Abstract
BACKGROUND
In normal cells, RAD51-mediated homologous recombination (HR) is a precise DNA repair mechanism which plays a key role in the maintenance of genomic integrity and stability. However, elevated (dysregulated) RAD51 is implicated in genomic instability and is a potential target for treatment of certain cancers, including Barrett’s adenocarcinoma (BAC). In this study, we investigated genomic impact and translational significance of moderate vs. strong suppression of RAD51 in BAC cells.
METHODS
BAC cells (FLO-1 and OE33) were transduced with non-targeting control (CS) or RAD51-specific shRNAs, mediating a moderate (40–50%) suppression or strong (80-near 100%) suppression of the gene. DNA breaks, spontaneous or following exposure to DNA damaging agent, were examined by comet assay and 53BP1 staining. Gene expression was monitored by microarrays (Affymetrix). Homologous recombination (HR) and single strand annealing (SSA) activities were measured using plasmid based assays.
RESULTS
We show that although moderate suppression consistenly inhibits/reduces HR activity, the strong suppression is associated with increase in HR activity (by ~15 – ≥ 50% in various experiments), suggesting activation of RAD51-independent pathway. Contrary to moderate suppression, a strong suppression of RAD51 is associated with a significant induced DNA breaks as well as altered expression of genes involved in detection/processing of DNA breaks and apoptosis. Stronger RAD51 suppression was also associated with mutagenic single strand annealing mediated HR. Suppression of RAD51C inhibited RAD51-independent (SSA-mediated) HR in BAC cells.
CONCLUSION
Elevated (dysregulated) RAD51 in BAC is implicated in both the repair of DNA breaks as well as ongoing genomic rearrangements. Moderate suppression of this gene reduces HR activity, whereas strong or near complete suppression of this gene activates RAD51C-dependent HR involving a mechanism known as single strand annealing (SSA). SSA-mediated HR, which is a mutagenic HR pathway, further disrupts genomic integrity by increasing DNA breaks in BAC cells.
Keywords: RAD51, RAD51C, Homologous Recombination, Single Strand Annealing, DNA Breaks, Genomic Integrity, Esophageal adenocarcinoma
INTRODUCTION
Genomic instability or vulnerability to acquire new genomic changes is a critical problem which not only enables cancer cells to acquire new features suited for disease progression[1,2] but also makes treatment and diagnosis much more difficult[3]. Genomic changes which are rarely seen in cells derived from normal healthy individuals are widely observed in tumor cells[4–6]. Importantly, the changes are not only present in pre-cancerous cells or in the cells derived from patients at diagnosis[7–11], they continue to evolve over time[5,12], leading to a clonal heterogeneity which is associated with cancer progression and treatment failure. Since genomic instability is associated with ongoing acquisition of genomic changes, it may serve as an important prognostic factor. Consistent with this, the investigation of mutational spectrum of myeloma patient samples indicates that increased number of mutations correlates with poor survival[12]. Mutational analyses of premalignant polyp and corresponding cancer specimens also suggest that genomic instability is associated with oncogenic transformation[13]. Mechanisms of genomic instability are not fully understood and their identification is essential to design new and better treatment strategies.
A variety of harmful agents, present either in environment (chemicals, viruses, radiation, heat) or inside our body (oxidative metabolites, acid, replication/recombination errors), cause damage to DNA on a regular basis[14]. The daily damage to DNA caused by oxidizing agents alone has been estimated to be quite substantial[15]. However, in a normal cellular environment, multiple repair systems work in a concerted and well-regulated manner to maintain genomic integrity of a cell. Double-strand breaks (DSBs), which can be induced by oxidizing agents, ionizing radiation, and stalled replication forks are probably the worst type of DNA damage in a cell; if left unrepaired in G2, they may recombine in subsequent G1 phase of cell cycle to cause unnecessary genomic rearrangements/changes. Moreover, the broken pieces of chromosomes which do not have centromeres, will not be able to segregate properly. It has been proposed that a single misrepaired or unrepaired DSB can potentially lead to oncogenic process or loss of cell viability. Therefore, damage to DNA, especially the DSBs, must be repaired before progression through cell cycle.
