, USP15 is required for cell migration (23). In a previous study by Eichhorn et al (24), the USP15 gene was revealed to be amplified in glioblastoma, breast cancer and ovarian cancer, and the depletion of USP15 decreased the oncogenic capacity of patient-derived glioma-initiating cells. The present study primarily explores USP15 at the genetic level with a focus on a particular glioma cell subpopulation. To the best of our knowledge, there has been no previous research regarding the general function of USP15 in glioma cells and therefore, the present study was a preliminary investigation into the role of USP15 in glioma cells. The U87-MG cell line was used as a general model of glioma in order to elucidate the general gene functions associated with glioma (25–27).
Materials and methods
Cell lines and cell culture
The glioma U251-MG and U87-MG cell lines were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and the American Type Culture Collection (ATCC; Manassas, VA, USA), respectively. The two human glioma cell lines were cultured at 37°C for 3 days in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 2 mM glutamine, 10% fetal bovine serum (FBS) (both from Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (both from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cells were maintained in an incubator at 37°C in a 5% CO2 atmosphere. Cells (293) were cultured at 37°C for 2 days in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 2 mM glutamine, 10% FBS (both from Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (both from Sigma-Aldrich; Merck KGaA). HEB cells were cultured at 37°C for 3 days in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 2 mM glutamine, 10% FBS (both from Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (both from Sigma-Aldrich; Merck KGaA).
Lentivirus production and transduction
The transduction system was conducted as previously described. Briefly, short hairpin RNA (shRNA) against USP15 (sequence, 5′-GCCAACTGAAGGTTGGAATAA-3′; USP15 KD) was generated and another construct expressing shRNA against a non-mammalian luciferase gene was used as a negative control (NC). These constructs were co-transfected with packaging plasmids (15 µg; Shanghai Hanyu Biotechnology Co., Ltd., Shanghai, China) into 293 cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions, and viral particles were harvested 48 h later. U87-MG and U251-MG cells were infected with a lentivirus containing 6 µg/ml polybrene (Sigma-Aldrich; Merck KGaA).
Transwell invasion assay
For the Transwell invasion assay, 2×104 U87-MG and U251-MG cells stably expressing USP15 shRNAs were plated into 24-well Boyden chambers (Sigma-Aldrich; Merck KGaA) with an 8-µm pore polycarbonate membrane, which was coated with 30 µg Matrigel (BD Biosciences, San Jose, CA, USA). Cells (1×104) were inserted into the upper chamber with 200 µl serum-free DMEM (Thermo Fisher Scientific, Inc.), and DMEM containing 20% FBS was added to the lower chamber to serve as a chemoattractant. Following incubation for 24 h at 37°C in a 5% CO2 atmosphere, cells were washed 3 times with phosphate buffered saline. Cells on the top surface of the insert were removed with a cotton swab. Cells adhering to the lower surface were fixed with 100% methanol at 4°C for 15 min, stained with Giemsa solution at room temperature for 30 sec and counted under a light microscope in five predetermined fields (magnification, ×200).
Cell proliferation
Cells were counted using a Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). A total of 1×103 GBM (U87 and U251 cells) were plated onto 96-well culture plates in triplicate and maintained at 37°C, while cell growth was determined daily for 5 days using a tetrazolium salt-based colorimetric assay (Dojindo Molecular Technologies, Inc.), according to the manufacturer's instructions. Absorbance was measured at 450 nm. The corresponding cells expressing shRNA against an irrelevant luciferase gene was used as a negative control (NC). Three independent experiments were performed.
