Abstract
Temozolomide (TMZ) chemotherapy, in combination with maximal safe resection and radiotherapy, is the current standard of care for patients with glioblastoma (GBM). Despite this multimodal approach, GBM inevitably relapses primarily due to resistance to chemo-radiotherapy, and effective treatment is not available for recurrent disease. In this study we identified TMZ resistant patient-derived primary and previously treated recurrent GBM stem cells (GSC), and investigated the therapeutic activity of a pro-apoptotic variant of oHSV (oHSV-TRAIL) in vitro and in vivo. We show that oHSV-TRAIL modulates cell survival and MAP Kinase proliferation signaling pathways as well as DNA damage response pathways in both primary and recurrent TMZ-resistant GSC. Utilizing real time in vivo imaging and correlative immunohistochemistry, we show that oHSV-TRAIL potently inhibits tumor growth and extends survival of mice bearing TMZ-insensitive recurrent intracerebral GSC tumors via robust and selective induction of apoptosis-mediated death in tumor cells, resulting in cures in 40% of the treated mice. In comparison, the anti-tumor effects in a primary chemoresistant GSC GBM model exhibiting a highly invasive phenotype were significant but less prominent. This work thus demonstrates the ability of oHSV-TRAIL to overcome the therapeutic resistance and recurrence of GBM, and provides a basis for its testing in a GBM clinical trial.
Keywords: Glioblastoma, Recurrence, Temozolomide, Oncolytic herpes simplex virus, TNF-related apoptosis inducing ligand (TRAIL)
Introduction
Glioblastoma (GBM) is the most common and aggressive primary brain cancer in adults 1. Temozolomide (TMZ) chemotherapy, in combination with maximal safe resection and radiotherapy, is the current standard of care for patients with GBM 2. Despite this multimodal approach, GBM inevitably relapses and the median survival of GBM patients remains only about 15 months 1, 3. The major reasons for tumor relapse include: 1) incomplete surgical resection due to the invasive nature of GBM, and 2) molecular deregulation of cell death pathways that contribute to intrinsic resistance to cytocidal therapies (recently reviewed in 4). The orally available alkylating agent TMZ induces O6-methyl guanine, causing accumulations of DNA mismatch, subsequent growth arrest, and ultimately apoptosis 5. Unfortunately, however, almost half of GBMs are TMZ resistant primarily due to the expression of O6-methytlguanine-DNA-methyltransferase (MGMT), which antagonizes the TMZ effect 6. There is no standard therapy for recurrent GBM and rigorous efforts to investigate molecular targeted agents have not achieved significant extension of overall survival in GBM patients 7. Thus developing novel therapeutics that successfully target therapeutically refractory subpopulations is urgently needed to improve the outcome of GBM.
GBM contains self-renewing, multipotent subsets of tumor cells termed GBM-initiating or stem-like cells (GSC) 8. Accumulating evidence indicates that GSC play a central role in evading conventional treatments and causing tumor relapse, and thus represent an important therapeutic target 9. In addition, GSC are able to recapitulate the genetic diversity seen in patients or the heterogeneity of tumor cells, and provide excellent preclinical GBM models, as opposed to traditional cell line based models that do not mirror clinical GBM 10, 11. We have established a large panel of GBM “neurosphere” cell (GSC) lines that we isolated from patients, both from newly diagnosed as well as post-TMZ recurrent tumors 12–15. These lines grown as neurospheres are enriched for GSC, and retain patient-specific oncogenic molecular alterations such as EGFR amplification and the phenotypic hallmarks of GBM such as extensive invasiveness and angiogenesis 12–15. These patient-derived newly diagnosed and recurrent GSC represent a unique resource that allows us to investigate the biology of therapeutic resistance and develop novel therapies to target GSC and overcome the challenge of tumor recurrence.
Oncolytic virus is genetically modified or naturally occurring virus that selectively replicates in and kills neoplastic cells while sparing normal cells. Genetically modified oncolytic herpes simplex virus (oHSV) is one of the most extensively investigated oncolytic viruses and the safety of administering oHSV in the human brain has been shown in clinical studies (reviewed in 16). Distinct mode of action renders oHSV a promising anti-cancer agent to overcome TMZ resistance; however, GBM cells differentially respond to oHSV-mediated oncolysis 17. To target GBM cells that are not permissive to oHSV killing, we created a recombinant variant of oHSV, oHSV-TRAIL 17. oHSV-TRAIL was engineered to express an anti-cancer protein, TNF-related apoptosis-inducing ligand (TRAIL). Providing multiple mechanisms of action, e.g., direct oncolysis and TRAIL-mediated apoptosis, oHSV-TRAIL showed potent anti-tumor activity in a mouse model of GBM 17, 18. However the role of oHSV-TRAIL in the context of TMZ resistance has not been tested previously.
In this study we first screened a cohort of primary and recurrent patient-derived GSC lines for their sensitivity to TMZ. We next determined the molecular mechanisms that underlie oHSV-TRAIL mediated killing of chemoresistant GSC, and characterized the efficacy of oHSV-TRAIL in mouse GBM models derived from chemoresistant primary and recurrent GSC.
Materials and Methods
Parental and engineered cell lines
Primary glioma neurosphere cell (GSC) lines (GSC4, GSC6, GSC8, GSC18, GSC23, GSC29, GSC32, GSC34, and GSC64) and recurrent GSC lines (GSC24R and GSC31) were all patient-derived and cultured in Neurobasal medium (Invitrogen, Carlsbad, CA) supplemented with 3 mmol/l l-glutamine (Mediatech, Manassas, VA), B27 (Invitrogen, Carlsbad, CA), 2 μg/ml heparin (Sigma-Aldrich, St Louis, MN), 20 ng/ml human EGF (R&D Systems, Minneapolis, MN), and 20 ng/ml human FGF-2 (Peprotech, Rocky Hills, NJ) as described previously 13, 14. Normal human astrocytes were purchased from ScienCell (Carlsbad, CA) and grown in DMEM supplemented with 10% fetal bovine serum. Lentiviral vector, Pico2-Fluc-mCherry, is a kind gift from Dr Andrew Kung (Dana Farber Cancer Institute; Boston, MA). Lentiviral packaging was performed by transfection of 293T cells as previously described 19. GSC23 and GSC31 were transduced with LV-Pico2-Fluc-mCherry at a MOI of 1 in medium containing protamine sulfate (2 μg/ml) and GSC23-Fluc-mCherry (GSC23-FmC) and GSC31-Fluc-mCherry (GSC31-FmC) lines were obtained after puromycin (1 μg/ml) selection in culture.
Recombinant oHSVs and viral growth assay
G47Δ-empty (referred to oHSV in this study), G47Δ-mCherry (oHSV-mCherry), and G47Δ-TRAIL (oHSV-TRAIL) are BAC-based recombinant oHSV vectors with the genomic backbone of G47Δ (γ34.5–, ICP6–, ICP47–) 17, 20–22. All of these oHSVs express E. coli lacZ driven by endogenous ICP6 promoter. oHSV bears no additional transgene sequences, while oHSV-mCherry and oHSV-TRAIL carry mCherry or S-TRAIL driven by the herpes simplex virus immediate early 4/5 promoter, respectively. S-TRAIL secretion from oHSV-TRAIL-infected Vero cells was confirmed by ELISA (26 ng/ml / 1×106 cells / 48 hours). For viral growth assay, cells plated on 12-well plates (80,000 cells) were infected with oHSV at MOI = 0.1. After virus adsorption, media was replaced and culture continued. Cells and culture supernatant were harvested at the indicated time points. Titers of infectious virus were determined by plaque assay on Vero cells (American Type Culture Collection, Manassas, VA).
