Abstract
Aberrant expression of Ras homolog enriched in brain (RHEB) has been observed in a variety of cancer tissues and is closely associated with clinicopathological features. However, the expression profile of RHEB in patients with hepatocellular carcinoma (HCC) and its clinical signature with underlying mechanisms have not been explored thus far. To analyze the association between RHEB expression and clinicopathological features, the RHEB expression levels were determined in the present study using gene microarrays, immunohistochemistry and western blotting in 60 liver cancer tissues and 35 normal liver tissues. Downregulation of RHEB expression in liver cancer cell lines was achieved by RNA interfering technology to explore its biological function in HCC. RHEB expression was high in liver cancer tissues, with an increase of 2.00±0.19-fold compared with normal tissues and of 2.00±0.27-fold compared with adjacent non-cancer tissues. RHEB expression increased along with the clinical staging of HCC, and the overall survival and mortality of patients were closely correlated to RHEB levels, micro-vascular invasion, hepatitis B virus-DNA titer, tumor differentiation and pathological satellites (P<0.05). After knocking down RHEB in SMMC-7721 cells, the growth of liver cancer cells was significantly reduced. The majority of cells were blocked in S-phase, and their colony-forming and proliferating abilities significantly decreased (P<0.05). In vivo, upon downregulation of RHEB expression, the tumorigenic ability of HCC significantly decreased (P<0.05). These data suggest that RHEB expression is a significant prognostic factor and may be important in HCC cell growth. The present study highlights the importance of RHEB as a novel prognostic marker of HCC.
Keywords: RHEB, HCC, metastasis, prognosis, gene microarray, RNA interference
Introduction
Primary liver cancer, which consists of 85–90% hepatocellular carcinoma (HCC), is the fifth and seventh most common cancer in men and women, respectively, worldwide, and ~85% of cases occur in developing countries (1–3). Liver cancer is the third most common cause of cancer-associated mortality worldwide, and in China, it accounts for half of the cancer cases and cancer-associated mortalities (1–3).
HCC is the leading cause of mortality from cancer in rural China, and the primary challenges for improving the prognosis of HCC are invasiveness, recurrence and metastasis (4–6). Surgical resection and liver transplantation are the main curative modalities for HCC (7,8). However, if the restrictive common international criteria were applied, only 5–10% of patients would be offered surgical resection (7). The long-term prognosis for HCC remains poor, with a high 5-year tumor recurrence rate and a low 5-year survival rate (7–9). Transcatheter arterial chemoembolization has increased the chances of conventional surgical treatments for HCC, and other clinical treatments, including radiofrequency ablation and microwave ablation, have been widely applied; however, improvements in the prognosis and survival rate remain limited and poor (10). Rapid advances in cellular and molecular techniques offer novel approaches for cancer treatments. Numerous molecular markers associated with recurrence, metastasis and invasiveness have been identified in tissue and serum, which exhibit a prognostic significance and may be promising therapeutic targets (8). However, the specificity and reliability of these markers is poor (8). Therefore, it is necessary to identify a novel reliable target molecule to improve the treatment of HCC.
In 1994, Ras homolog enriched in brain (RHEB) was first identified and cloned while screening genes that regulate neural activity (11). As a novel and unique member of the Ras superfamily of G proteins, RHEB is a conservative protein from yeast to humans, and plays a significant role in regulating growth and cell cycle (12). With its GTPase activity, RHEB can shift between the combination state with guanosine diphosphate and guanosine-5′-triphosphate (GTP), which determines its molecular biological function (11–13).
Previous studies demonstrated that RHEB is overexpressed in various malignant tumors (14,15). In breast cancer and in head and neck cancer, its overexpression correlates directly with a worse prognosis (14–16). RHEB is also overexpressed in skin cancer, and furthermore, depending on transgenic overexpression technology, the hyperplasia and canceration of the skin can be induced by increasing RHEB expression in mouse keratin cells (14). Prostate cancer studies revealed that RHEB is overexpressed in cancer tissues compared with adjacent non-cancer tissues, and in androgen-independent prostate cancer cell lines, RHEB overexpression is also observed (14–16). These studies suggest that RHEB has oncogene functions; however, whether RHEB plays an important function in the occurrence and progression of HCC remains currently unknown.
Therefore, in the present study, the association between RHEB expression and the clinicopathological features of HCC was determined. The present study provides a basis for elucidating the underlying molecular mechanisms of HCC.