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 5 mM DTT and 2% SDS) and protein concentration was determined using bicinchoninic acid assay (Beyotime Institute of Biotechnology, Haimen, China). Total protein (30 µg/lane) was resolved using a 10% SDS-PAGE gel and was electro-transferred to polyvinylidene fluoride membranes (Invitrogen; Thermo Fisher Scientific, Inc.), and blocked with 5% skimmed dry milk in Tris-buffered saline (pH 7.5) at room temperature for 1 h in Tris-buffered saline (pH 7.5). Membranes were immunoblotted overnight at 4°C with the following primary antibodies: N-cadherin (dilution 1:200; cat. no. 4061), vimentin (dilution 1:200; cat. no. 3932), E-cadherin (dilution 1:500; cat. no. 3195) (all from Cell Signaling Technology, Inc., Danvers, MA, USA) and β-actin (dilution 1:3,000; cat. no. HRP60008; ProteinTech Group, Inc., Chicago, IL, USA). A horseradish peroxidase-conjugated IgG secondary antibody (dilution 1:2,000; cat. no. ab6721, Abcam, Cambridge, MA, USA) was added and incubated at room temperature for 1 h. Bound antibodies were detected using the BeyoECL system (cat. no. P0018; Beyotime Institute of Biotechnology), and densitometry ImageQuant 5.2 software (GE Healthcare Life Sciences, Little Chalfont, UK) was used for analysis.
Statistical analysis
Data are presented in the graphs as the mean ± standard deviation of three independent experiments. The differences among groups were determined using analysis of variance with post hoc contrasts determined by Student-Newman-Keuls test and comparisons between two groups were analyzed using the Student's t-test. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
USP15 regulates glioma cell invasion in vitro
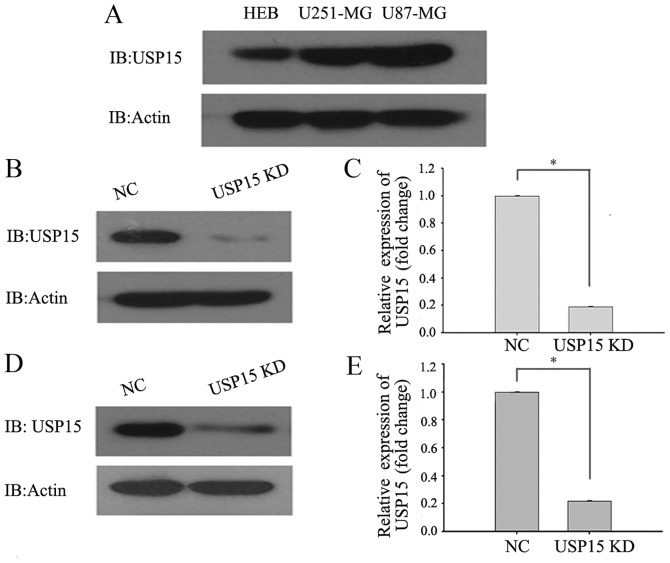
To directly investigate the role of USP15 in GBM cells, the protein levels of USP15 in the HEB cell line and the commonly used typical glioma cell lines (U87-MG and U251-MG) were determined using western blot analysis. The results demonstrated that the protein levels of USP15 in U87-MG and U251-MG cells were higher in the glioma cells than in the HEB cell line (Fig. 1A). RNA interference technology was subsequently used to deplete USP15 expression. U-87-MG and U251-MG cells were transduced with a lentivirus-encoding control or USP15 shRNA. At 3 days after transduction, cell lysates were subjected to western blot analysis to detect the protein level of USP15. Densitometric analysis of the western blotting results, by comparing USP15 protein levels in control shRNA-expressing cells with those expressing USP15 shRNA, revealed significant reductions in USP15 protein levels in the U87-MG (Fig. 1B and C) and U251-MG (Fig. 1D and E) glioma cells treated with lentivirus-mediated USP15 shRNA. These findings indicated that USP15 was successfully depleted in the two glioma cell lines.
Figure 1.
Knockdown of USP15 inhibits cell proliferation of glioma cells in Cell Counting Kit-8 assays in vitro. (A) Analysis of USP15 expression in a HEB cell line and typical glioma cell lines. Western blot analysis revealed that USP15 protein level was upregulated in U87-MG and U251-MG cells compared with expression in the HEB cell line. (B) USP15 was knocked down successfully in U87-MG cells, as revealed by western blot analysis. (C) Densitometry analysis demonstrated the efficiency of USP15 depletion in U87-MG cells (*P<0.001). (D) USP15 was knocked down successfully in U251-MG cells, as determined by western blot analysis. (E) Densitometry analysis demonstrated the efficiency of USP15 depletion in U251-MG cells (*P<0.001). USP15, ubiquitin specific peptidase 15; HEB, human normal glial; NC, negative control.