Immunocytochemistry
Differentiation of GSCs was induced by 7-day exposure to 5% fetal calf serum in DMEM. Staining for human nestin (Santa Cruz Biotechnology), GFAP (Sigma) and GalC (Chemicon) was performed as described previously 13.
In vitro cell viability assay
To determine the effects of TMZ, oHSV, and oHSV-TRAIL on cell viability, GBM cells or NHA were seeded on 96-well plates (0.5 × 104/well) and treated with different doses TMZ (0–1000 μM) or different MOIs of oHSV or oHSV-TRAIL 24 hours after plating. Cell viability was measured at indicated time points by determining the cell metabolic activity using an ATP-dependent luminescent reagent (CellTiter-Glo; Promega, Madison, WI) according to manufacturer’s instructions. All experiments were performed in triplicates.
Western blot analysis
GBM cells were seeded on 6-well plates (5.0 × 105/well) and treated with different doses TMZ (100 μM), S-TRAIL (200 ng/ml) or 1.0 MOI of oHSV or oHSV-TRAIL 24 hours after plating. 15 or 24 hours after treatment, GBM cells were lysed with NP40 buffer supplemented with protease (Roche, Indianapolis, IN) and phosphatase inhibitors (Sigma-Aldrich). Twenty micrograms of harvested proteins from each lysate were resolved on 10% SDS-PAGE, and immunoblotted with antibodies against cleaved caspase 3, cleaved PARP, PARP, p44/42MAPK (ERK 1/2), phospho-p44/42MAPK (ERK 1/2) (Thr202, Thr204), SAPK/JNK, phospho-SAPK/JNK (Thr183/Thr185), p38-MAPK, phospho-p38 MAPK, CHK1, phospho-CHK1 (all from Cell Signaling, Danvers, MA), MGMT (Sigma-Aldrich), β-actin (Cell Signaling) or α-tubulin (Sigma-Aldrich). Blots were then incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz, Santa Cruz, CA) and developed by chemiluminescence, followed by exposure to film.
Small Molecule Inhibition Assays
GBM cells were seeded on 96-well plates (0.5 × 104/well) and treated with different doses of U0126 (Promega Corporation, 20 μmol/l), CHK1 inhibitor (Selleck, 0.5 mmol/l), and S-TRAIL (100 ng/mL) 24 hours after plating. 48 hours later, cell viability was measured by using the CellTiter-Glo assay. All experiments were performed in triplicates.
Intracranial GBM cell implantation and in vivo bioluminescence imaging
GSC23-FmC cells (5 × 105 cells per mouse; n = 20) were stereotactically implanted into the brains (right striatum, 2.5 mm lateral from bregma and 2.5 mm deep) of athymic mice (6 weeks of age). Tumor growth was monitored 1–2 times a week by Fluc bioluminescence (BLI) as described before (23, 24). Tumor-bearing mice were intratumorally injected with 3 μl of 2.0 × 106 plaque-forming unit (pfu) of oHSV (n = 5), oHSV-TRAIL (n = 5), or PBS (n = 4) to study tumor regression and brain pathology. For survival study, tumor-bearing mice were intratumorally injected with 6 μl of 2.0 × 106 pfu of oHSV-TRAIL (n = 6), or PBS (n = 4) twice on days 14 and 26 after tumor cell implantation. Mice were imaged for Fluc BLI and followed for survival and sacrificed when neurological symptoms became apparent.
Similarly, GSC31 (5 × 105 cells per mouse; n = 10) were stereotactically implanted into the brains of athymic mice (6 weeks of age). Ten days post-implantation mice were injected intratumorally with 6 μl of 2.0 × 106 pfu of oHSV-TRAIL (n = 5) or PBS (n = 5). Mice were followed for survival and sacrificed when neurological symptoms became apparent. All in vivo procedures were approved by the institutional animal care and use committee (IACUC) at MGH.
Tissue processing and histopathological analysis
A set of mice with intracerebral GSC23-FmC and GSC31-FmC tumors were treated with intratumoral injections of PBS or oHSV-TRAIL on day 39 (for GSC23-FmC) or on day 59 (for GSC31-FmC) after tumor cell implantation (n=3 – 4 per group) as described above. 48 hours after treatment mice were perfused with cold 4% paraformaldehyde via the heart and the brains were fixed in 4% paraformaldehyde. Frozen sections were obtained for hematoxylin and eosin staining and immunohistochemistry. 5-Bromo-4-choloro-3-indolyl-β-d-galactopyranoside (X-gal) staining was performed to identify lacZ-expressing infected cells followed by hematoxylin counterstaining. For cleaved caspase 3 and cleaved PARP staining, sections were incubated for 1 hour in a blocking solution (0.3% bovine serum albumin, 8% goat serum, and 0.3% Triton-X100) at room temperature, followed by incubation at 4 °C overnight with anti-cleaved caspase 3 (Cell Signaling) diluted in blocking solution. Sections were incubated in Alexa Fluor 488 goat anti-rabbit secondary antibody (Invitrogen), and visualized using a confocal microscope (LSM Pascal; Zeiss, Oberkochen, Germany). DAPI (Vectashield) was used to stain nuclei. The percentage of cleaved caspase 3 positive cells was calculated by counting positive cells in 3 randomly chosen miscroscopic fields.
Immunohistochemistry was used to stain human GBM cells and mouse neurons with anti-human specific nestin (Santa Cruz Biotechnology) and NeuN (Millipore) antibodies, respectively. Paraformaldehyde-fixed frozen sections were treated with acetone for 3 minutes followed by washes with PBS. After blocking with 2.5% horse serum (Vector Laboratories), sections were incubated with diluted primary antibodies at 4 °C overnight. After PBS washes, sections were incubated with ImmPRESS HRP anti-mouse IgG polymer (Vector Laboratories) for 1 hour at room temperature, followed by PBS washes and color development with DAB (Dako). Nuclei were counterstained with hematoxylin.
Statistical analysis
Data were analyzed by Student t-test when comparing two groups. Data were expressed as mean ± SD and differences were considered significant at P < 0.05. Kaplan-Meier analysis was used for mouse survival studies, and the groups were compared using the log-rank test.