Materials and methods
Cell cultures
Human liver cancer cell lines Hep3B, HepG2 and Huh-7 were obtained from the American Type Culture Collection (Manassas, VA, USA), while SMMC-7721, MHCC-97-L and MHCC-97-H were a gift from the Liver Cancer Institute, Zhongshan Hospital, Fudan University (Shanghai, China). These cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin solution (Beyotime Institute of Biotechnology, Haimen, China) at 37°C in a 5% CO2 incubator.
Tumor formation assay
A total of 18 five-week-old male BALB/c nude mice, (weight, ~18 g) were obtained from the Shanghai Experimental Animal Center (Shanghai, China). The mice were kept under standard laboratory conditions: 21–23°C controlled temperature, 50–65% humidity and 12-h light-dark cycle lighting, with free access to drinking water and chow. The condition of the mice was monitored every other day. Tumor size was measured by a vernier caliper weekly and calculated as (length × width2)/2. All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (9).
HCC tissue collection and patient follow-up
The surgically resected tissue samples, including 60 cases of diagnosed liver cancer with matching adjacent tissue samples and 35 normal liver tissue samples from patients under liver transplantation, were collected from the Eastern Hepatobiliary Surgery Hospital, Second Military Medical University (Shanghai, China) between May 2010 and May 2011. Of the 60 HCC cases, 39 were males and 21 were females, with a median age of 52.0±8.9 years. Of these cases, 26 were clinical stage I, 15 were stage II and 19 were stage III–IV. All tissue samples were stored in liquid nitrogen within 30 min after surgical resection of the biopsy for later use.
Ethics statement
Written informed consent from all patients and approval from the Ethics Committee of the Eastern Hepatobiliary Surgery Hospital were obtained. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Second Military Medical University.
Gene microarray
Gene expression in cancer tissues and matching adjacent tissues was assayed using a DNA microarray, and the results were then analyzed using a gene map analysis software (Human CHIP version 1; DNA Chip Research Inc., Tokyo, Japan).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cultured cells, liver cancer and non-cancerous liver specimens using TRIzol regent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. RT for complementary DNA was achieved by a TaKaRa RNA PCR kit (Takara Biotechnology Co., Ltd., Dalian, China) and detected using an RT-qPCR kit and 7900HT Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers for the RHEB gene are as follows: Forward 5′-ACTCCTACGATCCAACCATAGA-3′ and reverse 5′-TGGAGTATGTCTGAGGAAAGATAGA-3′, and the probe was 6-carboxyfluorescein (6-FAM)-5′-AGACACAGCCGGGCAAGATGAATA-3′-minor groove binder (MGB). The GAPDH gene was used as an internal control, and the primers were as follows: Forward 5′-CATGGGTGTGAACCATGAGA-3′ and reverse 5′-GAGTCCTTCCACGATACCAAAG-3′, and the probe was FAM-5′-AGATCATCAGCAATGCCTCCTGCA-3′-MGB. The following cycling conditions were used: 94°C for 5 min, followed by 94°C for 1 sec, 65°C for 15 sec and 72°C for 30 sec for 35 cycles. RHEB messenger RNA (mRNA) from normal liver tissues and untreated cultured cells was used as the control for RT-qPCR.
Immunohistochemical analysis
Sections (4-µm) were cut from paraffin blocks for immunohistochemical staining. A rabbit anti-human RHEB monoclonal antibody (ab92313) was purchased from Abcam (Cambridge, MA, USA), and the PV-9000 immunohistochemical reagent kit was obtained from Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd. (Beijing, China). RHEB protein expression in liver cancer was detected by immunohistochemical staining with the PV-9000 kit according to the manufacturer's protocol. After conventional dewaxing and hydration, the sections were incubated in 3% H2O2 with deionized water to block the endogenous peroxidase activity. A microwave antigen retrieval procedure with ethylenediaminetetraacetic acid (pH 8.0) was used, and then the sections were cooled and incubated with rabbit anti-human RHEB antibody (dilution 1:100) at 4°C overnight. Next, the sections were incubated with a secondary anti-rabbit antibody (dilution 1:200; 8114; Cell Signaling Technology, Inc., Danvers, MA, USA) at 37°C for 30 min, and then rinsed with 1X PBS. Finally, 3,3′-diaminobenzidine was used to develop the sections. The sections were incubated at room temperature without light for 10 min, and the reaction was completed with distilled water. The tan or brown granules in the nucleus and cytoplasm represented positive RHEB protein expression.