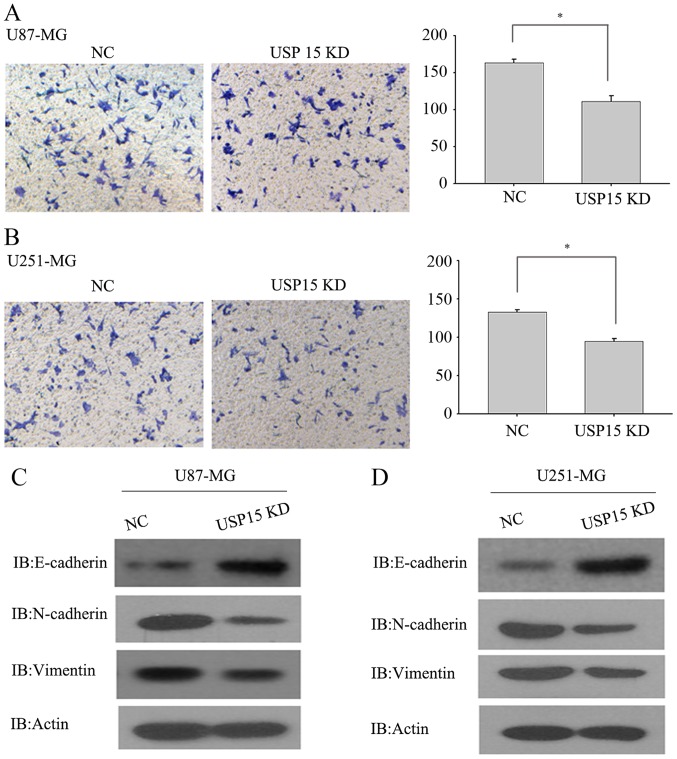
Eichhorn et al (24) observed that neurospheres expressing USP15 shRNA yielded fewer tumors than did the neurospheres expressing control shRNA. This result indicated that expression of USP15 may result in a loss of cell-cell contact and cell scattering, a potential consequence of cell invasion. We therefore hypothesized that USP15 may promote glioma cell invasion. To test this hypothesis, a Transwell invasion assay was conducted using U87-MG and U251-MG cells, which are invasive in vitro, to determine the invasive abilities of these cells. U87-MG and U251-MG cells transduced with lentivirus-encoding control or USP15 shRNA for 3 days were subjected to an invasion assay. Next, 24 h after incubation, USP15 depletion was observed using lentivirus-mediated USP15 shRNA in U87-MG cells, leading to a significant reduction in cell invasion compared with that in the cells treated with control shRNA (Fig. 2A; P=0.002). Furthermore, depletion of USP15 in U251-MG cells resulted in a significant reduction in invasion (P=0.012; Fig. 2B), albeit relatively moderate when compared with the extent of difference in U87-MG cells. These data indicate the role of USP15 in glioma cell invasion. Although the USP15 depletion efficiency in the U87-MG and U251-MG cells was virtually identical, as determined by western blot analysis (Fig. 1B-E), the degree of impairment of cell invasive capacities differed between the two cells types. This may be attributable to the effect of USP15 on the invasive capacity of different types of glioma cell.
Figure 2.
Knockdown of USP15 inhibits the invasiveness of glioma cells, as determined by Transwell invasion assay. (A) USP15 depletion led to a significant reduction in the invasive ability of U87-MG cells (*P=0.002). (B) USP15 depletion led to a significant reduction in the invasive ability of U251-MG cells (*P=0.012). Altered protein levels of endogenous E-cadherin and typical mesenchymal markers, N-cadherin and vimentin, in (C) U87-MG and (D) U251-MG were detected by western blot analysis. USP15, ubiquitin specific peptidase 15; NC, negative control; KD, knockdown.