Double-strand breaks are repaired by genetic recombination[16], which can either be homologous (HR) in which sequences to be recombined have to be strictly homologous or non homologous (known as non homologous end joining; NHEJ) which does not dependent on a strict homology[17,18]. Since HR uses the intact homologous chromosome or sister chromatid as template to repair the damage acquired by their counterparts, it is considered to be an accurate repair system[19]. This is probably why rate of HR per kbp of target DNA is much higher as compared to NHEJ. Besides DSBs, HR is also implicated in repair of certain other types of DNA damage including interstrand cross links[20]. A regulated and functional HR repair system is an absolute requirement for preservation of genomic integrity of a cell[21]. The process of HR includes detection of DNA damage, activation of signaling pathways leading to cell cycle arrest and recruitment of repair proteins, formation of repair foci (containing recombination/repair proteins) at the site of DNA damage, initial processing of damaged DNA strand, physical association of damaged DNA strand with homologous template DNA, repair of damaged DNA strand, separation of repaired and template DNA strands, and downregulation/inactivation of repair proteins[22]. The damage to DNA is detected by specific proteins such as RAD23B (HR23B)[23], RAD51B[24], and members of MRN complex (MRE11, RAD50, NBS1)[22]. Following detection of DNA damage, MRN complex is also implicated in the activation of ATM, ATR, and/or DNA-dependent protein kinases, which trigger a phosphorylation signaling cascade leading to activation of p53, H2AX, and other proteins implicated in cell cycle and DNA repair[22]. Activated p53 induces a cell cycle arrest or apoptosis depending on the extent of DNA damage, whereas phosphorylated H2AX initiates the recruitment of repair proteins leading to formation of DNA repair foci (containing MRE11/RAD50/NBS1, CHK2, BRCA1, RPA, and RAD 51) at site of DNA damage. Although H2AX seems to initiate the recruitment, several RAD51 paraolgs including XRCC2[25], XRCC3[26], and RAD51D[27] have been shown to be required for the formation of damage-induced foci.
Recombinase (hsRAD51), an important HR protein, plays a vital role in the repair of DNA damage and maintenance of genome stability. The protein can: (1) bind to both the double- or single-stranded DNA fragments; (2) mediate base pairing between homologous sequences; (3) initiate strand exchange between template and its damaged counterpart[28]. At least five paralogs of hsRad51 have been identified, including XRCC3[29], which seems to play an important role in keeping the repair process free of errors[30]. Although proteins like RAD51 are vital for the maintenance to genomic integrity and healthy survival of a living cell, an elevated or dysregulated expression/function of such proteins can also be harmful. Elevated RAD51 can potentially increase/dysregulate HR activity, leading to harmful genomic rearrangements and loss of genomic integrity. Consisitent with this, hsRAD51 has been shown to be mutated or overexpressed in a number of cancers[2,31–34] including BAC[35].
In our previous study we have shown that shRNAs mediating moderate (~50%) suppression of RAD51 in BAC cells, significantly reduce genomic instability[35]. Thus RAD51, one of the important proteins required for maintenance of genomic integrity of normal cells, is also a therapeutic target in cancer. In this manuscript, we usedspecific shRNAs to induce moderate or strong suppression of RAD51 in BAC cells and monitored the impact on DNA integrity, HR activity and cell survival. We show that whereas moderate suppression of RAD51 reduces HR activity, strong suppression increases this activity to varying levels in different experiments. This RAD51-independent HR is mediated through single strand annealing which depends on RAD51C and is associated with increased DNA breaks.
MATERIALS AND METHODS
Western-blot analyses
Cells, subjected to appropriate transgenic manipulations and/or treatments, were lysed and specific proteins measured by Western blotting as described previously[35]. Briefly, the extracts suspended in Laemmli’s sample buffer (0.1 M Tris-HCl buffer pH 6.8, 1% SDS, 0.05% β-mercaptoethanol, 10% glycerol, and 0.001% bromophenol blue), were boiled, fractionated by electrophoresis on 4–20% glycerol gradient SDS-polyacrylamide gel, and electroblotted onto Trans-Blot nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA) in a Trisglycine buffer system. The blots were treated with the primary antibody for 2 hrous at room temperature, washed, and incubated with horseradish peroxidase-conjugated secondary (anti-rabbit or anti-mouse) antibody for 2 hrous. The antigen-antibody complexes were detected using an enhanced chemiluminescence detection kit, as per manufacturer’s instructions (Amersham Life Sciences Inc., Arlington Heights, IL).
Lentiviruses and transductions
Lentivirus-based shRNAs, non-targeting control (C) or those targeting RAD51 and mediating moderate or strong suppression of this gene (RS and RS2), were obtained from Sigma Chemical Co., Saint Louis, MO. Lentiviral transductions were performed as described previously[35]. Briefly, the cells were plated into 24-well plates and after 24 hrs in culture, hexadimethrine bromide added to a final concentration of 8 μg/ml. Lentiviral particles were then added to each well and the cells incubated at 37°C in a humidified incubator with 5% CO2 for 16 hours. The medium was then replaced with fresh medium and cells continued in culture for another 24 hrs. At this point the cells were transferred to 25 cm2 flasks, cultured for another 48 hours, selected in puromycin (1 μg/mL) for seven days, and RAD51 suppression confirmed by western blotting.