Four different views were randomly observed with high magnification (×400), and the number of total cells and cells positive for nuclear staining were recorded. Based on the ratio of positive cells, the staining was scored as 1 (0–10 cells), 2 (11–50 cells), 3 (51–75 cells) and 4 (76–100 cells). The staining intensity was scored as 1 (negative staining), 2 (weak staining), 3 (moderate staining) and 4 (strong staining). The final score for RHEB protein expression was calculated as the product of the staining score and the intensity score. Based on the final score, 0–4 was (−), 5–8 was (+), 9–12 was (++) and 13–16 was (+++). For the purpose of statistical evaluation, (−) and (+) represented negative and weakly positive expression, respectively, while (++) and (+++) represented highly positive expression. All results were confirmed using a blind method by ≥2 pathologists.
Western blotting
After being cultured for 48–72 h, the adherent cells were rinsed in chilled PBS, and lysis buffer was added to cover the cells completely at room temperature. Then, the buffer was collected and the cells were centrifuged at 600 × g/min for 5 min at 4°C. Upon determination of protein concentration with the BCA Protein Assay kit (P0012; Beyotime Institute of Biotechnology), the remaining supernatant was boiled and placed on ice for 5 min. In total, 50 µg of sample was loaded in each well, and separated by 12% polyacrylamide gel electrophoresis at ~200 V. The separated proteins were electrophoretically transferred onto nitrocellulose membranes at 60 V. The blotted membrane was blocked with 5% skim milk in Tris-buffered saline with Tween 20 (TBST), and then washed with TBST. Anti-RHEB antibody (ab92313) was next added at a 1:1,000 dilution and incubated at 4°C overnight. The membrane was washed in TBST, and horseradish peroxidase-conjugated immunoglobulin G secondary antibody (7074; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) was added at a 1:1,000 dilution and incubated at room temperature for 2 h. The nitrocellulose membrane was developed with a chemiluminescent solution (Kangwei, Beijing, China), covered with plastic wrap and placed in a cassette. Finally, the blot was analyzed upon exposure.
RNA interference (RNAi)
The nucleotide sequences were designed for the RHEB mRNA sequence derived from the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) using the Ambion RNAi software version 3 (Thermo Fisher Scientific, Inc.). According to this nucleotide sequence, a pair of complementary oligonucleotide chains encoding the corresponding short hairpin (sh) small interfering (si) RNA that binds specifically to the RHEB mRNA were synthesized. The sequences were as follows: Sense strand 5′-GAAAGACCUGCAUAUGGAAAGGGTG-3′ and antisense strand 5′-CACCCUUUCCAUAUGCAGGUCUUUCUU-3′. Without any sequences matched to the human genome sequences, the siRNA-NC was used as the negative control, with sense strand 5′-UUCUCCGAACGUGUACGUTT-3′ and antisense strand 5′-ACGUGACACGUUCGGAGAATT-3′. The synthesis of these oligonucleotide chains was performed by Bioneer Corporation (Daejeon, Korea).
The pSilencer™ 2.1-U6neo plasmid (Ambion; Thermo Fisher Scientific, Inc.), which was digested with BamHI and HindIII endonucleases (New England BioLabs, Inc., Ipswich, MA, USA), was collected and purified with a plasmid DNA extraction kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol, to obtain the fragments of the target gene and carrier. The eukaryotic expression vector pU-RHEB-siRNA was constructed by ligating the oligonucleotide chains and carrier fragments into the pSilencer™ 2.1-U6neo plasmid, and was sequenced with the Sanger method by Thermo Fisher Scientific, Inc.
Prior to transfection (20 h), liver cancer cells were plated in 6-well plates (Corning Incorporated, Corning, NY, USA). Cells were transfected with the eukaryotic expression vector pU-RHEB-siRNA using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) when the cells reached 80–90% confluency. Both liver cancer cells, which were transfected with the empty vector, and untreated liver cancer cells served as the control groups. After transfection (24 h), the cells were subcultured in G418-free DMEM at a ratio of 1:20, and selective medium with G418 (400 µg/ml) (Sigma-Aldrich; Merck Millipore) was added the following day.
Cell cycle analysis
To analyze the cell cycle distribution, all the collected cells were fixed in 70% ethanol at −20°C overnight, stained with propidium iodide (36 mg/ml; Sigma-Aldrich; Merck Millipore) for 30 min and analyzed by flow cytometry (Beckman Coulter, Inc., Brea, CA, USA).
