Abstract
Bone mineral density (BMD) is a complex trait with high missing heritability. Numerous evidences have shown that BMD variation has a relationship with coronary artery disease (CAD). This relationship may come from a common genetic basis called pleiotropy. By leveraging the pleiotropy with CAD, we may be able to improve the detection power of genetic variants associated with BMD. Using a recently developed conditional false discovery rate (cFDR) method, we jointly analyzed summary statistics from two large independent genome wide association studies (GWAS) of lumbar spine (LS) BMD and CAD. Strong pleiotropic enrichment and 7 pleiotropic SNPs were found for the two traits. We identified 41 SNPs for LS BMD (cFDR<0.05), of which 20 were replications of previous GWASs and 21 were potential novel SNPs that were not reported before. Four genes encompassed by 9 cFDR-significant SNPs were partially validated in the gene expression assay. Further functional enrichment analysis showed that genes corresponding to the cFDR-significant LS BMD SNPs were enriched in GO terms and KEGG pathways that played crucial roles in bone metabolism (adjP < 0.05). In protein-protein interaction analysis, strong interactions were found between the proteins produced by the corresponding genes. Our study demonstrated the reliability and high-efficiency of the cFDR method on the detection of trait-associated genetic variants, the present findings shed novel insights into the genetic variability of BMD as well as the shared genetic basis underlying osteoporosis and CAD.
Keywords: Bone Mineral Density (BMD), Coronary Artery Disease (CAD), Pleiotropy, Genome Wide Association Study (GWAS)
1. Introduction
Bone mineral density (BMD) is a complex trait with high heritability. It’s a major risk factor for osteoporosis, the incidence of which increased dramatically in recent years due largely to the ageing of population worldwide. Nearly 54 million adults were affected by osteoporosis or osteopenia in 2010 in the US, and this number was estimated to increase to 70.8 million from 2010 to 2030 [1]. The incidence of fragility fracture – the most severe complication of osteoporosis, also increased proportionally with the high prevalence, leading to long term disability, high mortality to afflicted individuals and huge economic burden on their families and societies [2]. BMD is a heritable trait with the highest heritability of 83% at lumbar spine (LS) [3]. To date, genome-wide association studies (GWAS) have reported more than 200 SNPs associated with BMD (https://www.ebi.ac.uk/gwas, April 2017), yet only explained a small proportion of the whole heritability [4]. For better disease control and prevention, further studies are warranted to uncover novel BMD associated genetic variants.
Numerous epidemiological and clinical evidences have shown a relationship between BMD and coronary artery disease (CAD). CAD is the most common type of heart disease, which is the major cause of death worldwide, accounting for almost 1 of every 4 death in the United States [5]. BMD and CAD are both highly heritable traits [3,6], BMD decreasing diseases (such as osteoporosis and osteopenia) and CAD have high prevalence and occur concurrently in the elderly [1,5]. It has been reported that in healthy men, BMD was inversely associated with their 10-year CAD risk [7], while among women, slight increase of annual BMD loss was notably associated with higher coronary artery calcification - a main cause of plaques in inner lining of coronary artery which eventually leads to CAD [8]. Osteoporosis and CAD share common risk factors, such as dyslipidemia, estrogen and/or vitamin D deficiency [9], and GWAS analysis has revealed strong genetic overlap between BMD and CAD risk factors, such as lipoproteins cholesterol and blood pressure [10]. Molecular pathways involved in bone mineralization and coronary artery calcification are very similar [11], meanwhile bone marrow cells including osteoclasts and osteoblasts were identified in calcified plaques of vessels [12]. In addition, in mouse models, deletion of osteoprotegerin caused both osteoporosis and vascular calcification [13].
The underlying genetic mechanism of the relationship between BMD and CAD might partially come from the presence of pleiotropic effect, which means that one single gene could influence multiple traits. By leveraging pleiotropy with associated traits and using novel analytical methods, we may be able to explore more of the traits’ total heritability. Recently, Andreassen et al. proposed a novel, high-efficient and cost effective analysis method - pleiotropy informed conditional false discovery rate (cFDR) method on existing GWAS data and successfully identified potential novel loci for a number of traits as well as pleiotropic loci for multiple disorders [14,10]. cFDR method has the advantage to increase the effective sample size of existing GWAS data for individual traits/diseases and improve the detection of genetic variants without the need of larger datasets or new recruitments. According to the authors’ simulation, while compared with the unconditional FDR during a single trait analysis, using the cFDR method could increase 15–20 times the number of non-null SNPs discovered for a FDR smaller than 0.05 [14]. In our previous studies, we successfully implemented the cFDR analyses and identified potential novel and pleiotropic variants for type 2 diabetes and/or birth weight [15] as well as for BMD and height [16]. Here, we applied the cFDR method to two large independent GWAS data of LS BMD and CAD, aiming to detect more potential novel trait-associated single nucleotide polymorphisms (SNPs) and genes for BMD.
2 Method and materials
2.1 GWAS datasets
We obtained GWAS summary statistics of LS (lumbar spine) BMD from the Genetic Factors for Osteoporosis Consortium (GEFOS). This dataset contains summary statistics from meta-analysis of 35 individual GWAS studies (n=53,236) conducted by whole genome sequencing, whole exome sequencing and deep imputation of genotype data. To date, it is the largest published GWAS data of BMD and 10,582,866 SNPs were included [17]. The CAD GWAS summary statistics were obtained from the CARDIoGRAMplusC4D Consortium. It is a meta-analysis of 22 CAD GWAS studies with 22,233 cases and 64,762 controls (n=86,995), and 2,420,360 SNPs were included [18]. There were no overlapping subjects between the LS BMD and CAD GWAS datasets. The detailed phenotype characteristics and inclusion criteria for the two GWASs were described in the original publications [18,17].
2.2 Data processing
First, based on linkage disequilibrium (LD) SNP pruning method and using HapMap 3 genotypes as a reference, we performed pruning on LS BMD and CAD datasets respectively. The linkage disequilibrium (LD) between each pair of SNPs in the datasets was calculated by Plink, for those SNP pairs with r2 > 0.2, the SNP with the smaller allele frequency (MAF) was removed [19]. After pruning, there were 369,514 SNPs remained for LS BMD and 167,894 SNPs remained for CAD. Next, the two datasets were combined and there were 128,164 common SNPs remained for both phenotypes to be used in the subsequent analysis. Genomic control has already been applied in the two datasets to ensure that the variance estimates for each SNP were not inflated due to population structure [18,17].