As mesenchymal-associated genes are associated with glioma cell invasion, USP15 expression was depleted in U87-MG and U251-MG cells using lentivirus-encoding USP15-specific shRNA to examine changes to the expression of N-cadherin, vimentin and E-cadherin. As expected, USP15 depletion significantly diminished the expression of N-cadherin and vimentin, while the protein level of E-cadherin was increased upon USP15 depletion (Fig. 2C and D). These data provide further support for the hypothesis that USP15 expression is required for glioma cell invasion.
USP15 regulates glioma cell proliferation in vitro
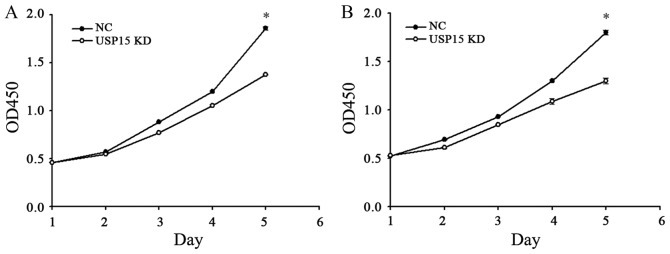
The fact that depletion of USP15 decreased the oncogenic capacity of patient-derived glioma-initiating cells indicated that cells with depleted expression of USP15 using lentivirus-encoding USP15 shRNA generated smaller tumors than cells expressing control shRNA. Therefore, further experimentation in the present study aimed to determine whether USP15 may promote glioma cell proliferation. To quantify cell proliferation, cell growth was examined at 0-, 1-, 2-, 3-, 4- and 5-day time-points using a CCK-8 assay. The depletion of USP15 did not lead to a significant reduction in U87-MG proliferation until day 4, with the peak reduction occurring at day 5 (P<0.001; Fig. 3), indicating that USP15 promotes glioma cell proliferation. A similar result was observed in U251-MG cells, thereby corroborating the aforementioned conclusion (Fig. 3B).
Figure 3.
Effects of USP15 depletion on U87-MG and U251-MG cell proliferation. (A) USP15 depletion resulted in significant reductions in U87-MG cell proliferation, particularly on day 5 (P<0.001), compared with NC. (B) USP15 depletion resulted in marked reductions in U251-MG cell proliferation, particularly on day 5 (P<0.001), compared with NC. USP15, ubiquitin specific peptidase 15; NC, negative control; KD, knockdown; OD, optical density.
Discussion
During tumor progression, cancer cells acquire capabilities that permit them to invade the neighboring tissues. The steps involved in the dispersion of cancer cells include the loss of cell-cell contact and increased invasiveness (28). In gliomas, tumor cells are known to exhibit a diffusely infiltrative invasion pattern, in which the tumor cells migrate as single cells, resulting in a reduced requirement for surgical and radiological interventions (29). Therefore, the identification of potent genes associated with glioma cell invasion is urgently required. Considering the fact that targeting existing invasion-associated gene targets has failed to significantly attenuate glioma cell invasiveness, the present study instead focused on the identification of novel targets that may exert suppressive effects on glioma cell invasion. Owing to the fact that inhibition of USP15 (a member of the USP family involved in deubiquitinization) caused a marked reduction in the number of glioma xenografts (24), it was speculated that the reduction may be attributed to the loss of cell-cell contact and cell scattering, a consequence of cancer cell invasion. The present study demonstrated that depletion of USP15 expression resulted in a reduction in the invasiveness of typical glioma cell models. These findings indicated that USP15 promotes the invasive capacity of glioma cells. Notably, a study by Eichhorn et al revealed that downregulation of USP15 decreased the number of tumors generated, indicating cancer cell scattering. However, cell scattering alone is unable to recapitulate the invasive behavior of cells. For instance, the overexpression of a truncated form of E-cadherin that lacks the ectodomain of the wild-type protein may result in loss of cell-cell contact and cell scattering. However, the truncated product failed to elicit cancer cell invasion (30). To the best of our knowledge, the results of the present study demonstrate for the first time that USP15 renders glioma cells capable of invasion and is involved in glioma progression.