Statistical analyses
The association between RHEB expression and clinicopathological features was evaluated with the χ2 test, or with the Fisher's exact test when the χ2 test was not suitable. The correlation between RHEB overexpression (as determined by immunohistochemistry and quantified by integral optical density) and tumor-node-metastasis (TNM) stages was evaluated by analysis of variance (ANOVA) and Spearman correlation analysis. Cox proportional hazards regression analysis of multiple factors was employed to assess the association between clinical data and survival or recurrence. All statistical analyses were performed using the SPSS version 12.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Aberrant expression of RHEB in liver cancer cell lines and tissues
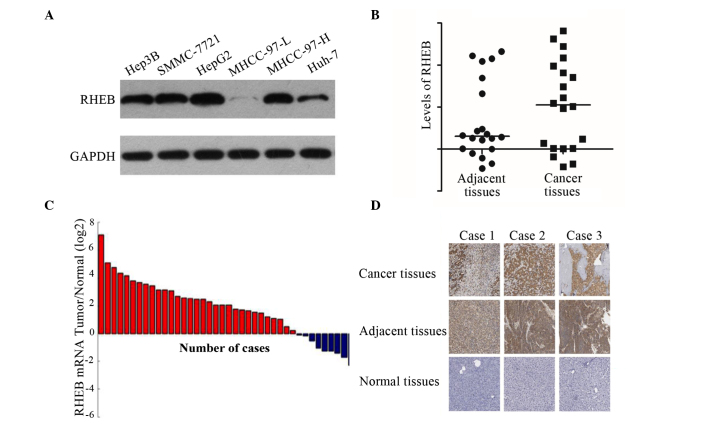
To investigate the RHEB expression in different liver cancer cell lines, a total of six strains of liver cancer cells were selected, including Hep3B, SMMC-7721, HepG2, MHCC-97-L, MHCC-97-H and Huh-7. Western blot analysis demonstrated that RHEB is overexpressed in all these cell lines, with the exception of MHCC-97-L (Fig. 1A).
Figure 1.
RHEB expression in liver cancer cell lines and tissues. (A) RHEB was overexpressed in Hep3B, SMMC-7721, HepG2 and MHCC-97-H cells, but not in MHCC-97-L cells, as demonstrated by western blot analysis. (B) RHEB expression in liver cancer relative to the control gene GAPDH (5.00±0.34-fold) was upregulated compared with that in adjacent tissues (2.00±0.27-fold) and normal tissues (2.00±0.19-fold), as evaluated by gene microarray analysis. (C) RHEB messenger RNA level in liver cancer samples was upregulated compared with that in non-cancerous liver samples, as demonstrated by reverse transcription-quantitative polymerase chain reaction analysis. (D) RHEB was overexpressed in liver cancer tissues, but little or no expression was detected in normal liver tissues, as analyzed by immunohistochemistry. Magnification, ×200. RHEB, Ras homolog enriched in brain; mRNA, messenger RNA.
To further study the RHEB expression in liver cancer tissue in vivo, 20 paired liver cancer tissues and adjacent tissue specimens were examined via gene microarray. The results revealed that the expression of RHEB in liver cancer compared with the control gene GAPDH (5.00±0.34-fold) was upregulated relative to adjacent tissues (2.00±0.27-fold; P=0.025) and normal tissues (2.00±0.19-fold; P=0.035) (Fig. 1B). Furthermore, the RHEB mRNA expression in the liver cancer tissue samples, as detected by RT-qPCR, was significantly upregulated compared with non-cancerous liver tissues (Fig. 1C), which was consistent with the results of gene microarray. The immunohistochemical results revealed that RHEB was expressed primarily in the nucleus and partially in the cytoplasm, and of 60 liver cancer tissues, 41 exhibited positive expression of RHEB, while little or no RHEB expression was observed in the adjacent or normal tissues (Fig. 1D).
Relative analysis between RHEB expression and clinicopathological features in patients with HCC
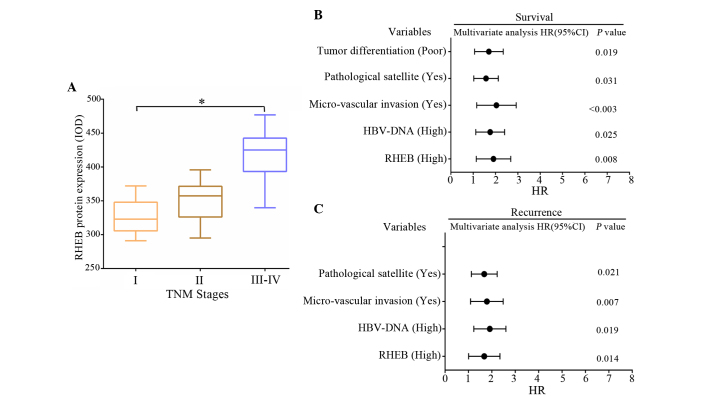
To further investigate the role of RHEB in patients with HCC, the association between the immunohistochemical results and the clinicopathological data was evaluated. According to the immunohistochemical results, of the 26 liver cancer samples at stage T1, positive RHEB expression occurred in 17. Of the 15 liver cancer samples at stage T2, positive RHEB expression occurred in 9. Of the 19 liver cancer samples at stage T3-T4, positive RHEB expression occurred in 13. Statistical analysis demonstrated that the differences in positive RHEB expression between T1 and the other stages were not significant (P=0.876). The differences in RHEB expression between liver cancer groups with different TNM stages were analyzed by ANOVA and were observed to be significant (P=0.020) (Fig. 2A). Spearman correlation analysis demonstrated that high expression of RHEB positively correlated with TNM staging in HCC (r=0.583, P=0.031). Survival analysis revealed that the survival time of HCC patients was closely correlated to RHEB expression level, hepatitis B virus (HBV)-DNA titer, micro-vascular invasion, pathological satellites and tumor differentiation (P<0.05) (Fig. 2B), and was not dependent on gender (P=0.231), age (P=0.064) or tumor size (P=0.157). According to the correlation analysis between clinicopathological data and postoperative recurrence, postoperative recurrence was closely correlated to RHEB expression level, HBV-DNA titer, micro-vascular invasion and pathological satellites (P<0.05) (Fig. 2C), and was not dependent on gender (P=0.165), age (P=0.073), tumor size (P=0.142) or tumor differentiation (P=0.351).