2.3 Statistical Analyses
2.3.1 Pleiotropic Enrichment Estimation
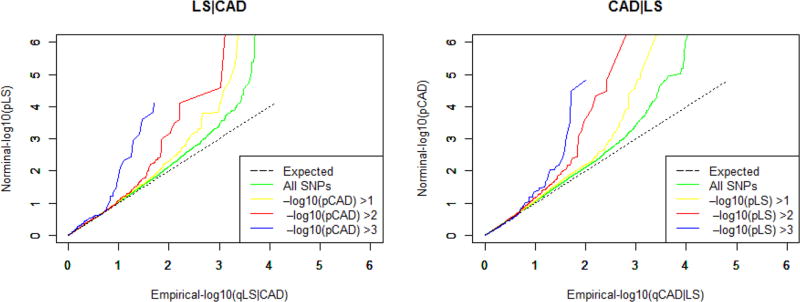
Quantile-quantile plot (Q-Q plot) is an exploratory tool implemented in R to evaluate whether the primary phenotype is related to the given phenotype under the null hypothesis, which is also termed as “enrichment”. The Q-Q curve was plotted for the quantile of nominal –log10 (p) values for association of the subset of variants that were below different significance threshold in the conditional trait. The empirical quantiles (-log10(q)) were plotted on x-axis and the nominal p-values (-log10 (p)) were plotted on y-axis. As the principal phenotype successively conditioned on more stringent significance criteria in the conditional phenotype, the pleiotropic enrichment could be observed intuitively by the degree of leftward deflection from the expected identity line.
2.3.2 The calculation of cFDR and conjunction cFDR (ccFDR)
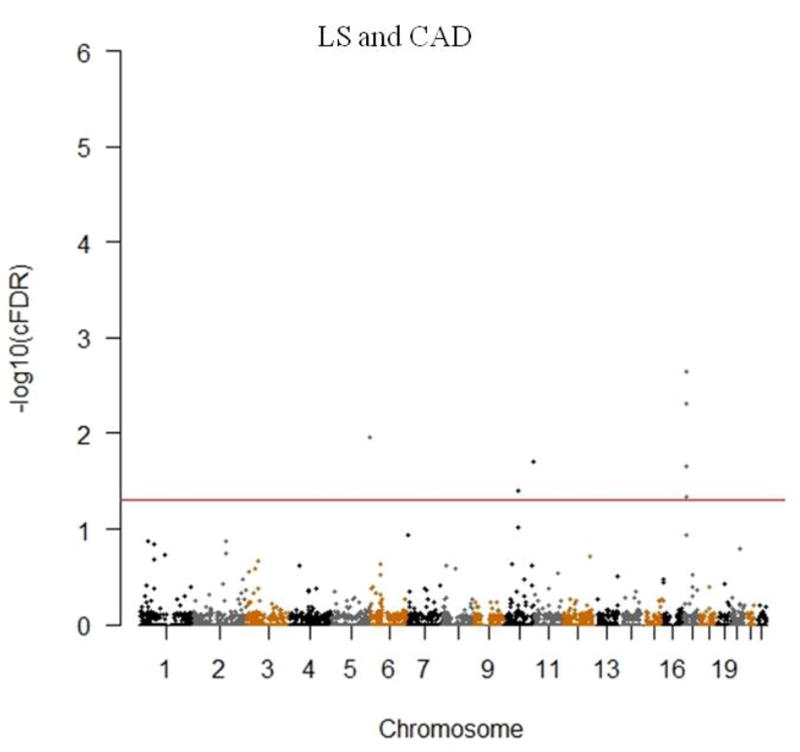
The cFDR method is regarded as an extension of the standard FDR framework. It represents the possibility that a random SNP is null for association with the principal phenotype given that the observed p-values for the principal and conditional phenotypes are both less than two pre-defined trait specific significance thresholds [14]. Using GWAS p-values of the two pruned datasets, we computed cFDR for every single SNP while LS BMD was the principal phenotype conditioned on the strength of its association with CAD (LS|CAD) and reversely (CAD|LS). To assess whether the cFDR method results in the enrichment of specific SNPs, the subset of SNPs was confined based on the level of significance for the association of each variant with the conditional trait using the following criteria: p < 1 (all SNPs), p < 0.1, p < 0.01 and p < 0.001. A threshold of cFDR less than 0.05 was used to determine whether the SNP was significantly associated with the principal phenotype. The procedure of this method has been described in detail by Jonathan et al [16]. ccFDR was computed to identify pleiotropic SNPs for both LS BMD and CAD after the calculation of cFDR. It refers to the possibility that a given SNP has a false positive association with the two traits, and is taken as the maximum cFDR value between them. We used a threshold of ccFDR less than 0.05 to identify whether the SNP was significantly associated with both traits and was a pleiotropic one. Manhattan plots of cFDR and ccFDR were constructed respectively by R to mark the chromosomal locations as well as significance of various SNPs. SNPs with –log10 cFDR value or –log10 ccFDR value more than 1.3 were regarded as significant.
2.4 Annotation of Potential Novel SNPs for LS BMD
To compare with the previous BMD GWAS findings and verify whether the cFDR-significant SNPs are potential novel ones, we assembled a confirmed_SNP set by downloading all the SNPs that were already confirmed to be associated with BMD in earlier GWASs from the web site https://www.ebi.ac.uk/gwas, April 2017 (Supplementary Table 1). Totally there were 251 SNPs that were confirmed by previous BMD GWASs. Then, based on HapMap genotype data and using LD threshold at 0.8 as a criteria, we input the set of cFDR-significant SNPs with p values > 5E-8 and the confirmed_SNP set into the SNPinfo Web Server to perform LD analysis (https://snpinfo.niehs.nih.gov/). Each LD block may contain one or more than one SNP, a SNP that is not clustered in the same LD block with SNPs of the confirmed_SNP set was regarded as a potential novel SNP. Meanwhile, SNPs that clustered in the same LD block with SNPs of the confirmed_SNP set were regarded as replication of the previous findings.
2.5 Gene Expressional Validation Analysis
Using gene expression profiling in iliac bone biopsies from 84 postmenopausal Caucasian women [20], we performed gene expressional partial validation analysis by calculating the correlation values between LS BMD and the mRNA levels of cFDR-significant LS BMD associated genes. The details of the gene expression profiling of iliac bone biopsies can be found in the original publication [20].
2.6 Functional Enrichment analysis and Protein-Protein Interaction analysis
To explore the biological roles of cFDR-significant SNPs/genes, we performed functional enrichment analysis and protein-protein interaction analysis. cFDR-significant SNPs were mapped to nearby genes by the online tool SNPinfo Web Server (https://snpinfo.niehs.nih.gov/). Using Web-Based Gene Set Analysis Toolkit [21], we input these genes into the WebGestalt system and performed functional term enrichment analysis. Gene ontology (GO) terms and KEGG canonical pathways with adjusted p value less than 0.05 were regarded as significant. Functional term enrichment analysis of the corresponding genes gave us the opportunity to further partially validate our results by identifying gene sets that play key roles in bone metabolism. Protein-protein interaction analysis for the corresponding genes were conducted using the online database STRING 10.0 (http://string-db.org/), it helped us explore the functional roles of the proteins produced by the identified LS BMD-associated genes in the context of biological mechanisms of bone metabolism.