Cadherins are critical players in a number of mechanisms with roles in tumor migration and invasion (9,31). These genes were initially identified owing to their roles in metaplasia, also known as EMT. During the EMT, upregulation of N-cadherin is associated with the suppression of E-cadherin, in a process known as the E- to N-cadherin switch (32). Although glioma cells are non-epithelial, with limited or non-existent E-cadherin expression, the cells with high invasive ability may employ a program similar to EMT (33). Therefore, the relative expression of E- and N-cadherin is regarded as a hallmark of examining glioma cell invasiveness. Furthermore, upon USP15 depletion, changes in the expression of E- and N-cadherin were associated with changes in the invasiveness of glioma cells. The acquisition of invasive ability prompts glioma cells to undergo a mesenchymal phenotypic shift, upregulating N-cadherin and vimentin, which is consistent with published findings that glioma cell invasion is associated with the increased expression of N-cadherin (14). Nonetheless, the association of E-cadherin expression with glioma cell invasiveness should be interpreted with caution, given that a number of glioma cell lines lack E-cadherin expression.
USP15 is known to be a tumor suppressor as it promotes apoptotic processes and is downregulated in drug-resistant ovarian cancer (22). Additionally, regulation of the Wnt signaling pathway is essential for a number of types of cancer, including glioma; USP15 acts as a negative mediator of this pathway by deubiquitylating and stabilizing the tumor suppressor adenomatous polyposis coli, thereby promoting the degradation of β-catenin by the proteasome (34). Furthermore, USP15 negatively regulates the nuclear factor NF-κB (NF-κB) signaling pathway, which is required for cancer cell invasion, by stabilizing IκB kinase-α, an inhibitor of NF-κB (35). However, evidence indicates that USP15 serves an opposing role in cancer cells, as exemplified by the USP15-dependent promotion of glioblastoma oncogenesis (24) and the USP-induced increase in MDA-MB-231 cell migration (23). With regards to the increase in migration, the present study revealed that USP15 promoted cell invasion and proliferation in glioma cells. The fact that USP15 was identified to serve a dual function was expected, as certain DUBs have already been documented to exhibit detrimental or protective dual functions in cancer, depending upon the context and cell type in which these specific DUBs function (21). For instance, USP2 rescues prostate cancer cells from apoptosis by stabilizing fatty-acid synthase (36) and deubiquitylates relevant apoptosis-inducing factors, thereby promoting cell death (37). These data reveal that the pro-cancer function of USP15 may contribute to an improved understanding of the gene, and may rationally delineate its complex interaction with key genes associated with cancer cell invasion and proliferation.
In conclusion, the present study identified a positive association between USP15 expression and cell invasion and proliferation in U87-MG and U251-MG cells, providing an insight into the importance of USP15 in a subset of high-grade gliomas. Future studies should focus on identifying the detailed molecular mechanisms underlying glioma invasion and proliferation. Since DUBs have been demonstrated to be targetable by small organic molecules, USP15 may be a potential therapeutic target and may contribute toward novel therapeutic interventions in GBM.
Acknowledgements
The present study was funded by the National Natural Science Foundation of China (grant no. 81660502).