Figure 2.
Correlation analysis between RHEB expression and clinicopathological data of patients with HCC. (A) The differences in RHEB expression between different tumor stages in liver cancer tissues was significant (P=0.020). (B and C) Correlation analysis between different clinicopathological data and (B) survival time and (C) postoperative recurrence of patients with HCC. RHEB, Ras homolog enriched in brain; IOD, integral optical density; HCC, hepatocellular carcinoma; HR, hazard ratio; CI, confidence interval; HBV, hepatitis B virus.
RHEB silencing downregulates the proliferation of a hepatoma cell line
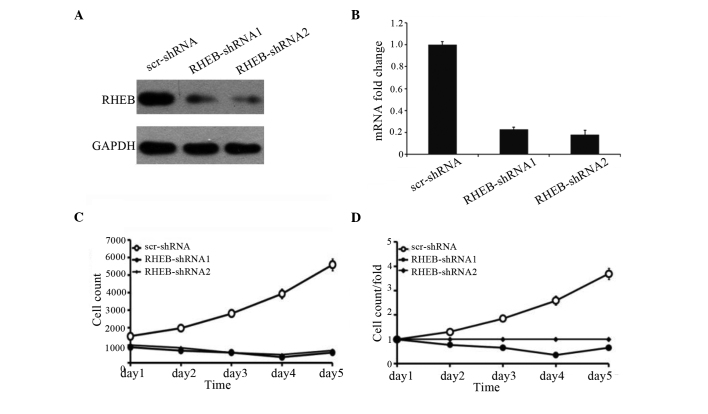
To identify the effects of RHEB expression on the growth of HCC cells, the SMMC-7721 liver cancer cell line with RHEB overexpression was selected and treated with shRNA-RHEB. shRNA-RHEB downregulated RHEB expression in SMMC-7721 cells, and its interference efficiency was detected by RT-qPCR and western blotting (Fig. 3A), which demonstrated 80% efficiency (P<0.0001; Fig. 3B). Upon downregulation of RHEB expression, the proliferation of the knocked down cells was significantly inhibited (P=0.025; Fig. 3C), and the inhibition was obvious on the third day post-transfection (P=0.03; Fig. 3D), which suggests that RHEB plays an important biological role in HCC.
Figure 3.
Influence of silencing RHEB on the growth of SMMC-7721 cells. (A) RHEB expression in SMMC-7721 cells treated with RHEB-shRNA according to by western blot analysis. (B) Variation in interference efficiency in SMMC-7721 cells treated with different shRNAs. (C and D) Changes in cell proliferation ability upon treatment with shRHEB were reported as (C) cell count and (D) cell count/fold. RHEB, Ras homolog enriched in brain; mRNA, messenger RNA; scr, scramble; shRNA, small hairpin RNA.