3 Results
3.1 Pleiotropic enrichment assessment and pleiotropic SNPs identified for LS BMD and CAD
Quantile-quantile plot (Q-Q plot) was constructed to assess the pleiotropic enrichment between LS BMD and CAD. As shown in Figure 1, strong enrichment could be observed for LS BMD associated SNPs across different levels of association with CAD (LS│CAD), the separation shown between different curves indicated the extent of pleiotropic SNPs shared between both traits. The earlier departure from the expected line indicated the greater proportion of true associations for a given CAD p-value. Similar patterns of strong enrichment could also be observed for CAD conditioning on LS BMD (CAD│LS) (Detailed information of the CAD cFDR-significant SNPs were presented in Supplementary Table 2). Totally we identified 7 pleiotropic SNPs mapping to 3 chromosomes reached the significance threshold of ccFDR < 0.05 (Figure 2 and Table 1). Genes encompassed by these pleiotropic SNPs were found to be associated with bone metabolism [22,23] and/or CAD pathology [24], as well as risk factors for both osteoporosis and CAD, such as obesity and insulin resistance [25,26].
Figure 1.
Quantile-quantile plots (Q-Q plots) of nominal versus empirical -log10(p) for LS BMD as a function of significance of the association with CAD (LS |CAD) at the level of -log10(p) > 0, -log10(p) > 1, -log10(p) > 2, -log10(p) > 3 corresponding to p < 1, p < 0.1, p < 0.01, p < 0.001 respectively; and reversely CAD as a function of significance of the association with LS BMD (CAD|LS).
Figure 2.
Conjunction Manhattan plot of conjunction –log10 FDR values for LS BMD and CAD. The red line marking the conditional –log10 FDR value of 1.3 corresponds to a ccFDR < 0.05. The figure shows the genomic locations of pleiotropic SNPs and further details are presented in Table 1.
Table 1.
Pleiotropic SNPs for LS BMD and CAD (ccFDR<0.05)
| SNP | Chr | Pos | p value | ccFDR | Nearby Gene | |
|---|---|---|---|---|---|---|
| LS | CAD | |||||
| rs10039254 | 5 | 178503520 | 1.49E-04 | 3.01E-04 | 1.08E-02 | ADAMTS2 |
| rs10765090 | 10 | 133238729 | 1.12E-03 | 1.17E-04 | 1.98E-02 | TCERG1L and FLJ46300 |
| rs11005240 | 10 | 57598774 | 7.41E-04 | 8.05E-04 | 4.02E-02 | LOC389970 and ZWINT |
| rs12449964 | 17 | 17485429 | 3.74E-03 | 4.02E-07 | 2.24E-02 | PEMT and RAI1 |
| rs12943500 | 17 | 17731313 | 7.69E-05 | 3.24E-05 | 2.23E-03 | TOM1L2 |
| rs6502629 | 17 | 17810367 | 2.57E-04 | 1.48E-05 | 4.88E-03 | TOM1L2 |
| rs7208561 | 17 | 17556253 | 5.92E-03 | 4.55E-05 | 4.59E-02 | RAI1 |
Column definition: SNP – single nucleotide polymorphisms; Chr – chromosome; Pos – chromosomal position; LS – lumbar spine bone mineral density; CAD – coronary artery disease; ccFDR – conjunction conditional false discovery rate.
3.2 LS BMD SNPs identified with cFDR
We identified 41 LS BMD SNPs mapping to 13 chromosomes conditioned on CAD with a significance threshold of cFDR < 0.05 (Supplementary Figure 1), all the details were presented in Table 2. Of these cFDR-significant SNPs, 16 have been discovered in the original GWAS to be significantly associated with LS BMD (p ≤ 5E-8 in the original GWAS), and 3 other SNPs with p values > 5E-8 for LS BMD (rs10464592, rs11753987 and rs2741856) have reached GWAS significance threshold of p ≤ 5E-8 for femoral neck BMD in the original GWAS [17]. SNP rs7683315 resides in the same LD block with two other SNPs (rs6532023, rs1471403) of the confirmed_SNP set (supplementary Table 1). These 20 SNPs were regarded as successful replication of previous BMD GWASs’ findings and well demonstrated the reliability of cFDR method in detecting trait-associated genetic variants. Totally we identified 21 potential novel SNPs for LS BMD (bold SNPs in Table2), which have been overlooked in the original GWAS analysis using standard statistical methods [17]. Of these potential novel SNPs, 9 out of 21 were located in or near genes (ESR1, FLJ42280, JAG1, MEPE, SOST, TNFRSF11B, TNFSF11, WNT4) that were confirmed in previous GWASs to be significantly associated with BMD [27], there were 12 potential novel SNPs located in or near genes which were not reported to be associated with BMD before.
Table 2.
LS BMD SNPs conditioned on CAD (cFDR<0.05)
| Expressed QTL
|
|||||||
|---|---|---|---|---|---|---|---|
| SNP | Chr | Pos | p (LS) | cFDR (LS│C AD) |
Nearby Gene | AffymetrixID | r |
| rs11209223 | 1 | 68373736 | 4.04E-06 | 2.04E-02 | WLS | NA | NA |
| rs1864569 | 1 | 68406845 | 2.80E-06 | 1.43E-02 | WLS | NA | NA |
| rs2473252 | 1 | 22411184 | 2.18E-05 | 4.53E-02 | WNT4/LOC343384 | 230751_at/NA | −0.04/NA |
| rs2566758 | 1 | 68405071 | 4.07E-08 | 6.02E-05 | WLS | NA | NA |
| rs7554551 | 1 | 68437611 | 1.29E-15 | 1.32E-11 | WLS | NA | NA |
| rs944082 | 1 | 68415679 | 3.36E-10 | 2.45E-06 | WLS | NA | NA |
| rs4854512 | 2 | 68857961 | 5.06E-06 | 1.22E-02 | ARHGAP25 | 1555076_at | −0.14 |
| rs7683315 | 4 | 88992119 | 2.91E-07 | 8.64E-04 | MEPE/HSP90AB3P | 221150_at/NA | *0.46/NA |
| rs10039254 | 5 | 178503520 | 1.49E-04 | 7.38E-03 | ADAMTS2 | 214454_at | −0.12 |
| rs2089190 | 5 | 171972970 | 8.69E-06 | 3.75E-02 | SH3PXD2B/LOC100130394 | 1562910_at/NA | −0.09/NA |
| rs1038304 | 6 | 151974868 | 1.05E-12 | 2.55E-08 | CCDC170 | 220581_at | *−0.25 |
| rs11753987 | 6 | 151893121 | 1.10E-07 | 7.90E-04 | CCDC170 | 220581_at | *−0.25 |
| rs12197879 | 6 | 151937470 | 7.31E-10 | 1.03E-05 | CCDC170 | 220581_at | *−0.25 |
| rs1293935 | 6 | 152059651 | 7.89E-07 | 4.57E-03 | CCDC170/ESR1 | 220581_at/211233_x_at | *−0.25/−0.09 |
| rs2504065 | 6 | 152136860 | 4.95E-07 | 2.88E-03 | CCDC170/ESR1 | 220581_at/211233_x_at | *−0.25/−0.09 |
| rs7746854 | 6 | 151970035 | 4.21E-06 | 1.87E-02 | CCDC170 | 220581_at | *−0.25 |
| rs10464592 | 7 | 96068622 | 5.08E-08 | 4.24E-04 | FLJ42280/LOC100128444 | NA/NA | NA/NA |
| rs10499926 | 7 | 96168243 | 2.55E-07 | 1.52E-03 | SHFM1 | 202276_at | 0.12 |