References
- 1.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: A social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Persano L, Rampazzo E, Basso G, Viola G. Glioblastoma cancer stem cells: Role of the microenvironment and therapeutic targeting. Biochem Pharmacol. 2013;85:612–622. doi: 10.1016/j.bcp.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the united states in 2006–2010. Neuro-Oncol. 2013;15(Suppl 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 5.Malzkorn B, Reifenberger G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr Opin Oncol. 2016;28:494–501. doi: 10.1097/CCO.0000000000000327. [DOI] [PubMed] [Google Scholar]
- 6.Colman H, Aldape K. Molecular predictors in gliobla=stoma: Toward personalized therapy. Arch Neurol. 2008;65:877–883. doi: 10.1001/archneur.65.7.877. [DOI] [PubMed] [Google Scholar]
- 7.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toki S, Kagaya S, Shinohara M, Matsumoto T, Takahata Y, Saito H, Matsumoto K. Genome-wide gene expression analysis of the human intestinal epithelial cells after stimulation with lactic acid bacteria. J Allerg Clin Immunol. 2006;117:S153. doi: 10.1016/j.jaci.2005.12.611. [DOI] [Google Scholar]
- 9.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 10.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–735. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 11.Anastasiadis PZ. p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim Biophys Acta. 2007;1773:34–46. doi: 10.1016/j.bbamcr.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 12.Gumbiner BM. Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/S0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 13.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D, Isachenko N, Fouse SD, Phillips JJ, Cheresh DA, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22:21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newcomb EW, Cohen H, Lee SR, Bhalla SK, Bloom J, Hayes RL, Miller DC. Survival of patients with glioblastoma multiforme is not influenced by altered expression of p16, p53, EGFR, MDM2 or Bcl-2 genes. Brain Pathol. 1998;8:655–667. doi: 10.1111/j.1750-3639.1998.tb00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacco JJ, Coulson JM, Clague MJ, Urbé S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62:140–157. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 19.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, Bianchi J, Aushev VN, Camonis J, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 20.Allende-Vega N, Sparks A, Lane DP, Saville MK. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. 2010;29:432–441. doi: 10.1038/onc.2009.330. [DOI] [PubMed] [Google Scholar]
- 21.Fraile JM, Quesada V, Rodríguez D, Freije JM, López-Otín C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 22.Xu M, Takanashi M, Oikawa K, Tanaka M, Nishi H, Isaka K, Kudo M, Kuroda M. USP15 plays an essential role for caspase-3 activation during Paclitaxel-induced apoptosis. Biochem Biophys Res Commun. 2009;388:366–371. doi: 10.1016/j.bbrc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, Enzo E, Moro S, Polo S, Dupont S, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 24.Eichhorn PJ, Rodón L, Gonzàlez-Juncà A, Dirac A, Gili M, Martínez-Sáez E, Aura C, Barba I, Peg V, Prat A, et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 25.Guo Z, Li G, Bian E, Ma CC, Wan J, Zhao B. TGF-β-mediated repression of MST1 by DNMT1 promotes glioma malignancy. Biomed Pharmacother. 2017;94:774–780. doi: 10.1016/j.biopha.2017.07.081. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y, Wang J, Yang J, Miao J. Synergistic antitumor effect of ING4/PTEN double tumor suppressors mediated by adenovirus modified with RGD on glioma. J Neurosurg Sci. 2017 Apr 12; doi: 10.23736/S0390-5616.17.03978-9. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 31.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 32.Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann Oncol. 2010;21(Suppl 7):vii89–vii92. doi: 10.1093/annonc/mdq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg R. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Huang X, Langelotz C, Hetfeld-Pechoc BK, Schwenk W, Dubiel W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J Mol Biol. 2009;391:691–702. doi: 10.1016/j.jmb.2009.06.066. [DOI] [PubMed] [Google Scholar]
- 35.Schweitzer K, Bozko PM, Dubiel W, Naumann M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J. 2007;26:1532–1541. doi: 10.1038/sj.emboj.7601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, Lechpammer M, Huesken D, Zimmermann J, Signoretti S, Loda M. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/S1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- 37.Oh KH, Yang SW, Park JM, Seol JH, Iemura S, Natsume T, Murata S, Tanaka K, Jeon YJ, Chung CH. Control of AIF-mediated cell death by antagonistic functions of CHIP ubiquitin E3 ligase and USP2 deubiquitinating enzyme. Cell Death Differ. 2011;18:1326–1336. doi: 10.1038/cdd.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]