Results
Screening of primary and recurrent patient-derived GSC lines identifies differential resistance to TMZ treatment
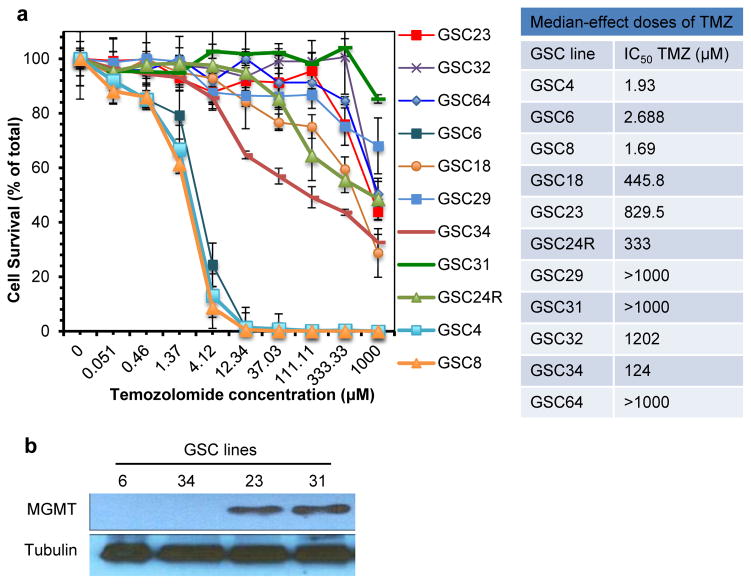
We first screened a panel of patient-derived GSC for their sensitivity to TMZ using in vitro cell viability assay. This panel of GSC includes 9 GSC lines that were established from primary (e.g., newly diagnosed) GBM (GSC4, 6, 8, 18, 23, 29, 32, 34, and 64), and 2 recurrent lines (GSC24R and 31) isolated from tumors that relapsed and progressed after the standard management regimens including TMZ chemotherapy. The ability of GSC to undergo multi-lineage differentiation was shown by immunofluorescence for GFAP (astrocyte marker) and GalC (oligodendrocyte marker)(Supplementary Figure S1). These GSC demonstrated variable sensitivities to TMZ, with 7 GSC lines (GSC18, 23, 24R, 29, 31, 32, and 64) being resistant with their IC50 values >300μM, 1 line (GSC34) being semi-resistant, and 3 lines (GSC4, 6, and 8) being sensitive (IC50 values <10μM) (Figure 1a). As expected, both of the recurrent GSC tested (GSC24R, GSC31) were TMZ resistant. Expression of the TMZ-antagonizing enzyme MGMT was confirmed in TMZ-resistant GSC23 and GSC31. In conjunction with our prior report, MGMT expression was not detected in TMZ-sensitive GSC4, GSC6, GSC8, and GSC34 as well as TMZ-resistant GSC18 and GSC29 (Figure 1b) 14. These data show that primary GSC lines have varying sensitivities to TMZ, and recurrent GSC that underwent standard-of-care (radiation and TMZ) in the patients are resistant to TMZ.
Figure 1. Screening of primary and recurrent patient-derived GSC lines for temozolomide sensitivities.
(a) Temozolomide (TMZ) sensitivity of patient derived GBM lines in vitro. TMZ dose response curves of primary GSC lines (GSC4, 6, 8, 18, 23, 29, 32, 34, and 64) and recurrent GSC lines (GSC24R and 31). Cells were treated with increasing concentrations of TMZ for 6 days, and cell viability was measured by CellTiter-Glo assay. The experiments were done in triplicate and repeated at least twice. Error bars, SD. IC50 (dose required for 50% effect) values were determined from dose-response curves. (b) Western blot analysis to detect expression of MGMT in representative GSC.
oHSV-TRAIL is more potent than oHSV against TMZ-resistant GSC lines
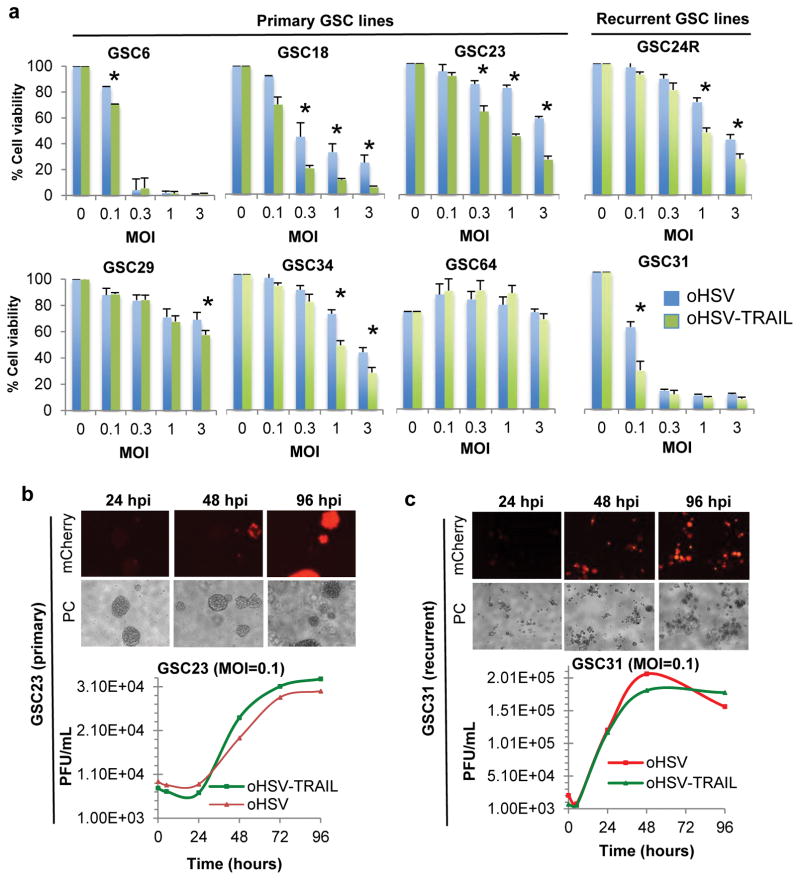
To assess the cytotoxic activity of a proapoptotic variant of oHSV (oHSV-TRAIL) against TMZ-resistant primary and recurrent tumors, we tested 6 primary (GSC6, 18, 23, 29, 34 and 64) and 2 recurrent (GSC24R and 31) GSC lines. Although dose (multiplicity of infection, MOI)-dependent cytotoxicity was typically seen, efficiency in oHSV killing of GSC lines was variable and 5 of 8 GSC lines tested showed IC50 < MOI 1.0 (Figure 2a). oHSV-TRAIL was more efficacious than oHSV in 7 out of the 8 lines tested including the recurrent GSC, 24R and 31 (Figure 2a). Treatment of TMZ-resistant GSC with an imaging variant of oHSV (oHSV-mCherry (mCh)) showed an MOI-dependent increase in infectability (Supplementary Fig S2a), and oHSV-mCh spread over time within and between GSC spheres at low MOI=0.1 (Figure 2bc). oHSV-mCh spread poorly in GSC29 (Supplementary Fig S2b), and oHSV and oHSV-TRAIL killing was modest in this line. GSC64 was the only line that did not respond to both oHSV and oHSV-TRAIL, and was found very poorly infectable (Supplementary Fig S2c). oHSV and oHSV-TRAIL replicated similarly in GSCs, indicating that the expression of TRAIL does not compromise the replicative capability of oHSV (Figure 2bc). Replication of oHSV and oHSV-TRAIL was poor in normal human astrocytes (NHA)(Supplementary Fig S3a), and NHA were resistant to both viruses (Supplementary Fig S3b). These data demonstrate the potent cytotoxic activity of oHSV-TRAIL in vitro against TMZ-resistant GSC derived from primary as well as recurrent GBM. Based on these studies, we selected two TMZ-resistant GSC lines, one primary and invasive (GSC23 14) and one recurrent (GSC31), for further studies.
Figure 2. Characterizing oHSV-mediated effects in primary and recurrent patient-derived GSC lines.