| rs1376264 | 7 | 37924814 | 9.30E-09 | 8.49E-05 | SFRP4/EPDR1 | 204052_S_at/223253_at | *0.3/0.05 |
| rs10955924 | 8 | 120122524 | 7.26E-19 | 7.54E-14 | TNFRSF11B/COLEC10 | 204933_s_at/207420_at | −0.08/−0.13 |
| rs12375331 | 8 | 120120103 | 1.52E-07 | 1.02E-03 | TNFRSF11B/COLEC10 | 204933_s_at/207420_at | −0.08/−0.13 |
| rs1905776 | 8 | 120088833 | 2.42E-07 | 1.78E-03 | TNFRSF11B/COLEC10 | 204933_s_at/207420_at | −0.08/−0.13 |
| rs1948467 | 8 | 119905753 | 1.77E-09 | 1.78E-05 | LOC441377/TNFRSF11B | NA/204933_s_at | NA/−0.08 |
| rs2055101 | 8 | 119956104 | 9.70E-12 | 2.82E-07 | LOC441377/TNFRSF11B | NA/204933_s_at | NA/−0.08 |
| rs3134063 | 8 | 120028838 | 1.85E-14 | 6.23E-10 | TNFRSF11B | 204933_s_at | −0.08 |
| rs1007865 | 10 | 124040120 | 2.60E-05 | 2.76E-02 | BTBD16 | 1552566_at | 0.05 |
| rs10765090 | 10 | 133238729 | 1.12E-03 | 1.98E-02 | TCERG1L/FLJ46300 | 231257_at/NA | 0.07/NA |
| rs11005240 | 10 | 57598774 | 7.41E-04 | 3.52E-02 | LOC389970/ZWINT | NA/204026_s_at | NA/0.14 |
| rs2513277 | 11 | 68117463 | 5.42E-11 | 1.16E-06 | SAPS3 | NA | NA |
| rs7936582 | 11 | 68049506 | 3.18E-08 | 2.82E-04 | SAPS3 | NA | NA |
| rs12430303 | 13 | 41930027 | 8.30E-09 | 7.76E-05 | FABP3P2/TNFSF11 | 216569_at/210643_at | 0.14/−0.12 |
| rs4454853 | 13 | 41871297 | 4.74E-09 | 5.42E-05 | FABP3P2/TNFSF11 | 216569_at/210643_at | 0.14/−0.12 |
| rs9594738 | 13 | 41850145 | 3.63E-20 | 3.95E-15 | FABP3P2/TNFSF11 | 216569_at/210643_at | 0.14/−0.12 |
| rs724404 | 16 | 49515626 | 3.22E-07 | 1.81E-03 | LOC727992/LOC728654 | NA/NA | NA/NA |
| rs12449964 | 17 | 17485429 | 3.74E-03 | 2.24E-02 | PEMT/RAI1 | 207621_s_at/226143_at | −0.16/−0.1 |
| rs12943500 | 17 | 17731313 | 7.69E-05 | 2.23E-03 | TOM1L2 | 214840_at | −0.06 |
| rs2741856 | 17 | 39182365 | 9.59E-08 | 6.41E-04 | LOC100128016/SOST | NA/223869_at | NA/*0.5 |
| rs6502629 | 17 | 17810367 | 2.57E-04 | 4.88E-03 | TOM1L2 | 214840_at | −0.06 |
| rs7208561 | 17 | 17556253 | 5.92E-03 | 4.59E-02 | RAI1 | 226143_at | −0.1 |
| rs8073599 | 17 | 62797556 | 6.05E-06 | 2.93E-02 | PSMD12/LOC729822 | 202352_s_at/NA | −0.13/NA |
| rs6040061 | 20 | 10588306 | 5.94E-09 | 3.91E-05 | JAG1 | 216268_s_at | 0.1 |
Column definition: LS BMD – lumbar spine bone mineral density; SNP – single nucleotide polymorphisms; bold SNPs represent potential novel SNPs identified by cFDR; CAD – coronary artery disease; Chr – chromosome; Pos – chromosomal position; p (LS) – p value in the original BMD GWAS for lumbar spine; cFDR – conditional false discovery rate; LS│CAD –lumbar spine bone mineral density as the principal phenotype conditioned on the strength of its association with coronary artery disease; r – correlation coefficient; NA – not applicable (undetected);
r values represent nominally significant (p < 0.05) Pearson correlations, that means its corresponding gene was partially validated in the gene expression assay.
3.3 Gene expressional validation analysis for cFDR-significant genes associated with LS BMD
Using gene expression profiling in iliac bone biopsies from 84 postmenopausal Caucasian women [20], the correlation values between LS BMD and the mRNA levels of cFDR-significant genes were calculated and presented in the rightmost columns of Table 1. mRNA transcripts of 4 genes (MEPE, CCDC170, SFRP4 and SOST) annotated by 9 LS BMD cFDR-significant SNPs (with *r values) were significantly correlated with LS BMD (p value < 0.05, this threshold of p value was used here for the purpose of partial validation).
3.4 Functional enrichment analysis for LS BMD cFDR-significant genes
To explore the functional role of LS BMD associated genes in bone metabolism, we performed a series of analysis. Firstly, we input the corresponding genes into WebGestalt system for GO term analysis and KEGG pathway analysis. The corresponding genes were enriched in as much as 47 GO terms (Supplementary Table 3), many of which were significantly associated with bone metabolism (Table 3). As shown in Table 3, “Ossification” (adjP = 2.30E-03) was one of the most significantly enriched GO terms for LS BMD associated genes. It is the process of bone formation and a major part of bone metabolism. Other GO terms which obviously played important roles in bone were also found for the corresponding genes, such as “negative regulation of canonical Wnt receptor signaling pathway” (adjP = 3.70E-03), “regulation of bone resorption” (adjP = 7.30E-03), “regulation of bone remodeling” (adjP = 8.00E-03), “negative regulation of ossification” (adjP = 9.00E-03). Interestingly, LS BMD associated genes were also enriched in GO terms related to estrogen, such as “Response to estrogen stimulus” (adjP = 2.40E-03) and “Cellular response to estrogen stimulus” (adjP = 3.70E-03). BMD is believed to be a marker of women’s lifetime exposure to estrogen, estrogen helps maintain bone mass and its deficiency plays a key role in the process of osteoporosis among menopausal women [28]. This interesting GO term result further validated the functional roles of LS BMD associated genes in the estrogen related bone metabolism. The significant KEGG pathways were presented in Table 4, of which “proteasome” (adjP = 1.20E-02) was the most significantly enriched. Protein degradation mediated by ubiquitin-proteasome is very crucial to both bone formation and resorption [29]. TNFRSF11B and TNFSF11 were enriched in “osteoclast differentiation pathway” (adjP = 4.30E-02), which was crucial to the resorption and remodeling of bone. WNT4 and SFRP4 were enriched in “Wnt signaling pathway” (adjP = 4.30E-03). Activation of Wnt signaling pathway increases bone mass while inhibition of it decreases bone mass and promotes osteoporosis in mice [30], and polymorphisms in Wnt pathway have been identified to be associated with altered BMD variation in GWAS and meta-analysis [31].