Gene Expression Analysis and Biostatistics
Impact of moderate and strong suppression of RAD51 on genomewide expression was demonstrated as described previously [35]. Transduced cells were harvested at day seven after selection, total RNA isolated, and expression profile evaluated using Gene 1.0 ST Arrays (Affymetrix, Santa Clara, CA). Gene arrays were scanned on an Affymetrix scanner to obtain expression values, which were normalized by dChip Analyzer, the software which uses “Invariant Set” normalization and “model-based” approaches to make arrays comparable and calculate normalized expression values[36, 37].
Estimation of DNA breaks
Spontaneous and UV-induced DNA breaks in transduced cells were estimated by comet assay, a gel-based method for detection of DNA damage at the level of individual cell as well as from levels of 53BP1, a marker for DNA breaks. For comet assay, the cells encapsulated in a low melting point agarose gel were lysed at neutral pH and suspended nucleoids were then subjected to electrophoreseis. The gel was stained and comet tail moments (representing tail length as well as the intensity of broken DNA pieces in the tail) were calculated using “OpenComet” software. 53BP1 was detected by immunofluorescence.
Homologous recombination assay
Homologous Recombination (HR) activity was monitored using either a luminescence based HR assay[35] and/or a commercially available fluorescence-based assay, as reported by us previously [38]. In luminescence based assay, HR in a substrate plasmid generates a functional firefly luciferase gene, whereas another (gaussia) luciferase serves as an internal control; HR is then calculated from the ratio of two activities. In fluorescence-based HR assay substrate (pDRGFP; a gift from Maria Jasin, Addgene, plasmid # 26475)[30], the introduction of I-Sce I induced DNA break initiates HR, generating a functional GFP gene. HR in this assay is calculated from fluorescence intensity divided by total number of cells in of each microscopic field.
Single strand annealing assay
Homologous recombination mediated by a mechanism involving single strand annealing (SSA) was assessed using a plasmid (hprtSAGFP) substrate, a gift from Maria Jasin (Addgene plasmid # 41594)[39]. Briefly, the plasmid has two fragments of a GFP gene separated by a drug resistance gene. One of the fragments has a SCEI recognition sequence incorporated in it. Induction of DNA break by SCEI leads to SSA between homologous sequences in two fragments generating a functional GFP gene. Homologous recombination (RAD51 dependent) within this substrate does not produce functional GFP.
RESULTS
Impact of moderate vs. strong suppression of RAD51 on HR activity in BAC cells
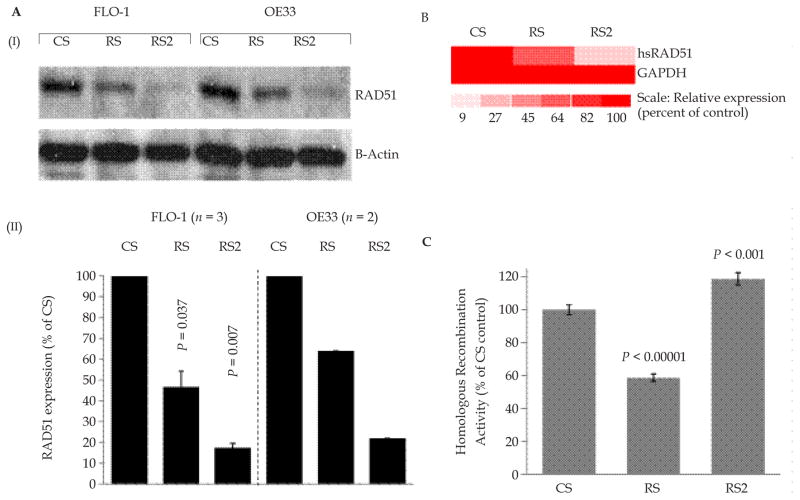
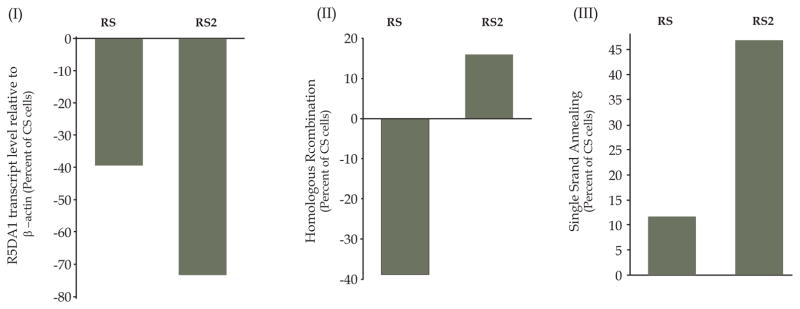
BAC cells were transduced with lentiviruses producing control shRNA (CS) or two different RAD51-targeting shRNAs “RS” and “RS2”. Puromycin selected cells were examined for RAD51 protein by Western blotting. Lentivirus “RS” produced a moderate (~40–60%) suppression of RAD51 in both BAC cell lines, whereas transduction of the same cell lines with “RS2” consistently caused a strong (80-near 100%) suppression of the gene, in multiple independent experiments (Figure 1A). Consistent with these observations, transcript levels of RAD51 were also suppressed moderately (by 36%) in RS cells and strongly (by 81%) in RS2 cells (Figure 1B). Consistent with our previous observations[35], moderate suppression of RAD51 (by RS shRNA) in this study was also associated with a moderate (41%) reduction (P < 0.0000002) in HR activity, whereas a stronger suppression (by RS2 shRNA) did not inhibit but caused a 19% increase in HR activity (P < 0.001; Figure 1C) in this experiment. In various independent experiments the stronger suppression of RAD51 (by RS2) was associated with ~ 15% – ≥50% increase in HR activity (Figures 1, 5,6 and unpublished data), indicating possible activation of RAD51-independent HR activity. These data suggest that loss or low levels of RAD51 may lead to activation of RAD51-independent HR in BAC cells.