Influence of RHEB silencing on the biological behavior of a liver cancer cell line
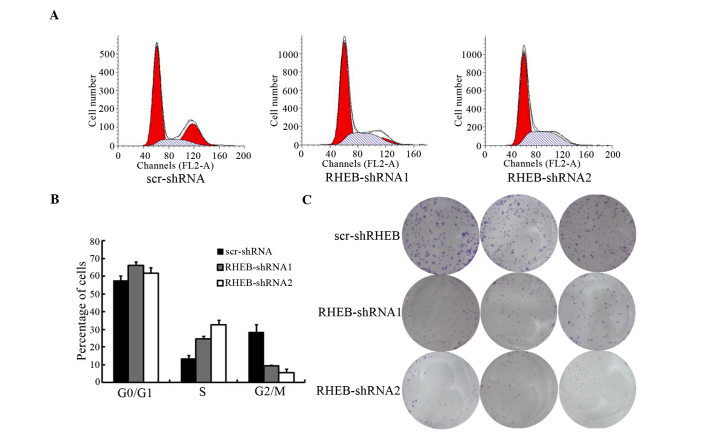
To evaluate the biological functions of RHEB, the cell cycle of SMMC-7721 cells treated with shRNA-RBEB was analyzed, and was observed to change significantly, according to the results of flow cytometry[scramble (scr)-shRNA vs. RHEB-shRNA1, P=0.041 and scr-shRNA vs. RHEB-shRNA2, P=0.021; Fig. 4A]. The number of S-phase cells increased significantly, while the number of G2/M-phase cells decreased significantly (scr-shRNA vs. RHEB-shRNA1, P=0.016 and scr-shRNA vs. RHEB-shRNA2, P=0.011), with the majority of cells arrested in S phase (Fig. 4B). In soft agar colony-forming assay, the colony-formation and proliferation abilities of the groups treated with shRNA-RHEB decreased significantly by 6.0±0.5-fold (scr-shRNA vs. RHEB-shRNA1, P=0.004 and scr-shRNA vs. RHEB-shRNA2, P=0.003; Fig. 4C).
Figure 4.
Influence of silencing RHEB on the biological behavior of the liver cancer cell line SMMC-7721. (A) Cell cycle changes of SMMC-7721 cells with low expression of RHEB were analyzed by flow cytometry. (B) Comparison of the cell cycle changes in SMMC-7721 cells treated with different shRNAs. (C) Analysis of soft agar colony-formation ability of SMMC-7721 cells with downregulated RHEB expression. The experiments were performed in triplicate, and representative images are shown. RHEB, Ras homolog enriched in brain; scr, scramble; FL2-A, fluorescence 2-area; shRNA, small hairpin RNA.
RHEB silencing suppresses the growth of HCC cells in vivo
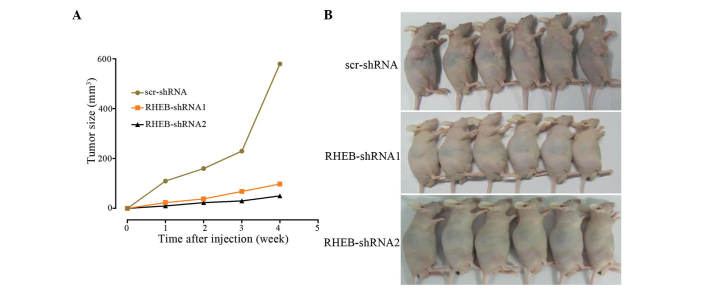
In the tumorigenicity assay, SMMC-7721 cells were subcutaneously injected into nude mice, and knocked down RHEB in SMMC-7721 cells was observed to decelerate cancer growth, with no difference in tumor growth between the RHEB-shRNA1 and the RHEB-shRNA2 groups (RHEB-shRNA1 vs. RHEB-shRNA2, P=0.620), although there was a significant difference between the downregulated groups and the scr-shRNA group (scr-shRNA vs. RHEB-shRNA1, P=0.003 and scr-shRNA vs. RHEB-shRNA2, P=0.006; Fig. 5A). In addition, the tumorigenic ability of the control group was higher than that of the RHEB-shRNA1 and RHEB-shRNA2 groups (Fig. 5B), and the difference in tumorigenic ability between the experimental and control groups was significant (scr-shRNA vs RHEB-shRNA1, P=0.003 and scr-shRNA vs. RHEB-shRNA2, P=0.006).
Figure 5.
Influence of RHEB silencing on the biological behavior of nude mice. Influence of low RHEB expression on (A) tumor growth and (B) nude mice tumorigenic ability. RHEB, Ras homolog enriched in brain; scr, scramble; shRNA, small hairpin RNA.
Discussion
The RHEB protein is an extremely important activator in cell signal transduction mediated by insulin, and previous studies have demonstrated that insulin elevates its GTP-binding state (17,18). GTP-loaded RHEB binds directly to the catalytic domain of mechanistic target of rapamycin (mTOR)to activate its serine/threonine kinase activity and to increase cancer cell growth, which can be blocked by rapamycin (17–19). In numerous tissues, RHEB overexpression can promote cancer cell growth and change the cell cycle kinetics through accelerating the G1-S phase, while the cell division rates are not affected (20).