Table 3.
Bone associated GO term results for LS BMD cFDR-significant genes (adjP < 0.05)
| GO ID | adjP | GO Term | Genes |
|---|---|---|---|
| 1503 | 2.30E-03 | ossification | WNT4,TNFSF11,MEPE,SOST,ESR1 |
| 43627 | 2.40E-03 | response to estrogen stimulus | WNT4,ESR1,SFRP4 |
| 71391 | 3.70E-03 | cellular response to estrogen stimulus | WNT4,ESR1 |
| 90090 | 3.70E-03 | negative regulation of canonical Wnt receptor signaling pathway | WNT4,SOST,SFRP4 |
| 60828 | 7.30E-03 | regulation of canonical Wnt receptor signaling pathway | WNT4,SOST,SFRP4 |
| 45124 | 7.30E-03 | regulation of bone resorption | TNFSF11,TNFSF11B |
| 30178 | 7.30E-03 | negative regulation of Wnt receptor signaling pathway | WNT4,SOST,SFRP4 |
| 46850 | 8.00E-03 | regulation of bone remodeling | TNFSF11,TNFSF11B |
| 30279 | 9.00E-03 | negative regulation of ossification | MEPE,SOST |
| 30278 | 1.08E-02 | regulation of ossification | WNT4,MEPE,SOST |
Column definition: GO – gene ontology term; adjP – p value adjusted by the multiple test adjustment.
Table 4.
KEGG pathway analysis for LS BMD cFDR-significant genes (adjP < 0.05)
| Pathway Name | adjP | Genes |
|---|---|---|
| Proteasome | 1.20E-03 | SHFM1, PSMD12 |
| Wnt signaling pathway | 4.30E-03 | WNT2, SFRP4 |
| Osteoclast differentiation | 4.30E-03 | TNFRSF11B, TNFSF11 |
| Cytokine-cytokine receptor interaction | 9.50E-03 | TNFRSF11B, TNFSF11 |
Column definition: adjP – p value adjusted by the multiple test adjustment.
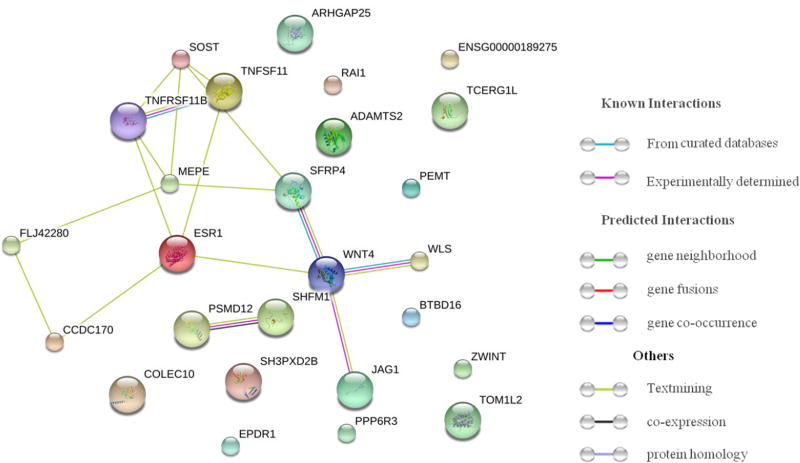
3.5 Protein-protein interaction analysis for LS BMD cFDR-significant genes
Based on connection evidence with STRING 10.0 summary score above 0.4, protein-protein interaction network was constructed to explore the biological function of the cFDR-significant genes (Figure 3). Strong interactions were detected between proteins produced by cFDR-significant genes, especially among WNT4, SFRP4, WLS, JAG1, MEPE, ESR1, TNFRSF11B, TNFSF11 and SOST. For example, protein produced by SFRP4 regulates Wnt signaling through direct interaction [32], JAG1 encodes Jagged1 protein, Jagged1-Notch1 signaling induces the expression of WNT4 in encocardial cells [33]. SOST inhibits Wnt signaling and negatively regulates bone formation [34], TNFRSF11B is the decoy receptor for TNFSF11 and neutralizes its function in osteoclastogenesis [35]. Interestingly, though some genes encompassed by cFDR-significant SNPs did not have any interaction with other cFDR-significant genes in this network, they were found in studies to be associated with bone tumors (TOM1L2 and ADAMTS2) [22,23] and risk factors to osteoporosis, such as insulin resistance (TCERG1L) [36] and obesity (RAI1) [25].
Figure 3.
Protein-protein interaction analysis for LS BMD cFDR-significant genes. Connections are based on evidence with a STRING 10.0 summary score above 0.4. Network nodes represent proteins produced by the corresponding genes, edges between nodes indicate protein-protein associations, edge color indicates the type of interaction and was specified on the right in the figure.
4 Discussion
Our study demonstrated a strong pleiotropy between LS BMD and CAD, 7 pleiotropic SNPs were identified and validated the hypotheses of the shared genetic basis between the two complex traits. More importantly, by leveraging power from the epidemiologically and clinically related trait CAD, we successfully identified 41 LS BMD-associated SNPs with cFDR < 0.05, of which, 20 SNPs have been identified in the original and other BMD GWAS analyses to be significantly associated with BMD [17,37]. These 20 SNPs were considered as successful replication of the previous BMD GWASs and demonstrated the reliability of cFDR method on detecting trait-associated genetic variants for complex phenotypes. Totally we identified 21 potential novel LS BMD associated SNPs without any further recruitment or new study participants, some of these SNPs were located in or near genes that reached detection level in our gene expression validation assay, and most of their corresponding genes were enriched in GO terms and KEGG pathways that have close relationship with bone metabolism. Our findings strongly highlighted the reliability, cost-effectiveness and high-efficiency of the current cFDR method on its enhanced power of detecting potential novel potential genetic variants in complex traits and diseases.