Figure 1. Impact of strong vs. moderate suppression of RAD51 on HR activity in BAC cells.
(A) shRNA “RS2” consistently mediates a stronger suppression of RAD51 than “RS” in BAC cells. FLO-1 and OE33 cells were transduced with lentiviruses, expressing control shRNA (CS) or two different RAD51-targeting shRNAs “RS” or “RS2”. Following recovery and selection in puromycin, the cells were evaluated for levels of RAD51 protein by Western blotting. Panels: (I) Representative image of Western blot; (II) Bar graph showing relative expression of RAD51 following normalization with β-actin; error bars indicate SEMs of three independent experiments, each involving a new transduction of FLO-1 cells. P values indicate significance of change relative to control (CS) cells. Data for OE33 is mean of 2 experiments. (B) Transcript levels of RAD51 in CS, RS and RS2 cells evaluated using Human Gene 1.0 ST Arrays. The color scale at the bottom of the figure represents % change in expression of RAD51 relative to control (CS) cells. (C) HR activity was assessed at day six after transduction, using a plasmid based assay. Relative HR activity in RAD51-suppressed RS and RS2 cells is shown as percent of activity in control (CS) cells.
Impact of moderate vs. strong suppression of RAD51 on spontaneous and induced DNA breaks in BAC cells
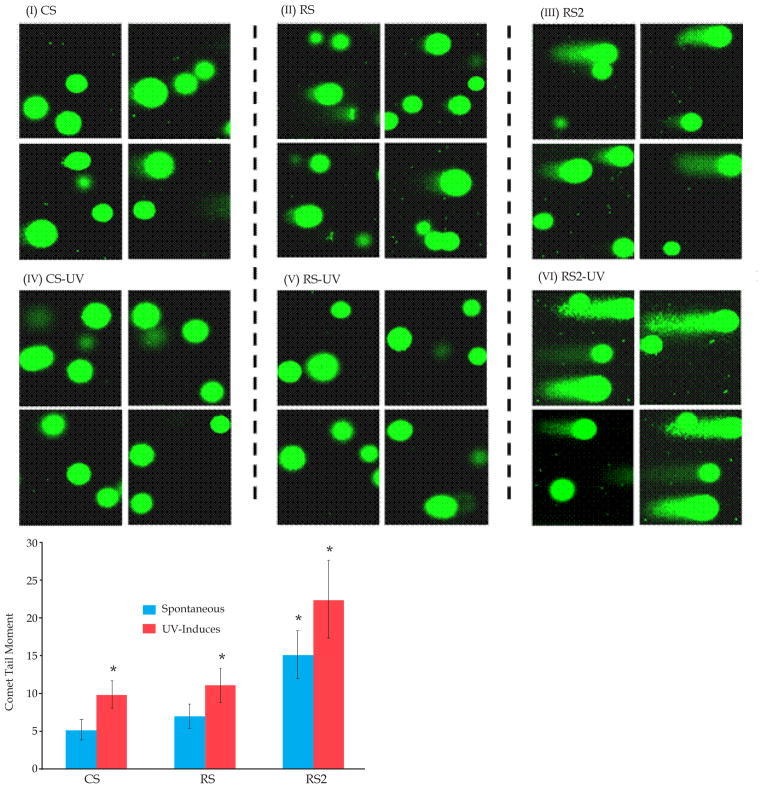
BAC cells were transduced with lentiviruses producing control (CS) and RAD51-specific shRNAs (RS, mediating moderate suppression; or RS2, mediating strong suppression). DNA breaks in these cells were detected, either spontaneously or following exposure to ultraviolet (UV) light, using comet assay. For UV exposure, cells were exposed to 25j/m2 UV and incubated for 24 hrs prior to evaluation of DNA breaks. Fraction of cells with comets were counted in different microscopic fields and tail moment, a quantitative measure of DNA breaks which incorporates length as well intensity of comet tail (representing broken DNA), was calculated as described in Methods. Figure 2A shows representative images of comets (indicating) DNA breaks under spontaneous condition (panels I–III) as well as following exposure to UV light (panels IV–VI). Under spontaneous condition, comets were observed in 30 ± 9% control (CS), 27 ± 9% RS, and 67 ± 11% RS2 cells, indicating ~ 2-fold (P < 0.03) increase in percentage of cells with DNA breaks in RS2 whereas no increase in RS cells (representative images shown in Figure 1A, I–III). As shown in Figure 2B, tail moments were not significantly different in RS but increased in RS2 relative to CS cells, indicating more DNA breaks in RS2 cells. Similarly, following UV exposure, an increase in the fraction of cells with total as well as very large comets was seen in RS2 and not in RS, relative to control (CS) cells (representative images shown in Figure 1A, IV–VI and bar graph of tail moments shown in Figure 2B).
Figure 2. Impact of RAD51 amount on DNA breaks in BAC cells.
FLO-1 cells were treated either with control shRNA (CS) or RAD51-targeting (RS, RS2, shRNAs) described in Figure 2 and following selection, cells evaluated for impact on DNA breaks by Comet assay. (A) Images of Comet assay showing impact of these shRNAs on DNA breaks under spontaneous condition (I-III) or following exposure to UV (IV-VI). (B) Bar graph shows comet tail moments in CS, RS and RS2 cells under spontaneous condition as well as following UV treatment; * indicates a significant change (P < 0.05) relative to control (CS) cells.
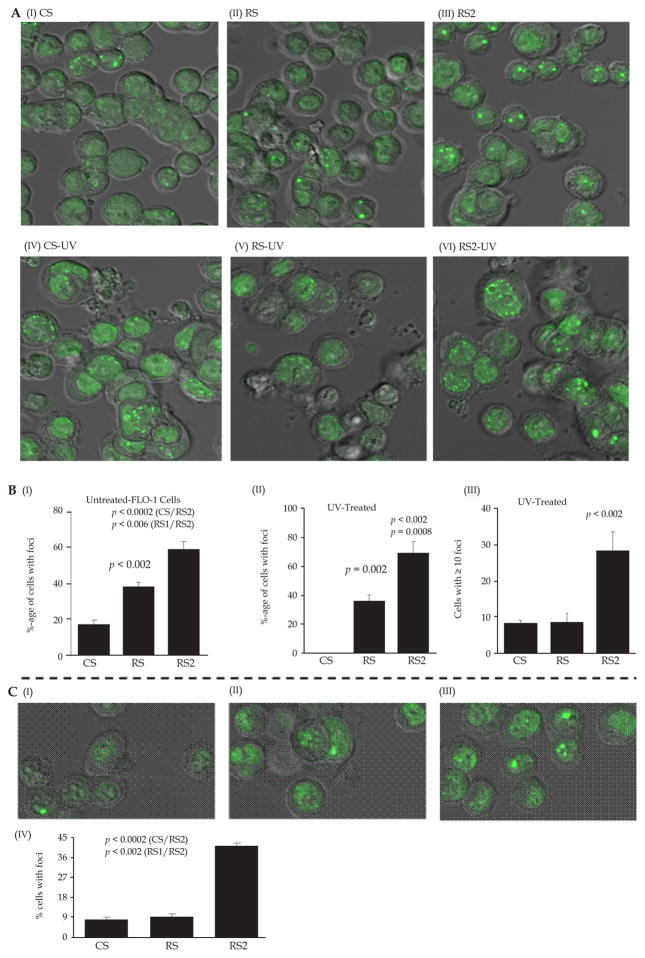
Similar observations were made by investigating 53BP1 in these cells. 53BP1 is a p53 binding protein, which following DNA damage, associates with DNA breaks[40] and mediates G2M and S-phase checkpoint arrests[41]. In FLO-1 cell line, transduction with CS, RS, and RS2 lentivirus shRNAs was associated with spontaneous detection of 53BP1 foci in 17 ± 3%, 38 ± 3%, and 59 ± 5% cells, respectively (images shown in Figure 3A, panels I–III and bar graph in 3B, panel I). Although 53BP1 was also higher in RS relative to control cells, the fraction of cells staining positive for 53BP1 was 21% (P < 0.006) higher in RS2 relative to RS cells (Figure 3B, panel I). Exposure to UV increased the fraction of cells with multiple foci, leading to detection of 8 ± 1% CS, 9 ± 3% RS and 28 ± 5% of RS2 cells with ≥ 10 foci per cell, thus indicating a significant (20%; P = 0.01) increase in 53BP1 foci in cells with strong suppression of RAD51 (Figure 3A, panels IV–VI and 3B, panels II–III). RS2 cells, in another cell line OE33, also had significantly higher spontaneous levels of 53BP1 foci (Figure 3C, panels I–IV).