In the present study, RHEB overexpression in liver cancer tissues was observed to be significant, as demonstrated by gene microarrays and verified by RT-qPCR. According to the results of immunohistochemistry, RHEB protein displayed positive expression in HCC tissues compared with normal liver tissues, and was expressed in the cytoplasm of liver cancer cells. Furthermore, RHEB overexpression was observed in the majority of liver cancer lines in the present study, which suggests that RHEB serves an important role in the occurrence and development of HCC. In addition, analysis of the association between RHEB expression and clinicopathological data in patients with HCC demonstrated that RHEB expression increased with clinical staging, which further suggests that RHEB is implicated in the development and progression of HCC. These results are consistent with those from previous studies on head and neck, breast and prostate cancer, which further indicates that RHEB overexpression is common in the development and progression of numerous cancers (14,15,16).
Survival analysis revealed that the overall survival time was closely correlated to the RHEB expression level, and was decreased in patients with high RHEB expression. Further analysis of the association between clinicopathological data and recurrence upon resection revealed that a high RHEB expression level was closely correlated to overall survival time. Based on these findings, overexpression of RHEB in patients with HCC appears to result in increased rates of recurrence following resection and in decreased overall survival time. These results further suggest that RHEB is involved in the metastasis, recurrence and prognosis of patients with HCC.
Due to its high RHEB expression, the liver cancer cell line SMMC-7721 was selected and treated with siRNA-RHEB in the present study. Inhibition of its cell growth could be observed, with the majority of cells being blocked in S phase, which led to an increase in the number of cells in S phase, while the number of cells in G2/M phase was decreased, which suggests that RHEB is involved in regulating the synthesis of DNA in S phase. Additionally, the decrease in soft agar colony-forming ability demonstrated a decrease in cancer cell invasiveness, which suggests that RHEB may be involved in the regulation of invasiveness and metastasis in patients with HCC. Tumor growth also decreased subsequent to downregulation of RHEB expression in the animal experiments, and the tumorigenesis of the nude mice used in the study also decreased, which further suggests that the RHEB gene participates in the regulation of tumor growth.
However, the critical role of RHEB in the occurrence, development, metastasis and recurrence of HCC remains unknown. Previous studies have demonstrated that RHEB can activate the S6 kinase (S6K) transcription factor through the regulation of the mTOR signaling pathway and androgen receptor trans-activity, thus regulating the proliferation of prostate cancer cells (15,21). RHEB is located downstream of the tumor inhibitors tuberous sclerosis (Tsc)1 and Tsc2, and upstream of mTOR in the mTOR-S6K signaling pathway (19). The GTP-bound activated state of RHEB could activate the mTOR signaling pathway and increase the phosphorylation of S6K and 4E-binding protein 1, which promotes mRNA translation and protein synthesis, thus enhancing cancer cell growth and inhibiting autophagy (19,22,23). A similar function of RHEB in head and neck cancer, breast cancer and lymphoma was also reported in previous studies (16,24,25), which suggests that RHEB potentially promotes the proliferation, invasiveness and metastasis of HCC via the mTOR signaling pathway, thus affecting the development, progression and prognosis of patients with HCC. Further studies on the underlying mechanism of RHEB in HCC should be conducted.
RHEB overexpression also occurs in non-small cell lung cancer and lymphoma (24,26). Previous studies have suggested that RHEB is likely to be a therapeutic target (24,26). The present study demonstrates the significance of RHEB in HCC and provides a basis for non-operative treatment of patients with HCC (27).
In conclusion, the present study demonstrated that RHEB overexpression is closely correlated to the clinicopathological features of patients with HCC, and is involved in the proliferation, invasiveness, metastasis, recurrence and prognosis of HCC. RHEB plays a significant role in the development and progression of HCC, and is an important prognostic marker and likely to be a therapeutic target for patients with HCC.