Of the 21 potential novel LS BMD SNPs identified by cFDR method, there were 8 SNPs located in or near 5 genes which were confirmed before to be GWAS-associated with BMD. These 5 previously confirmed genes were WNT4, WLS, CCDC170, ESR1 and TNFRSF11B. rs11209223 and rs1864569 were located in WLS, rs2473252 was located in WNT4. Transmembrane protein produced by WLS can regulate WNT proteins’ sorting and secretion in a feedback regulation [38]. Meanwhile, as an important member of WNT family, WNT4 was the key gene in the enrichment analysis of our study, it was included in nearly all the GO terms with adjP < 0.05, many of which have been proved to be very important to bone metabolism, such as “Ossification”, “regulation of canonical Wnt receptor signaling pathway” and “regulation of bone remodeling”. In osteoporosis mouse models, WNT4 decreases osteoclast formation as well as bone resorption via inhibiting nuclear factor-kappaB (NF-kappaB), and its expression on osteoblast in transgenic mice can greatly promote bone formation [39]. rs7746854 was located in CCDC170, rs1293935 and rs2504065 were located near CCDC170 and ESR1. CCDC170 was one of the genes that were validated in our gene expressional validation assay, its genetic variation was proved to be significantly associated with BMD after adjustment of age and gender [40]. ESR1 is a crucial gene involved in hormone-associated diseases, it encodes estrogen receptor and regulates the function of estrogen on its target tissues including bone. rs12375331 and rs1905776 were located near TNFRSF11B. TNFRSF11B encodes osteoprotegerin (OPG), which is a decoy receptor for receptor activator of nuclear factor kappa-B ligand (RANKL), after binding to RANKL, OPG can prevent osteoclast maturation and reduce bone resorption [41]. Considering the functional roles these 5 genes have played in bone metabolism, we believed that these 8 potential novel SNPs represent important genetic variants to LS BMD, further replication studies and biological function research may follow their attention on them.
In addition to the 8 potential novel SNPs mentioned above, there were other 13 potential novel SNPs located in or near genes that were not found to be GWAS-associated with BMD before, including 7 pleiotropic SNPs (rs11005240, rs12449964, rs12943500, rs6502629, rs10039254, rs10765090 and rs7208561). Some genes annotated by these SNPs were found in studies to be associated with bone and/or CAD. rs12449964 was located near PEMT and RAI1, rs7208561 was located in the intron region of RAI1. PEMT expression could be positively activated by estrogen [42], which was also a crucial regulator to bone metabolism. RAI1 was confirmed to be a CAD significant gene in a previous GWAS [24], in mice and humans, haploinsufficiency of RAI1 was revealed to be significantly associated with obesity - a risk factor for both osteoporosis and CAD [25]. TOM1L2 was encompassed by rs12943500 and rs6502629, it was reported in a previous study that TOM1L2 was associated with 17p amplicons in osteosarcoma – a fatal malignant tumor of bone [23]. rs10039254 was located in the intron region of ADAMTS2, which was associated with the invasion and metastasis of chondrosarsoma – a common bone tumor [22], increased ADAMTS2 expression was detected in coronary plaques causing myocardial infarction [43]. rs10765090 was located near TCERG1L, in African Americans, TCERG1L was confirmed to be significantly associated with insulin resistance [26], which negatively affects bone mass and is an important risk factor for CAD. To date, studies of these potential novel LS BMD associated SNPs (including the 7 pleiotropic SNPs) were very limited, there is still much to learn about the precise mechanism how they affect BMD and/or CAD, yet taken the various relationship between their corresponding genes and bone and/or CAD mentioned above, we inferred that these cFDR-significant SNPs might be potential novel SNPs associated with BMD and/or CAD, RAI1, TOM1L2, ADAMTS2 and TCERG1L might play key roles in the pleiotropy of LS BMD and CAD. More studies were warranted to follow up with them and explore their biological function in bone metabolism and/or pathophysiology of CAD.
In addition to LS BMD, we also performed the pleiotropy analysis on CAD GWAS data with GEFOS data for femoral neck BMD and forearm BMD respectively. However, no significant pleiotropy was found between them. The results may not be surprising, since different skeleton sites do not have all the same genetic determination and the degree of their pleiotropic determination with CAD may also be different [44].
There may be some limitations of our present study. First, cFDR method were not able to identify causal variants for the interested trait, the aim of the present study was to provide more potential novel BMD associated genetic variants, so that replication studies or fine mapping studies could follow up with them in the future. Second, the contribution of our findings to the proportion of BMD’s variability could not be estimated, since we only analyzed GWAS summary statistics, and the raw genotype data was unavailable.
5 Conclusion
cFDR method increased the effective sample size of existing GWAS data and greatly improved the detection of trait-associated genetic variants. By leveraging the pleiotropy with the associated phenotype CAD, we identified 21 potential novel SNPs associated with LS BMD, genes encompassed by these SNPs played important roles in bone metabolism. Our findings shed novel insight into the genetic variability of BMD as well as the shared genetic basis underlying osteoporosis and CAD.
Supplementary Material
Highlights.
Applied cFDR method to GWAS data to identify potential novel SNPs for complex trait
Identified pleiotropic SNPs for BMD and coronary artery disease
Identified 21 potential novel loci for lumbar spine BMD
Acknowledgments
Hong-Wen Deng was partially supported by grants from the National Institutes of Health [R01AR057049, R01AR059781, D43TW009107, P20 GM109036, R01MH107354, R01MH104680, R01GM109068], the Edward G. Schlieder Endowment fund to Tulane University. We acknowledge GEFOS-seq Consortium, AOGC Consortium, UK10K Consortium, CARDIoGRAMplusC4D Consortium for their GWAS summary statistics posted online. Chun-ping Zeng was partially supported by Medical Research Fund of Guangdong Province, Guangdong, China (A2017575). Cheng Peng was partially supported by Guangzhou Planed Project of Science and Technology, Guangzhou, China [201704020105] and Guangzhou First People’s Hospital during this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: None
Contributors:
Hong-Wen Deng conceived and initiated the development of this study, he is responsible for general development and design of the study and contributed to critical revisions and finalization of the manuscript, he is guarantor. Cheng Peng contributed to the acquisition and analysis of the data and drafted the manuscript. Kuan-Jui Su, Wei Zhu contributed to data analysis. Jonathan Greenbaum, Wei-Feng Deng, Chun-Ping Zeng contributed to critical revisions. Hui-Ling Lou, Feng Liu and Jie Shen contributed to the general study design and development. All authors have given approval to the final version of the manuscript. All authors agree to be accountable for the work and ensure that any questions relating to the accuracy and integrity of the paper are investigated and properly resolved.