Figure 3. Impact of RAD51 amount on 53BP1 in BAC cells.
FLO-1 cells were treated either with control shRNA (CS) or RAD51-targeting (RS, RS2, shRNAs) described in Figure 2 and following selection, cells evaluated for impact on DNA breaks by by investigating levels of 53BP1. (A) Images of immunofluorescence showing impact of these treatments under spontaneous condition (I–III) and following exposure to UV (IV–VI) are shown. (B) Bar graphs showing percentage of cells with any number of foci (I–II) or those with ≥ 10 foci (III) are shown. (C) Impact of RAD51 amount on 53BP1 in OE33 cells treated as described in Panel A. Images of immunofluorescence (I–III) Bar graphs showing percentage of cells with any number of foci (IV) are shown.
These data indicate that strong suppression of RAD51 (in RS2 cells) leads to significant increase in spontaneous as well as UV-induced DNA breaks, whereas moderate suppression does not increase the damage.
Impact of moderate vs. strong RAD51-suppression on expression profile in FLO-1 cells
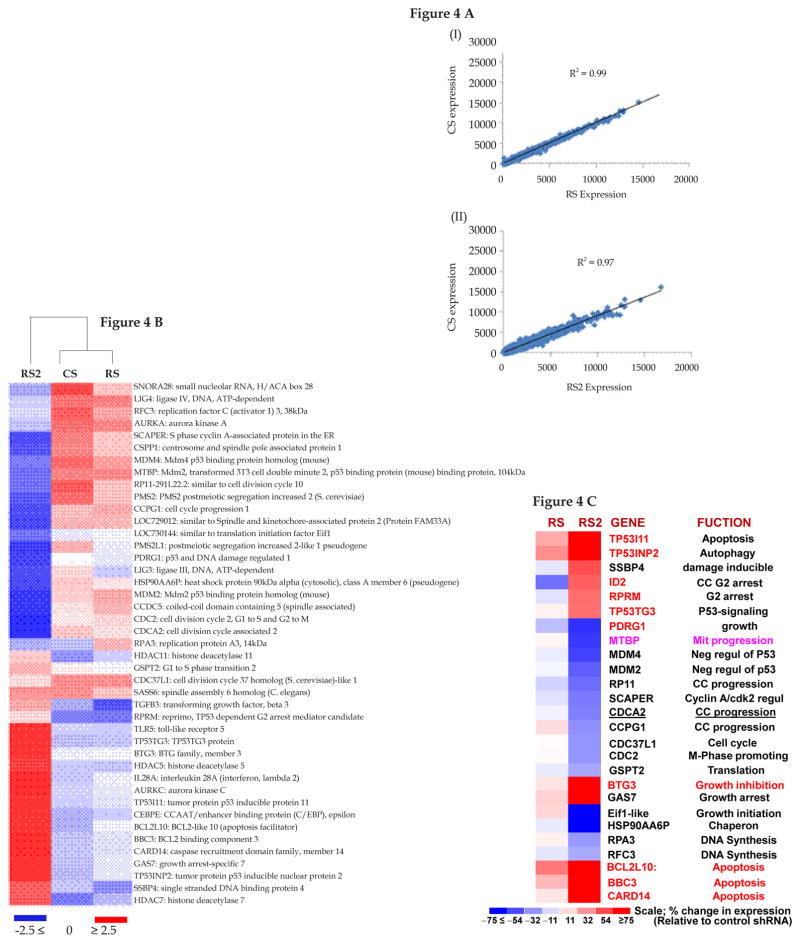
To evaluate and compare the impact of moderate (RS) vs. strong suppression of RAD51 (RS2) on expression profile, we harvested transduced cells at day seven after selection and conducted genomewide expression profile, using Agilent whole Human Genome arrays (4 × 44K format). Genomewide expression profiles of both RS and RS2 correlated strongly (r2 = ≥ 0.97) with control (CS) cells (Figure 4A), indicating that these shRNAs were not associated with widespread/nonspecific expression changes. Although overall expression profile of RS2 was similar to RS and CS, a cluster analysis based on genes implicated in DNA break/damage processing and related processes (cell cycle, apoptosis), clearly separated RS2 from RS and CS (Figure 4B). Figure 4C shows a subset of DNA damage repair, cell cycle and/or apoptosis genes differentially expressed in RS2 and RS, relative to CS cells. For example, DNA damage inducible SSBP4, growth and cell cycle arrest genes (ID2, RPRM, BTG3, GAS7), and p53/apoptosis-related genes (TP53I11, TP53INP2, TP53TG3, BCL2L10, BBC3, CARD14) are upregulated whereas several cell cycle and growth promoting genes including negative regulators of p53 (MDM2, MDM4) are downregulation in RS2 cells. These gene expression data are consistent with significantly more spontaneous DNA breaks and 53BP1 foci in RS2 cells (shown in Figures 2–3).