(a) Differential sensitivities of primary and recurrent GSC to oncolytic herpes simplex virus (oHSV)-mediated oncolysis. GSC lines were infected with oHSV (Blue bars) and oHSV-TRAIL (Green bars) at multiplicities of infection (MOIs)=0.1, 0.3, 1 and 3, and assayed for cell viability at 5 days post-infection using CellTiter-Glo assay. Relative cell viability to untreated control (MOI=0) + SD (bars) is shown. The experiments were done in triplicate and repeated at least twice. *P <0.05, oHSV-TRAIL vs oHSV. (b, c) oHSV infection, spread and replication in TMZ-resistant primary GSC23 (b) and recurrent GSC31 (c). For each GSC, the top panels show fluorescent (mCherry) and phase-contrast (PC) microscopic images showing infection and spread of oHSV-mCh. The lower panels show virus yield assay showing comparable replication of oHSV and oHSV-TRAIL in GSC. Also see Supplementary Figure S3.
oHSV-TRAIL targets cell proliferation, death and check point kinase signaling pathways in TMZ-resistant GSC
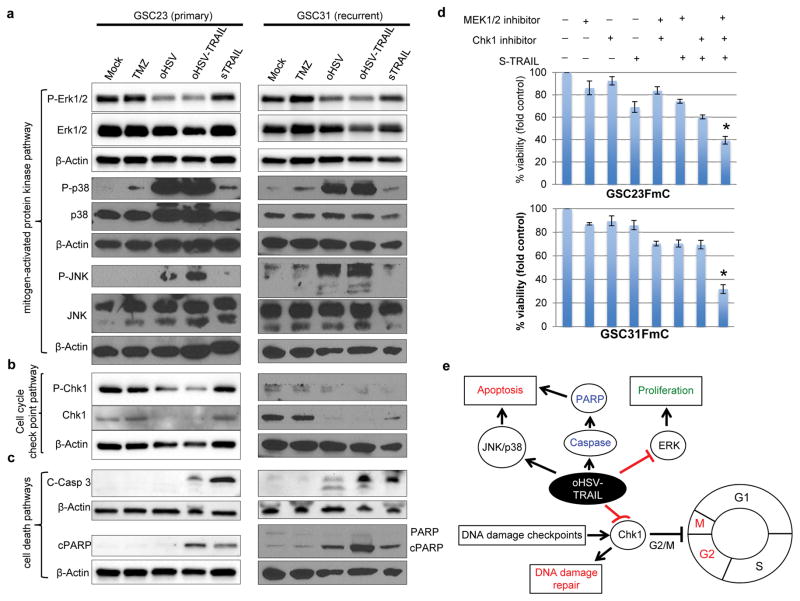
Next, to assess whether oHSV-TRAIL induces perturbation of cell proliferation pathways in TMZ-resistant GSC, GSC23 and GSC31 were treated with oHSV or oHSV-TRAIL and analyzed. Western blot analysis revealed that oHSV and oHSV-TRAIL potently induced phosphorylation of JNK and p38 while the impacts of TMZ or TRAIL on these MAPK pathways were minimal (Figure 3a). TRAIL and TMZ activated ERK1/2 phosphorylation in GSC23 and GSC31, respectively, however, oHSV and oHSV-TRAIL suppressed ERK1/2 phosphorylation in both GSC23 and GSC31 (Figure 3a).
Figure 3. oHSV and oHSV-TRAIL alter MAPK and DDR signaling pathways.
a–c, Western blot showing changes in MAPK pathways (a), cell cycle checkpoint pathways (b), and cell death pathways (c) in GSC23 (left) and GSC31 (right). Treatments include TMZ (100 μM), oHSV (MOI=1.0), oHSV-TRAIL (MOI=1.0), and secretory (s) TRAIL (200 ng/mL) for 24 hours. β-actin was used as loading control. Same β-actin is presented when different pathways were examined with same membranes. d, Cell viability assays after treatment of GSC23FmC and GSC31FmC cells with different combinations of MEK inhibitor U0126 (20μmol/l), Chk1 inhibitor (0.5 mmol/l) and S-TRAIL (100ng/ml) for 48 hours. Relative cell viability to control + SD (error bars) is shown. *P <0.05, Triple combination group in comparison with control groups. e, Proposed mechanism of oHSV-TRAIL-mediated killing of TMZ-resistant GSC.
Since oHSV can provoke DNA damage response (DDR) 25, we assessed the activation of checkpoint kinase-1 (Chk1), a key effector in the cell cycle checkpoint pathway, in GSC. Strikingly, oHSV and oHSV-TRAIL treatment markedly decreased the levels of the Chk1 protein compared to TMZ-treatment (Figure 3b). Importantly, greater induction of caspase 3 and PARP cleavage was seen in oHSV-TRAIL-treated GSC than in oHSV-treated GSC (Figure 3c). These results indicate that both oHSV and oHSV-TRAIL target cell proliferation pathway in a similar manner, but oHSV-TRAIL, not oHSV, induces caspase-3 cascade which results in efficient killing of both primary and recurrent patient-derived GSCs.
Determining critical pathways for oHSV-TRAIL killing effects through pharmacological small molecule inhibitors
We next sought to determine key signaling pathways or crosstalk that oHSV-TRAIL modifies to promote killing of GSC. We investigated whether targeted inhibition of MEK/ERK and/or Chk1 with highly selective inhibitor in conjunction with exogenous recombinant soluble TRAIL (S-TRAIL) could mimic the oHSV-TRAIL-mediated killing of GSC. Cell viability analysis using small molecule targeted agents against MEK/ERK and Chk1 individually, or in combination with S-TRAIL showed that triple combined treatment with MEK/ERK and Chk1 inhibitors and s-TRAIL effectively decreased cell viability in TMZ-resistant GSC23 and GSC31 compared with single treatment or the combination of any two (Figure 3d). These results reveal that oHSV-TRAIL-mediated inhibition of the ERK-MAPK and Chk1 signaling may contribute to oHSV-TRAIL-induced apoptotic cell death in chemo-resistant GBM cells. Thus oHSV-TRAIL as a single agent acts on and alters multiple pathways: (i) ERK1/2 inhibition (inhibition of proliferation), (ii) p38/JNK MAPK stimulation (stimulation of cell death/differentiation) and (iii) degradation of Chk1 preventing cell-cycle arrest and DNA damage repair pathways. Our results suggest that this simultaneous multi-pathway inhibition underlies the oHSV-TRAIL-mediated killing of TMZ-resistant GSC (Figure 3e).
oHSV-TRAIL inhibits GBM growth in vivo and prolongs survival of mice bearing primary invasive GBM derived with chemoresistant GSC
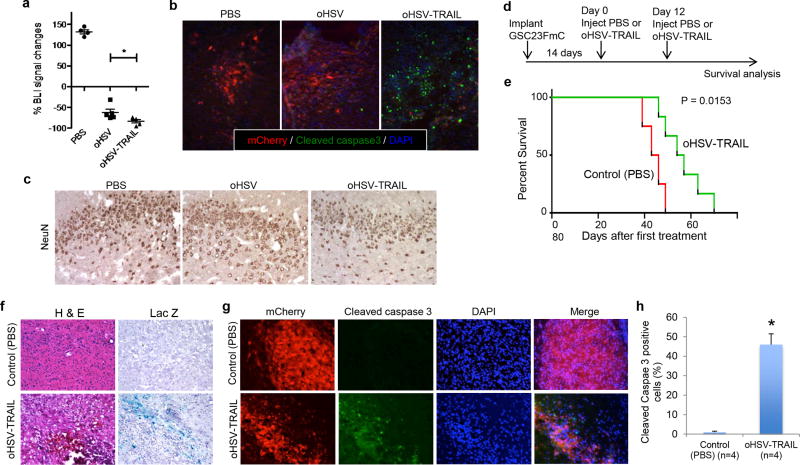
To non-invasively monitor tumor progression and therapeutic effects in vivo, TMZ-resistant primary GSC23 was lentivirally engineered to express firefly luciferase (F) and mCherry (mCh). This genetic manipulation retained the ability of GSC to form neurospheres (Supplementary Figure S4a), and did not alter sensitivity to TMZ (Supplementary Figure S4b) or oHSV-TRAIL-mediated killing efficiency in vitro (Supplementary Figure S4c). In vivo, using an orthotopic tumor model in mice generated with GSC23-FmC, we first examined the anti-GBM activity of oHSV and oHSV-TRAIL. Bioluminescence imaging revealed that single intratumoral injections of oHSV and oHSV-TRAIL both mediated rapid regression of tumors, but with oHSV-TRAIL producing more robust effects (p < 0.05, Figure 4a, Supplementary Figure S5a). This difference of anti-GBM activity between oHSV and oHSV-TRAL was associated with increased induction of cleaved caspase 3 by oHSV-TRAIL (Figure 4b). In oHSV-TRAIL-treated tumors, we observed areas of densely populated cleaved caspase 3 positive cells without mCherry expression, and confirmed that these were apoptotic and dying GBM cells by showing the immunopositivity of human nestin (Supplementary Figure S5b). Cleaved PARP, downstream of the caspase-mediated apoptosis pathway, was detected in oHSV-TRAIL treated tumors, but not in PBS and oHSV treated tumors (Supplementary Figure S5c). Importantly, we did not find neurotoxicity caused by injections of these viruses as there was neither noticeable change in the neuronal structure nor loss of neurons in the cerebral cortex of treated mice (Figure 4c). These observations provide a rationale to further investigate the molecular mechanisms and survival effects of oHSV-TRAIL therapy in vivo.