Acknowledgements
This work was supported by the grants from International Science and Technology Cooperation Program of the Ministry of Science and Technology (2011DFA32980), National Natural Science Foundation of China (81772529, 81271694, 81372652, 81572336), The Innovation Program of Shanghai Municipal Education Commission (2013ZZ060), One Hundred Person Project of the Shanghai Health (XBR2013117) and National Key Basic Research Program of China 973 Program (2012CB526706).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Song P, Feng X, Zhang K, et al. Screening for and surveillance of high-risk patients with HBV-related chronic liver disease: Promoting the early detection of hepatocellular carcinoma in China. Biosci Trends. 2013;7:1–6. [PubMed] [Google Scholar]
- 5.Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye QH, Wang L, Zhou J, Qiu SJ, Li Y, et al. A decade's studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130:187–196. doi: 10.1007/s00432-003-0511-1. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka M, Katayama F, Kato H, Tanaka H, Wang J, Qiao YL, Inoue M. Hepatitis B and C virus infection and hepatocellular carcinoma in China: A review of epidemiology and control measures. J Epidemiol. 2011;21:401–416. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 8.Singhal A, Jayaraman M, Dhanasekaran DN, Kohli V. Molecular and serum markers in hepatocellular carcinoma: Predictive tools for prognosis and recurrence. Crit Rev Oncol Hematol. 2012;82:116–140. doi: 10.1016/j.critrevonc.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Feng YX, Wang T, Deng YZ, Yang P, Li JJ, Guan DX, Yao F, Zhu YQ, Qin Y, Wang H, et al. Sorafenib suppresses postsurgical recurrence and metastasis of hepatocellular carcinoma in an orthotopic mouse model. Hepatology. 2011;53:483–492. doi: 10.1002/hep.24075. [DOI] [PubMed] [Google Scholar]
- 10.Wu KT, Wang CC, Lu LG, Zhang WD, Zhang FJ, Shi F, Li CX. Hepatocellular carcinoma: Clinical study of long-term survival and choice of treatment modalities. World J Gastroenterol. 2013;19:3649–3657. doi: 10.3748/wjg.v19.i23.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Im E, von Lintig FC, Chen J, Zhuang S, Qui W, Chowdhury S, Worley PF, Boss GR, Pilz RB. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene. 2002;21:6356–6365. doi: 10.1038/sj.onc.1205792. [DOI] [PubMed] [Google Scholar]
- 12.Aspuria PJ, Tamanoi F. The Rheb family of GTP-binding proteins. Cell Signal. 2004;16:1105–1112. doi: 10.1016/j.cellsig.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu ZH, Shvartsman MB, Lee AY, Shao JM, Murray MM, Kladney RD, Fan D, Krajewski S, Chiang GG, Mills GB, Arbeit JM. Mammalian target of rapamycin activator RHEB is frequently overexpressed in human carcinomas and is critical and sufficient for skin epithelial carcinogenesis. Cancer Res. 2010;70:3287–3298. doi: 10.1158/0008-5472.CAN-09-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T, Shimizu Y, Terada N, Yamasaki T, Nakamura E, Toda Y, Nishiyama H, Kamoto T, Ogawa O, Inoue T. Regulation of androgen receptor transactivity and mTOR-S6 kinase pathway by Rheb in prostate cancer cell proliferation. Prostate. 2010;70:866–874. doi: 10.1002/pros.21120. [DOI] [PubMed] [Google Scholar]
- 16.Wazir U, Newbold RF, Jiang WG, Sharma AK, Mokbel K. Prognostic and therapeutic implications of mTORC1 and Rictor expression in human breast cancer. Oncol Rep. 2013;29:1969–1974. doi: 10.3892/or.2013.2346. [DOI] [PubMed] [Google Scholar]
- 17.Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- 18.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 21.Inoue T, Yoshida T, Shimizu Y, Kobayashi T, Yamasaki T, Toda Y, Segawa T, Kamoto T, Nakamura E, Ogawa O. Requirement of androgen-dependent activation of protein kinase Czeta for androgen-dependent cell proliferation in LNCaP Cells and its roles in transition to androgen-independent cells. Mol Endocrinol. 2006;20:3053–3069. doi: 10.1210/me.2006-0033. [DOI] [PubMed] [Google Scholar]
- 22.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 23.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 24.Mavrakis KJ, Zhu H, Silva RL, Mills JR, Teruya-Feldstein J, Lowe SW, Tam W, Pelletier J, Wendel HG. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev. 2008;22:2178–2188. doi: 10.1101/gad.1690808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen ZH, Su YC, Lai PL, Zhang Y, Xu YF, Zhao A, Yao GY, Jia CH, Lin J, Xu S, et al. Critical role of arachidonic acid-activated mTOR signaling in breast carcinogenesis and angiogenesis. Oncogene. 2013;32:160–170. doi: 10.1038/onc.2012.47. [DOI] [PubMed] [Google Scholar]
- 26.Zheng H, Liu A, Liu B, Li M, Yu H, Luo X. Ras homologue enriched in brain is a critical target of farnesyltransferase inhibitors in non-small cell lung cancer cells. Cancer Lett. 2010;297:117–125. doi: 10.1016/j.canlet.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Mazhab-Jafari MT, Marshall CB, Ho J, Ishiyama N, Stambolic V, Ikura M. Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR) J Biol Chem. 2014;289:12195–12201. doi: 10.1074/jbc.C113.543736. [DOI] [PMC free article] [PubMed] [Google Scholar]