References
- 1.Wright NC, Looker AC, Saag KG, Curtis JR, Delzell ES, Randall S, Dawson-Hughes B. The recent prevalence of osteoporosis and low bone mass in the United States based on bone mineral density at the femoral neck or lumbar spine. J Bone Miner Res. 2014;29(11):2520–2526. doi: 10.1002/jbmr.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curtis EM, Moon RJ, Harvey NC, Cooper C. The impact of fragility fracture and approaches to osteoporosis risk assessment worldwide. Bone. 2017 doi: 10.1016/j.bone.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Videman T, Levälahti E, Battié MC, Simonen R, Vanninen E, Kaprio J. Heritability of BMD of femoral neck and lumbar spine: a multivariate twin study of Finnish men. Journal of Bone and Mineral Research. 2007;22(9):1455–1462. doi: 10.1359/jbmr.070606. [DOI] [PubMed] [Google Scholar]
- 4.Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat Rev Genet. 2012;13(8):576–588. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 5.Writing Group M. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jimenez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER, 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB, American Heart Association Statistics, Stroke Statistics S Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133(4):e38–360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 6.McPherson R, Tybjaerg-Hansen A. Genetics of Coronary Artery Disease. Circ Res. 2016;118(4):564–578. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- 7.Lee HT, Shin J, Min SY, Lim YH, Kim KS, Kim SG, Kim JH, Lim HK. Relationship between bone mineral density and a 10-year risk for coronary artery disease in a healthy Korean population: the Korea National Health and Nutrition Examination Survey 2008–2010. Coron Artery Dis. 2015;26(1):66–71. doi: 10.1097/MCA.0000000000000165. [DOI] [PubMed] [Google Scholar]
- 8.Campos-Obando N, Kavousi M, Roeters van Lennep JE, Rivadeneira F, Hofman A, Uitterlinden AG, Franco OH, Zillikens MC. Bone health and coronary artery calcification: The Rotterdam Study. Atherosclerosis. 2015;241(1):278–283. doi: 10.1016/j.atherosclerosis.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Marini F, Brandi ML. Genetic determinants of osteoporosis: common bases to cardiovascular diseases? Int J Hypertens 2010. 2010 doi: 10.4061/2010/394579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reppe S, Wang Y, Thompson WK, McEvoy LK, Schork AJ, Zuber V, LeBlanc M, Bettella F, Mills IG, Desikan RS, Djurovic S, Gautvik KM, Dale AM, Andreassen OA, Consortium G. Genetic Sharing with Cardiovascular Disease Risk Factors and Diabetes Reveals Novel Bone Mineral Density Loci. PLoS One. 2015;10(12):e0144531. doi: 10.1371/journal.pone.0144531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anagnostis P, Karagiannis A, Kakafika AI, Tziomalos K, Athyros VG, Mikhailidis DP. Atherosclerosis and osteoporosis: age-dependent degenerative processes or related entities? Osteoporos Int. 2009;20(2):197–207. doi: 10.1007/s00198-008-0648-5. [DOI] [PubMed] [Google Scholar]
- 12.Eastell R, Newman C, Crossman DC. Cardiovascular disease and bone. Arch Biochem Biophys. 2010;503(1):78–83. doi: 10.1016/j.abb.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Bucay N, Sarosi I, Dunstan CR, Morony S, Tarpley J, Capparelli C, Scully S, Tan HL, Xu W, Lacey DL, Boyle WJ, Simonet WS. osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Genes Dev. 1998;12(9):1260–1268. doi: 10.1101/gad.12.9.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, Kelsoe JR, Kendler KS, O’Donovan MC, Rujescu D, Werge T, Sklar P, Psychiatric Genomics C. Bipolar D, Schizophrenia Working G. Roddey JC, Chen CH, McEvoy L, Desikan RS, Djurovic S, Dale AM. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9(4):e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng CP, Chen YC, Lin X, Greenbaum J, Chen YP, Peng C, Wang XF, Zhou R, Deng WM, Shen J, Deng HW. Increased identification of novel variants in type 2 diabetes, birth weight and their pleiotropic loci. J Diabetes. 2016 doi: 10.1111/1753-0407.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenbaum J, Wu K, Zhang L, Shen H, Zhang J, Deng HW. Increased detection of genetic loci associated with risk predictors of osteoporotic fracture using a pleiotropic cFDR method. Bone. 2017;99:62–68. doi: 10.1016/j.bone.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ, Dahia CL, Park-Min KH, Tobias JH, Kooperberg C, Kleinman A, Styrkarsdottir U, Liu CT, Uggla C, Evans DS, Nielson CM, Walter K, Pettersson-Kymmer U, McCarthy S, Eriksson J, Kwan T, Jhamai M, Trajanoska K, Memari Y, Min J, Huang J, Danecek P, Wilmot B, Li R, Chou WC, Mokry LE, Moayyeri A, Claussnitzer M, Cheng CH, Cheung W, Medina-Gomez C, Ge B, Chen SH, Choi K, Oei L, Fraser J, Kraaij R, Hibbs MA, Gregson CL, Paquette D, Hofman A, Wibom C, Tranah GJ, Marshall M, Gardiner BB, Cremin K, Auer P, Hsu L, Ring S, Tung JY, Thorleifsson G, Enneman AW, van Schoor NM, de Groot LC, van der Velde N, Melin B, Kemp JP, Christiansen C, Sayers A, Zhou Y, Calderari S, van Rooij J, Carlson C, Peters U, Berlivet S, Dostie J, Uitterlinden AG, Williams SR, Farber C, Grinberg D, LaCroix AZ, Haessler J, Chasman DI, Giulianini F, Rose LM, Ridker PM, Eisman JA, Nguyen TV, Center JR, Nogues X, Garcia-Giralt N, Launer LL, Gudnason V, Mellstrom D, Vandenput L, Amin N, van Duijn CM, Karlsson MK, Ljunggren O, Svensson O, Hallmans G, Rousseau F, Giroux S, Bussiere J, Arp PP, Koromani F, Prince RL, Lewis JR, Langdahl BL, Hermann AP, Jensen JE, Kaptoge S, Khaw KT, Reeve J, Formosa MM, Xuereb-Anastasi A, Akesson K, McGuigan FE, Garg G, Olmos JM, Zarrabeitia MT, Riancho JA, Ralston SH, Alonso N, Jiang X, Goltzman D, Pastinen T, Grundberg E, Gauguier D, Orwoll ES, Karasik D, Davey-Smith G, Consortium A, Smith AV, Siggeirsdottir K, Harris TB, Zillikens MC, van Meurs JB, Thorsteinsdottir U, Maurano MT, Timpson NJ, Soranzo N, Durbin R, Wilson SG, Ntzani EE, Brown MA, Stefansson K, Hinds DA, Spector T, Cupples LA, Ohlsson C, Greenwood CM, Consortium UK, Jackson RD, Rowe DW, Loomis CA, Evans DM, Ackert-Bicknell CL, Joyner AL, Duncan EL, Kiel DP, Rivadeneira F, Richards JB. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526(7571):112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Cardiogenics, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O’Donnell CJ, McPherson R, Erdmann J, Consortium CA, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, Rujescu D, Werge T, van de M, Morris AP, McCarthy MI, International Consortium for Blood Pressure G, Diabetes Genetics R, Meta-analysis C, Psychiatric Genomics Consortium Schizophrenia Working G. Roddey JC, McEvoy LK, Desikan RS, Dale AM. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92(2):197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reppe S, Refvem H, Gautvik VT, Olstad OK, Hovring PI, Reinholt FP, Holden M, Frigessi A, Jemtland R, Gautvik KM. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone. 2010;46(3):604–612. doi: 10.1016/j.bone.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 41 (Web Server issue) 2013:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akyol S, Comertoglu I, Firat R, Cakmak O, Yukselten Y, Erden G, Ugurcu V, Demircan K. Effect of insulin on the mRNA expression of procollagen N-proteinases in chondrosarcoma OUMS-27 cells. Oncol Lett. 2015;10(2):1091–1096. doi: 10.3892/ol.2015.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atiye J, Wolf M, Kaur S, Monni O, Bohling T, Kivioja A, Tas E, Serra M, Tarkkanen M, Knuutila S. Gene amplifications in osteosarcoma-CGH microarray analysis. Genes Chromosomes Cancer. 2005;42(2):158–163. doi: 10.1002/gcc.20120. [DOI] [PubMed] [Google Scholar]