Figure 4. oHSV-TRAIL induces apoptosis and prolongs survival in invasive orthotopic GBM models generated with primary GSC23FmC.
a, Plot showing the percentage of tumor volume changes measured by BLI 48 hours after treatment with PBS (N=4), oHSV (3 μl of 2.0 × 106 pfu, N=5) or oHSV-TRAIL (3 μl of 2.0 × 106 pfu, N=5) in the orthotopic GSC23FmC model. Individual values as well as mean + SD are shown. *P = 0.049 (oHSV vs. oHSV-TRAIL). b, Representative immunofluorescent merged images showing mCherry (red), cleaved caspase 3 (green) and DAPI (blue) in GSC23FmC tumors at 5 days after treatment with PBS, oHSV and oHSV-TRAIL. Original magnification, ×20. c, Immunohistochemistry for NeuN (brown) in the cerebral cortex on representative brain sections 5 days after PBS, oHSV and oHSV-TRAIL treatment of intracerebral GSC23FmC tumors. Original magnification, ×20. d, Experimental schema and timeline. e, Kaplan-Meier survival curves of tumor-bearing mice treated with oHSV-TRAIL or control (PBS). P = 0.0153 by log-rank test. f, H & E staining (left) and X-gal staining (right) of tumor sections from control (PBS)- (upper panels) and oHSV-TRAIL-injected (lower panels) tumors. Infected cells stain blue. Original magnification, ×10. g, Immunofluorescence of cleaved caspase 3 staining (green) on brain sections from mice injected with control (PBS) (upper panels) or oHSV-TRAIL (lower panels). mCherry-positive tumor cells and DAPI (nuclei) of the same slides and the merged images (mCherry+cleaved caspase 3+DAPI) are also shown. Original magnification, ×10. h, Plot showing the percentage of cleaved caspase 3-positive tumor cells on brain sections. *P = 0.007. N = 4 in each group. Error bars indicate SD.
We next determined the therapeutic efficacy and underlying molecular mechanisms of oHSV-TRAIL. Mice bearing GSC23-FmC intracerebral tumors were treated with intratumoral injections of PBS or oHSV-TRAIL on days 14 and 26 after implantation (Figure 4d). BLI-assisted noninvasive tumor volume monitoring demonstrated that oHSV-TRAIL injections resulted in a significant decrease in tumor volumes compared to control (PBS) injections (Supplementary Figure S6a, P=0.01). Accordingly, oHSV-TRAIL treatment significantly prolonged the survival of animals; after treatment initiation, the median survival of oHSV-TRAIL-treated mice was 55 days, compared to 44 days with PBS-treated mice (Figure 4e; P = 0.015, log-rank test). Histopathological evaluation on brain sections collected 48 hours post PBS or oHSV-TRAIL injection showed massive cell death within the tumors treated with oHSV-TRAIL injections (Figure 4f). X-gal staining on adjacent sections showed a distribution of reporter β-galactosidase positivity, which overlapped with the tumor area, indicating the spread of oHSV-TRAIL (Figure 4f). Immunofluorescence analysis showed a striking increase in cleaved caspase 3 positivity in the sections prepared from oHSV-TRAIL-treated tumors as compared to PBS-treated tumors (Figure 4gh, P=0.007). In both treatment groups cleaved caspase 3 staining was confined to tumor areas and not seen in host mouse cells, revealing tumor specificity of oHSV-TRAIL treatment (Figure 4g). Thus direct injections of oHSV-TRAIL mediated apoptotic cell death in chemoresistant GSC-derived invasive GBM in vivo, and provided substantive therapeutic benefits.
oHSV-TRAIL mediates robust tumor apoptosis and markedly prolongs survival of mice bearing orthotopic tumors generated with recurrent GSC
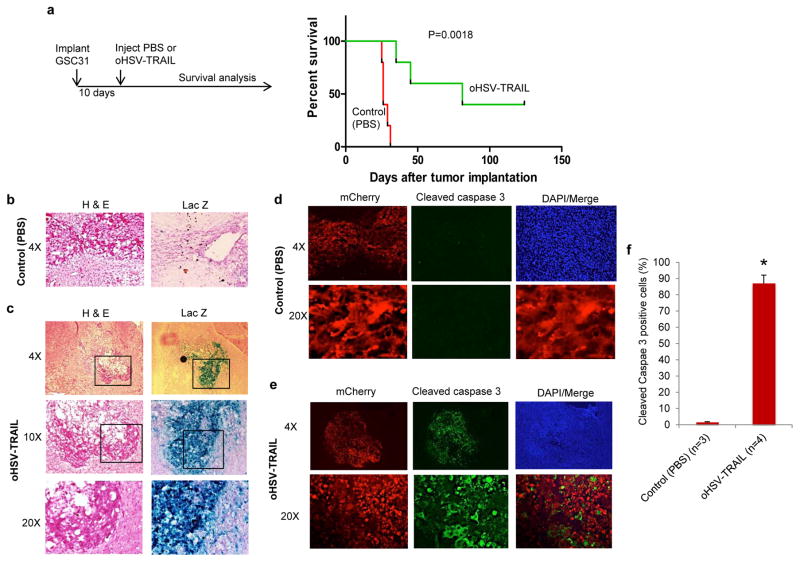
Nearly 100% of GBM relapse after the current standard of care, and no effective therapy exists for recurrent GBM, which is typically associated with acquired resistance. Preclinical development of novel strategies for recurrent GBM is a challenge, and we took advantage of our unique GSC model isolated from recurrent GBM. Intracerebral tumors were generated with the recurrent GSC31 in athymic mice and were treated with intratumoral injections of PBS or oHSV-TRAIL on day 10 (n=5 per group) (Figure 5a). Subsequent follow-up and survival analysis showed that all mice in the PBS group succumbed to tumor growth within 32 days with median survival of 26 days, revealing the malignant nature of the model (Figure 5a). In stark contrast, oHSV-TRAIL treatment resulted in much prolonged survival of GBM-bearing mice with the median survival of 81 days (P = 0.0018, log-rank test) (Figure 5a). On day 125 post implantation, 2 long-term surviving mice (40%) with oHSV-TRAIL treatment were euthanized and tumor cures were histologically confirmed.