Figure 4. Impact of strong vs. moderate suppression of RAD51 on expression profile in BAC cells.
(A) FLO-1 cells were transduced with lentiviruses, expressing control shRNA (CS) or those inhibiting RAD51 expression, either moderately (RS) or strongly (RS2). Following recovery and selection in puromycin, the cells were evaluated for impact on expression profile, using Human Gene 1.0 ST Arrays. (A) Regression plots show that global gene expression patterns of RS (panel I) and RS2 (panel II) were similar to CS. (B) Cluster analysis using a subset of DNA damage and repair genes shows that CS and RS cluster together, whereas RS2 shows variation. (C) Expression of selected growth and apoptosis related genes in RS and RS2 cells. The color scale at the bottom of the figure represents fold change in expression, relative to control (CS) cells.
RAD51-independent HR involves single strand annealing and depends on RAD51C
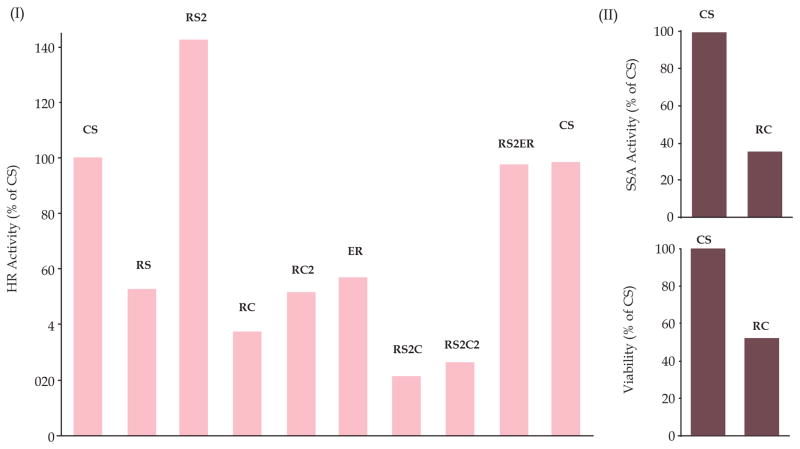
In absence or low RAD51, a less precise and mutagenic homologous recombination pathway known as single strand annealing (SSA)[39] can be activated. To investigate if stronger suppression of RAD51 (by RS2 shRNAs) in BAC cells is associated with increased SSA, we assessed both the HR and SSA activities in control and knockdown cells using plasmid based assays described in Methods. Moderate RAD51 suppression (by RS) in FLO-1 cells was associated with ~ 40% reduction in HR and slight increase (by ~ 11%) in SSA activity (Figure 5, panel II), whereas strong suppression led to ~ 15% increase in HR and ~ 48% increase in SSA activity (Figure 5, panel III). These data suggest that BAC cells depend on homologous recombination and, therefore, suppression of RAD51 leads to activation of a less precise SSA-mediated HR. We also found that suppression of RAD51C in RS2 cells inhibits RAD51-independent HR by >70% of control cells (Figure 6, panel I). Suppression of RAD51C was also associated with inhibition of spontaneous SSA (Figure 6, panel II) as well as reduced cell viability (Figure 6, panel III) in BAC cells. These data show that RAD51C contributes to both HR and SSA in BAC cells and in absence of RAD51, RAD51C shifts the balance of HR towards SSA pathway.
Figure 5. Impact of strong vs. moderate suppression of RAD51 on HR and SSA activities in BAC cells.
FLO-1 cells were transduced with lentiviruses, expressing shRNAs (CS, control; RS, suppressing RAD51 moderately or RS2, suppressing RAD51 strongly. Following puromycin selection, cells were evaluated for transcript levels of RAD51 using real time PCR (I) or homologous recombination (II) and single strand annealing (III) activities, using plasmid based assays described in Methods.