- 24.Dichgans M, Malik R, Konig IR, Rosand J, Clarke R, Gretarsdottir S, Thorleifsson G, Mitchell BD, Assimes TL, Levi C, O’Donnell CJ, Fornage M, Thorsteinsdottir U, Psaty BM, Hengstenberg C, Seshadri S, Erdmann J, Bis JC, Peters A, Boncoraglio GB, Marz W, Meschia JF, Kathiresan S, Ikram MA, McPherson R, Stefansson K, Sudlow C, Reilly MP, Thompson JR, Sharma P, Hopewell JC, Chambers JC, Watkins H, Rothwell PM, Roberts R, Markus HS, Samani NJ, Farrall M, Schunkert H, Consortium M, Consortium CA, Consortium CD, International Stroke Genetics C Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke. 2014;45(1):24–36. doi: 10.1161/STROKEAHA.113.002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns B, Schmidt K, Williams SR, Kim S, Girirajan S, Elsea SH. Rai1 haploinsufficiency causes reduced Bdnf expression resulting in hyperphagia, obesity and altered fat distribution in mice and humans with no evidence of metabolic syndrome. Hum Mol Genet. 2010;19(20):4026–4042. doi: 10.1093/hmg/ddq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, Bentley A, Adeyemo A, Shriner D, Zhou J, Doumatey A, Huang H, Ramos E, Erdos M, Gerry N, Herbert A, Christman M, Rotimi C. Genome-wide association study identifies novel loci association with fasting insulin and insulin resistance in African Americans. Hum Mol Genet. 2012;21(20):4530–4536. doi: 10.1093/hmg/dds282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YJ, Zhang L, Papasian CJ, Deng HW. Genome-wide Association Studies for Osteoporosis: A 2013 Update. J Bone Metab. 2014;21(2):99–116. doi: 10.11005/jbm.2014.21.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen TV, Jones G, Sambrook PN, White CP, Kelly PJ, Eisman JA. Effects of estrogen exposure and reproductive factors on bone mineral density and osteoporotic fractures. J Clin Endocrinol Metab. 1995;80(9):2709–2714. doi: 10.1210/jcem.80.9.7673413. [DOI] [PubMed] [Google Scholar]
- 29.Vriend J, Reiter RJ. Melatonin, bone regulation and the ubiquitin-proteasome connection: A review. Life Sci. 2016;145:152–160. doi: 10.1016/j.lfs.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 30.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19(2):179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 31.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, Gonzalez-Macias J, Kahonen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren O, Lorenc RS, Marc J, Mellstrom D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NL, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gomez C, Palsson ST, Reppe S, Rotter JI, Sigurdsson G, van Meurs JB, Verlaan D, Williams FM, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw KT, Lehtimaki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HA, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AW, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JP, Kiel DP, Rivadeneira F. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, Wan R, Yu G, Shen J, Ni J, Yin G, Xing M, Chen C, Fan Y, Xiao W, Xu G, Wang X, Hu G. Imbalance of Wnt/Dkk negative feedback promotes persistent activation of pancreatic stellate cells in chronic pancreatitis. PLoS One. 2014;9(4):e95145. doi: 10.1371/journal.pone.0095145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Wu B, Chamberlain AA, Lui W, Koirala P, Susztak K, Klein D, Taylor V, Zhou B. Endocardial to myocardial notch-wnt-bmp axis regulates early heart valve development. PLoS One. 2013;8(4):e60244. doi: 10.1371/journal.pone.0060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005;280(29):26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 35.Liu W, Zhang X. Receptor activator of nuclear factor-kappaB ligand (RANKL)/RANK/osteoprotegerin system in bone and other tissues (review) Mol Med Rep. 2015;11(5):3212–3218. doi: 10.3892/mmr.2015.3152. [DOI] [PubMed] [Google Scholar]
- 36.Srikanthan P, Crandall CJ, Miller-Martinez D, Seeman TE, Greendale GA, Binkley N, Karlamangla AS. Insulin resistance and bone strength: findings from the study of midlife in the United States. J Bone Miner Res. 2014;29(4):796–803. doi: 10.1002/jbmr.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, Zhang B, Zhu J, Liu D, Guan F, He X. 4q22.1 contributes to bone mineral density and osteoporosis susceptibility in postmenopausal women of Chinese Han population. PLoS One. 2013;8(11):e80165. doi: 10.1371/journal.pone.0080165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Port F, Basler K. Wnt trafficking: new insights into Wnt maturation, secretion and spreading. Traffic. 2010;11(10):1265–1271. doi: 10.1111/j.1600-0854.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 39.Yu B, Chang J, Liu Y, Li J, Kevork K, Al-Hezaimi K, Graves DT, Park NH, Wang CY. Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-kappaB. Nat Med. 2014;20(9):1009–1017. doi: 10.1038/nm.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullin BH, Walsh JP, Zheng HF, Brown SJ, Surdulescu GL, Curtis C, Breen G, Dudbridge F, Richards JB, Spector TD, Wilson SG. Genome-wide association study using family-based cohorts identifies the WLS and CCDC170/ESR1 loci as associated with bone mineral density. BMC Genomics. 2016;17:136. doi: 10.1186/s12864-016-2481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khosla S. Minireview: the OPG/RANKL/RANK system. Endocrinology. 2001;142(12):5050–5055. doi: 10.1210/endo.142.12.8536. [DOI] [PubMed] [Google Scholar]
- 42.Resseguie M, Song J, Niculescu MD, da Costa KA, Randall TA, Zeisel SH. Phosphatidylethanolamine N-methyltransferase (PEMT) gene expression is induced by estrogen in human and mouse primary hepatocytes. FASEB J. 2007;21(10):2622–2632. doi: 10.1096/fj.07-8227com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CW, Hwang I, Park CS, Lee H, Park DW, Kang SJ, Lee SW, Kim YH, Park SW, Park SJ. Expression of ADAMTS-2, −3, −13, and −14 in culprit coronary lesions in patients with acute myocardial infarction or stable angina. J Thromb Thrombolysis. 2012;33(4):362–370. doi: 10.1007/s11239-011-0673-7. [DOI] [PubMed] [Google Scholar]
- 44.Paternoster L, Lorentzon M, Lehtimaki T, Eriksson J, Kahonen M, Raitakari O, Laaksonen M, Sievanen H, Viikari J, Lyytikainen LP, Mellstrom D, Karlsson M, Ljunggren O, Grundberg E, Kemp JP, Sayers A, Nethander M, Evans DM, Vandenput L, Tobias JH, Ohlsson C. Genetic determinants of trabecular and cortical volumetric bone mineral densities and bone microstructure. PLoS Genet. 2013;9(2):e1003247. doi: 10.1371/journal.pgen.1003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.