Figure 5. oHSV-TRAIL mediated robust apoptosis and markedly prolonged survival of mice bearing orthotopic tumors generated with recurrent GSC31.
a, Experimental schema and Kaplan-Meier survival curves of GSC31 tumor-bearing mice treated with oHSV-TRAIL or control (PBS). P = 0.0018, log-rank test. b, c, H & E staining (left panels) and X-gal staining (right panels) of tumor sections from control (PBS)-injected (b) and oHSV-TRAIL (c)-injected tumors. Infected cells stain blue with X-gal. Original magnification is indicated. In c, boxed areas are magnified in the lower panels. d, e, Lower (4×, upper) and higher (20×, lower) magnification confocal microscopic images of cleaved caspase 3 immunofluorescence (green) on brain sections from control (PBS) (d) or oHSV-TRAIL (e)-injected mice. mCherry-positive tumor cells and DAPI (nuclei) of the same slides and the merged images of mCherry and cleaved caspase 3 are also shown. f, Plot showing the percentage of cleaved caspase 3 positive tumor cells on brain sections. *P = 0.0015. Error bars indicate SD.
We next generated intracerebral GSC31FmC xenografts, lentivirally expressing firefly luciferase (F) and mCherry (mC), in athymic mice, and serially monitored tumor growth with BLI. Mice were then randomized and treated with intratumoral injections of PBS (n=3) or oHSV-TRAIL (n=4) and 48h later the brains were harvested. In some of the mice subjected to BLI, we observed a drastic and rapid reduction of tumor signals following oHSV-TRAIL treatment (Supplementary Figure S6b). X-gal staining of the brain sections showed an extensive distribution of reporter β-galactosidase positivity within tumor areas, revealing tumor-specific spread of oHSV-TRAIL (Figure 5bc). Immunofluorescence and confocal microscopy analysis demonstrated wide-spread positivity of cleaved caspase 3 staining on sections from oHSV-TRAIL-treated tumors as opposed to nearly no staining in PBS-treated tumors (P = 0.0015), highlighting the highly efficient tumor apoptosis induction by oHSV-TRAIL (Figure 5d–f). Furthermore, no cleaved caspase 3 staining was seen in mCherry-negative normal brain cells confirming tumor-specific effect (Figure 5de). The extensive, tumor selective apoptosis induction and robust therapeutic activity observed in the aggressive recurrent GSC31 model suggest apoptosis induction as key to the efficacy of oHSV-TRAIL therapy. Our results thus demonstrate potent therapeutic activity of oHSV-TRAIL in vivo in a highly invasive primary GBM as well as an aggressive recurrent GBM model that represent chemoresistant tumors.
Discussion
In this study, we identified patient-derived primary and recurrent GSC lines that are resistant to TMZ, and showed a potent therapeutic activity of oHSV-TRAIL against these refractory subsets of GBM cells. oHSV-TRAIL simultaneously alters cell proliferation, death and DDR pathways. In vivo, direct injections of oHSV-TRAIL prolong survival of mice bearing intracranial tumors derived from TMZ-resistant primary and recurrent GSC through robust induction of apoptotic cell death.
A body of evidence suggests that GSC contribute to therapeutic resistance and recurrence of GBM, and initial studies showed GSC are more resistant to TMZ than the bulk of tumor cells 26. However, heterogenous responses of GSC to TMZ have been reported, and whether GSC generally resist chemotherapeutics remains controversial 25, 27–29. Our screening of 11 patient-derived GSC lines revealed 7 as TMZ resistant. The recurrent GSC, 24R and 31, were TMZ resistant, consistent with the fact that these were established from tumors that failed TMZ treatment in the patients. Six of the 7 TMZ-resistant GSC lines responded to oHSV in vitro, indicating that TMZ-resistance does not render GSC resistant to oHSV. Of note, oHSV-TRAIL exerted superior cytotoxic effects against these GSC lines including recurrent GSC. However, 1 GSC (GSC64) was resistant and another GSC (GSC29) was only marginally susceptible to oHSV-TRAIL, most likely due to poor viral infectivity and/or replication as shown in Figure S2. Targeting these GSC with oHSV-TRAIL might require combinatorial approaches with agents such as chemotherapeutics or epigenetic modifiers (e.g., histone deacetylase inhibitors) 16, 30.
In our earlier study we showed that oHSV-TRAIL targets MAPK signaling pathways in established GBM lines resistant to oHSV and TRAIL 17. Here we confirmed oHSV and oHSV-TRAIL targeting of MAPK signaling pathways in TMZ-resistant primary and recurrent GSC. Interesting new finding is that, in addition to MAPK signaling pathways, oHSV and oHSV-TRAIL infection markedly downregulates Chk1, a critical cell cycle checkpoint kinase that gets activated in response to DNA damage, in GSC. Previous studies have shown that HSV-1 phosphorylates and activates check kinase 2 (Chk2) and blocks the cell cycle in the G2/M phase to enhance viral replication 31, 32. HSV-1 replication proteins disable ATR, inhibiting its phosphorylation of Replication protein A (RPA) and Chk1 33, 34. On the other hand, HSV-1 activates the ATM signaling pathway, and the activated ATM-MRN complex co-localizes with HSV replication compartments 25, 31, 35. However, HSV-induced depletion of the Chk1 protein has not been described, and this could be due to the degradation activity of HSV-encoded proteins. Indeed, the ubiquitin ligase activity of HSV proteins degrades the catalytic subunit of ATM through proteasome-dependent mechanisms 36, 37.
Our studies revealed an ability of oHSV-TRAIL to simultaneously inactivate MEK/ERK and Chk1 signaling pathways. Using highly selective inhibitors for MEK and Chk1, our studies support that the blockade of these pathways underlies the anti-GSC activity of oHSV-TRAIL. Active MEK/ERK signaling has been shown to interrupt Chk1 inhibitor activity against different tumors, and combination of MEK/ERK and Chk1 inhibitors was more effective than single inhibitors in treating multiple myeloma and GBM 38, 39. Interestingly, adding TRAIL to this combination was co-operative in killing human leukemia cells as co-administration of TRAIL promoted mitochondrial dysfunction and apoptosis induced by Chk1 and MEK inhibitors 40. This is in line with our data showing the triple combination of MEK/ERK inhibitor, Chk1 inhibitor, and S-TRAIL was more cytotoxic to GSC than any other combinations or monotherapies. These observations support that the multi-mechanistic action of oHSV-TRAIL underlies its apoptosis induction and efficient therapeutic targeting of refractory GBM cells. The oHSV-TRAIL disruption of multiple signaling pathways has promise in the treatment of GBM given GBM cells and GSC display sustained activation of multiple non-overlapping signaling pathways 41–44 which might reflect disappointing results of many clinical trials testing specific targeted agents as monotherapy 45. The detailed molecular mechanisms of oHSV-TRAIL will need to be validated by studies involving genetic silencing of Chk1 or ERK1/2 or overexpression of p38/JNK.
To assess the therapeutic efficacy of oHSV-TRAIL in orthotopic xenografts of chemoresistant GBM, we employed two models that each represents one of the two major challenges in clinical management of GBM, invasiveness and recurrence. Using a highly invasive model of GSC23 we showed that the treatment with oHSV-TRAIL significantly increased survival of tumor-bearing mice, which was consistent with our previous work that used the similarly invasive but TMZ-sensitive GSC8 model 13. However, the therapeutic benefit was not robust and cures were not achieved. Insufficient oHSV delivery and spread to widely dispersed tumor cells represents a problem that could limit oHSV efficacy in highly invasive GBM. Repeated injections or convection-enhanced delivery of the virus might enhance the infection of tumor cells exhibiting extensive infiltration 46. Extracellular matrix (ECM) within GBM can hamper intratumoral spread of oncolytic viruses by presenting physical barriers 47. We recently demonstrated that an important component of ECM, hyaluronan, is abundant in GBM and an oncolytic virus expressing hyaluronidase mediates increased intratumoral spread and anti-GBM potency 48. Simultaneous expression of TRAIL and hyaluronidase in the context of oHSV might overcome both resistance and delivery problems associated with oHSV therapy of highly invasive GBM.
The second model we used to evaluate oHSV-TRAIL efficacy was derived from the recurrent GSC31. GSC31 is very resistant to TMZ (Figure 1) and generates aggressive orthotopic tumors in mice that are non-invasive. Direct injection of oHSV-TRAIL had remarkable effects in the GSC31 model with a 40% cure rate, which was associated with extensive but tumor-specific induction of apoptosis as illustrated by cleaved caspase 3 staining (Figure 5). The particularly prominent therapeutic benefits of oHSV-TRAL seen in the recurrent GSC31 model may be associated with its non-invasive phenotype. Meisen et al reported that oHSV therapy of GBM elicited TNF secretion by macrophages and microglia, which led to GBM apoptosis, suppressed oHSV replication and reduced efficacy 49. However, we did not observe differences in the ability of oHSV and oHSV-TRAIL to replicate in GSC in vitro despite oHSV-TRAIL induction of apoptosis, which may reflect the in vivo efficacy of oHSV-TRAIL.
Recurrent GBM that progressed after radiation and TMZ chemotherapy has been shown to have distinct molecular characteristics that include increased expression of stemness or mesenchymal markers 50, 51, and a hypermutator phenotype caused primarily by TMZ 52. Therefore, newly diagnosed and recurrent GBM can exhibit differential response to treatment. Since most clinical trials testing investigational agents including oncolytic virus are initially designed for recurrent disease, use of recurrent GBM model for preclinical assessment is relevant and can be of predictive value. Thus, demonstration of strong efficacy of oHSV-TRAIL in a patient-derived recurrent GSC-based intracerebral model provides immediate translational implications. We have established a cohort of patient-derived recurrent GSC lines that display diverse biological phenotypes (e.g., invasive and nodular) and genotypes (12, 15, unpublished work), offering a useful platform to test experimental therapeutics. Studying oHSV-TRAIL in these models will allow us to better understand its mechanisms, efficacy and limitations in clinically representative settings. These efforts could lead us to a clinical trial testing oHSV-TRAIL for GBM patients, which ultimately may change the therapeutic paradigm for this devastating malignancy.
Supplementary Material
Novelty and Impact.
Resistance to temozolomide chemotherapy causes tumor recurrence and ultimately leads to mortality in patients with glioblastoma. This work demonstrates potent therapeutic efficacy of an apoptotic variant of oncolytic herpes simplex virus in preclinical glioblastoma models that recapitulate chemo-resistance and recurrence, and warrants a clinical trial testing of this strategy.
Acknowledgments
We acknowledge Shah lab members for helpful discussion and technical advice. This work was supported by JM Foundation (to KS) and ABTA (to HW). We have no conflicts of interest to declare.
Footnotes
Conflict of Interest and Financial Disclosure: Khalid Shah (KS) owns equity in and is a member of the Board of Directors of, AMASA Technologies, a company developing stem cell based therapies for cancer. KS’s interests were reviewed and are managed by Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict of interest policies. The other authors declare that there is no conflict of interest. This work was supported by NIH R01CA204720 (KS), JSMF (KS) and ABTA (HW).
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The Lancet Oncology. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Wojton J, Meisen WH, Kaur B. How to train glioma cells to die: molecular challenges in cell death. J Neurooncol. 2016;126:377–84. doi: 10.1007/s11060-015-1980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Molecular cell. 2006;22:501–10. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 7.Venur VA, Peereboom DM, Ahluwalia MS. Current medical treatment of glioblastoma. Cancer treatment and research. 2015;163:103–15. doi: 10.1007/978-3-319-12048-5_7. [DOI] [PubMed] [Google Scholar]
- 8.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–46. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 9.Park DM, Rich JN. Biology of glioma cancer stem cells. Molecules and cells. 2009;28:7–12. doi: 10.1007/s10059-009-0111-2. [DOI] [PubMed] [Google Scholar]
- 10.Gunther HS, Schmidt NO, Phillips HS, Kemming D, Kharbanda S, Soriano R, Modrusan Z, Meissner H, Westphal M, Lamszus K. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–909. doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Nigim F, Cavanaugh J, Patel AP, Curry WT, Jr, Esaki S, Kasper EM, Chi AS, Louis DN, Martuza RL, Rabkin SD, Wakimoto H. Targeting Hypoxia-Inducible Factor 1alpha in a New Orthotopic Model of Glioblastoma Recapitulating the Hypoxic Tumor Microenvironment. Journal of neuropathology and experimental neurology. 2015;74:710–22. doi: 10.1097/NEN.0000000000000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakimoto H, Kesari S, Farrell CJ, Curry WT, Jr, Zaupa C, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu TC, Jeyaretna DS, Debasitis J, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–81. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakimoto H, Mohapatra G, Kanai R, Curry WT, Jr, Yip S, Nitta M, Patel AP, Barnard ZR, Stemmer-Rachamimov AO, Louis DN, Martuza RL, Rabkin SD. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol. 2012;14:132–44. doi: 10.1093/neuonc/nor195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, Fulci G, Wakimoto H, Cheema TA, Buhrman JS, Jeyaretna DS, Stemmer Rachamimov AO, Rabkin SD, Martuza RL. Combination of oncolytic herpes simplex viruses armed with angiostatin and IL-12 enhances antitumor efficacy in human glioblastoma models. Neoplasia. 2013;15:591–9. doi: 10.1593/neo.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ning J, Wakimoto H. Oncolytic herpes simplex virus-based strategies: toward a breakthrough in glioblastoma therapy. Frontiers in microbiology. 2014;5:303. doi: 10.3389/fmicb.2014.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Wakimoto H, Agarwal AS, Rabkin SD, Bhere D, Martuza RL, Kuroda T, Kasmieh R, Shah K. Multimechanistic tumor targeted oncolytic virus overcomes resistance in brain tumors. Mol Ther. 2013;21:68–77. doi: 10.1038/mt.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duebgen M, Martinez-Quintanilla J, Tamura K, Hingtgen S, Redjal N, Wakimoto H, Shah K. Stem cells loaded with multimechanistic oncolytic herpes simplex virus variants for brain tumor therapy. J Natl Cancer Inst. 2014;106:dju090. doi: 10.1093/jnci/dju090. [DOI] [PubMed] [Google Scholar]
- 19.Shah K, Hingtgen S, Kasmieh R, Figueiredo JL, Garcia-Garcia E, Martinez-Serrano A, et al. Bimodal viral vectors and in vivo imaging reveal the fate of human neural stem cells in experimental glioma model. J Neurosci. 2008;28:4406–13. doi: 10.1523/JNEUROSCI.0296-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheema TA, Wakimoto H, Fecci PE, Ning J, Kuroda T, Jeyaretna DS, Martuza RL, Rabkin SD. Multifaceted oncolytic virus therapy for glioblastoma in an immunocompetent cancer stem cell model. Proc Natl Acad Sci U S A. 2013;110:12006–11. doi: 10.1073/pnas.1307935110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroda T, Martuza RL, Todo T, Rabkin SD. Flip-Flop HSV-BAC: bacterial artificial chromosome based system for rapid generation of recombinant herpes simplex virus vectors using two independent site-specific recombinases. BMC Biotechnol. 2006;6:40. doi: 10.1186/1472-6750-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todo T, Martuza RL, Rabkin SD, Johnson PA. Oncolytic herpes simplex virus vector with enhanced MHC class I presentation and tumor cell killing. Proc Natl Acad Sci USA. 2001;98:6396–401. doi: 10.1073/pnas.101136398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasportas LS, Kasmieh R, Wakimoto H, Hingtgen S, van de Water JA, Mohapatra G, Figueiredo JL, Martuza RL, Weissleder R, Shah K. Assessment of therapeutic efficacy and fate of engineered human mesenchymal stem cells for cancer therapy. Proc Natl Acad Sci U S A. 2009;106:4822–7. doi: 10.1073/pnas.0806647106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Water JA, Bagci-Onder T, Agarwal AS, Wakimoto H, Roovers RC, Zhu Y, Kasmieh R, Bhere D, Van Bergen en Henegouwen PM, Shah K. Therapeutic stem cells expressing variants of EGFR-specific nanobodies have antitumor effects. Proc Natl Acad Sci U S A. 2012;109:16642–7. doi: 10.1073/pnas.1202832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanai R, Rabkin SD, Yip S, Sgubin D, Zaupa CM, Hirose Y, Louis DN, Wakimoto H, Martuza RL. Oncolytic virus-mediated manipulation of DNA damage responses: synergy with chemotherapy in killing glioblastoma stem cells. J Natl Cancer Inst. 2012;104:42–55. doi: 10.1093/jnci/djr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Molecular cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beier D, Rohrl S, Pillai DR, Schwarz S, Kunz-Schughart LA, Leukel P, Proescholdt M, Brawanski A, Bogdahn U, Trampe-Kieslich A, Giebel B, Wischhusen J, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68:5706–15. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 28.Blough MD, Westgate MR, Beauchamp D, Kelly JJ, Stechishin O, Ramirez AL, Weiss S, Cairncross JG. Sensitivity to temozolomide in brain tumor initiating cells. Neuro Oncol. 2010;12:756–60. doi: 10.1093/neuonc/noq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong X, Schwartz PH, Linskey ME, Bota DA. Neural stem/progenitors and glioma stem-like cells have differential sensitivity to chemotherapy. Neurology. 2011;76:1126–34. doi: 10.1212/WNL.0b013e318212a89f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanai R, Rabkin SD. Combinatorial strategies for oncolytic herpes simplex virus therapy of brain tumors. CNS oncology. 2013;2:129–42. doi: 10.2217/cns.12.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirata N, Kudoh A, Daikoku T, Tatsumi Y, Fujita M, Kiyono T, Sugaya Y, Isomura H, Ishizaki K, Tsurumi T. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J Biol Chem. 2005;280:30336–41. doi: 10.1074/jbc.M500976200. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Baskaran R, Krisky DM, Bein K, Grandi P, Cohen JB, Glorioso JC. Chk2 is required for HSV-1 ICP0-mediated G2/M arrest and enhancement of virus growth. Virology. 2008;375:13–23. doi: 10.1016/j.virol.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohni KN, Livingston CM, Cortez D, Weller SK. ATR and ATRIP are recruited to herpes simplex virus type 1 replication compartments even though ATR signaling is disabled. J Virol. 2010;84:12152–64. doi: 10.1128/JVI.01643-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohni KN, Smith S, Dee AR, Schumacher AJ, Weller SK. Herpes simplex virus type 1 single strand DNA binding protein and helicase/primase complex disable cellular ATR signaling. PLoS Pathog. 2013;9:e1003652. doi: 10.1371/journal.ppat.1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci U S A. 2005;102:5844–9. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, Spencer CA. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkinson J, Lees-Miller SP, Everett RD. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–7. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai Y, Chen S, Pei XY, Almenara JA, Kramer LB, Venditti CA, Dent P, Grant S. Interruption of the Ras/MEK/ERK signaling cascade enhances Chk1 inhibitor-induced DNA damage in vitro and in vivo in human multiple myeloma cells. Blood. 2008;112:2439–49. doi: 10.1182/blood-2008-05-159392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Y, Dai Y, Grant S, Dent P. Enhancing CHK1 inhibitor lethality in glioblastoma. Cancer biology & therapy. 2012;13:379–88. doi: 10.4161/cbt.19240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai Y, Dent P, Grant S. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) promotes mitochondrial dysfunction and apoptosis induced by 7-hydroxystaurosporine and mitogen-activated protein kinase kinase inhibitors in human leukemia cells that ectopically express Bcl-2 and Bcl-xL. Mol Pharmacol. 2003;64:1402–9. doi: 10.1124/mol.64.6.1402. [DOI] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark PA, Iida M, Treisman DM, Kalluri H, Ezhilan S, Zorniak M, Wheeler DL, Kuo JS. Activation of multiple ERBB family receptors mediates glioblastoma cancer stem-like cell resistance to EGFR-targeted inhibition. Neoplasia. 2012;14:420–8. doi: 10.1596/neo.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, Leversha MA, Mikkelsen T, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–6. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S, Bronson RT, Pfannl R, White F, Housman DE, Iyer L, Whittaker CA, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31:3039–50. doi: 10.1038/onc.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reardon DA, Wen PY, Mellinghoff IK. Targeted molecular therapies against epidermal growth factor receptor: past experiences and challenges. Neuro Oncol. 2014;16(Suppl 8):viii7–13. doi: 10.1093/neuonc/nou232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogelbaum MA, Aghi MK. Convection-enhanced delivery for the treatment of glioblastoma. Neuro Oncol. 2015;17(Suppl 2):ii3–ii8. doi: 10.1093/neuonc/nou354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nature biotechnology. 2012;30:658–70. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Quintanilla J, He D, Wakimoto H, Alemany R, Shah K. Encapsulated Stem Cells Loaded With Hyaluronidase-expressing Oncolytic Virus for Brain Tumor Therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2015;23:108–18. doi: 10.1038/mt.2014.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meisen WH, Wohleb ES, Jaime-Ramirez AC, Bolyard C, Yoo JY, Russell L, Hardcastle J, Dubin S, Muili K, Yu J, Caligiuri M, Godbout J, et al. The Impact of Macrophage- and Microglia-Secreted TNFalpha on Oncolytic HSV-1 Therapy in the Glioblastoma Tumor Microenvironment. Clin Cancer Res. 2015;21:3274–85. doi: 10.1158/1078-0432.CCR-14-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K, Aoyagi M, Ando N, Ogishima T, Wakimoto H, Yamamoto M, Ohno K. Expansion of CD133-positive glioma cells in recurrent de novo glioblastomas after radiotherapy and chemotherapy. Journal of neurosurgery. 2013;119:1145–55. doi: 10.3171/2013.7.JNS122417. [DOI] [PubMed] [Google Scholar]
- 51.Mahabir R, Tanino M, Elmansuri A, Wang L, Kimura T, Itoh T, Ohba Y, Nishihara H, Shirato H, Tsuda M, Tanaka S. Sustained elevation of Snail promotes glial-mesenchymal transition after irradiation in malignant glioma. Neuro Oncol. 2014;16:671–85. doi: 10.1093/neuonc/not239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.