Figure 6. Role of RAD51C in RAD51-independent HR (SSA) in BAC cells.
FLO-1 cells were transduced with lentiviruses which include: CS, control; R1, suppressing RAD51 moderately; R2, suppressing RAD51 strongly; RC1 and RC2, two different shRNAs targeting RAD51C; ER, targeting ERCC1; R2C1, cells co-transduced with R2 and RC1; R2C2, cells co-transduced with R2 and RC2; and R2ER, cells co-transduced with R2 and ER. (I) Cells were selected in puromycin and evaluated for HR activity using a plasmid based assay; (II) CS and RC cells were evaluated for SSA activity immediately following selection; (III) CS and RC cells were evaluated for viability immediately following selection.
DISCUSSION
Under existing therapeutic strategies most of the malignancies remain incurable. The explanation for the lack of our ability to treat cancer is the ability of most cancer cells to genomically evolve, leading to emergence of new clones[12], progression to advanced and more aggressive phenotype, and development of resistance to therapy[2]. The problem is further intensified used in cancer treatment are damaging to DNA and thus mutagenic. It is therefore extremely important to develop new drugs that may reduce genomic instability in cancer cells, whether intrinsic or that caused by certain treatments. We have previously shown that homologous recombination, the most accurate means of repairing double strand breaks (DSBs) in the genome, is elevated/dysregulated in BAC cells and contributes to genomic instability; and suppression of RAD51 significantly reduces acquisition of new genomic changes in BAC cells. These and the data from other laboratories indicate that elevated RAD51 is an important target for prevention of genomic instability and treatment of cancer.
In this manuscript, we investigated the impact of moderate vs. strong suppression of RAD51 on spontaneous and induced DNA breaks and other mechanisms underlying genomic instability, using BAC as a model system.
We utilized two different shRNAs (RS and RS2) targeting RAD51; evaluation of transcript as well as protein levels confirmed that RS shRNAs consistently mediate a moderate (~ 40 to 50%) suppression whereas RS2 cause a stronger (~ 80% to near complete) inhibition of RAD51 in different cancer cell lines including BAC as well as multiple myeloma (not shown). Consistent with our previous observations [35], moderate RAD51-suppression (by RS shRNA) consistently reduced HR activity in this study as well. Contrary to our expectations, the stronger RAD51 suppression (by RS2 shRNA) did not inhibit but led to ~ 15% to ≥ 50% increase in HR activity in various experiments, indicating activation of RAD51-independent HR pathway. Overall expression profile of both shRNAs was very similar to control shRNA (r ≥ 0.97), indicating that these shRNAs were not associated with wide-spread nonspecific changes in gene expression. Both the evaluation of physical DNA breaks by comet assay[42] and 53BP1 (a DNA break-associated protein)[40] showed that amount of spontaneous DNA breaks was not significantly different in control and RS cells (with moderate RAD51-suppression), whereas significantly increased in RS2 cells (with strong RAD51-suppression). When DNA breaks were induced with UV and cells evaluated after 24 hrs, the number of DNA breaks were not significantly changed in RS whereas significantly increased in RS2, relative to control cells. These data demonstrate that in the absence or low levels of RAD51, an alternate pathway of HR is activated as a compensatory mechanism, which is associated with more DNA breaks or inefficient DNA break repair.
It has been shown that in absence of RAD51, an error prone pathway is activated in which HR between two homologous sequences is initiated by a double strand break (DSB). The break is followed by resection of DNA ends. Homologous sequences within single strand overhangs anneal with each other to assist recombination process. This pathway, known as single strand annealing (SSA), involves repair of double-strand breaks between two repeat sequences in a same chromosome causing intra-chromosomal deletions and is considered to be error prone[43] and associated with genomic rearrangements[44]. As our HR assay plasmid “FG1” contains two consecutive repeats interrupted by a spacer, the assay detects both the classical HR and SSA. To investigate if RAD51-suppression in FLO1 cells is associated with activation of SSA, we assessed SSA activity in these cells following moderate or strong suppression of RAD51 using the plasmid substrate specific for SSA. Moderate RAD51 suppression increased SSA by 11%, whereas strong suppression led to 48% increase in this pathway. These observations clearly demonstrate that loss of RAD51 in BAC cells activates an alternate HR pathway which requires DSBs for its initiation and is error prone and thus further disrupts genomic integrity and stability. Consistent with these observations, FLO-1 cells with strong RAD51-suppression had increased number of spontaneous as well as UV-induced DNA breaks.
Pathway intermediates of SSA (including the nuclease activity required for generation of DSBs) have been identified and well-studied in yeast[45]. However, the SSA pathway in human cells, especially in cancer background, is not so well-defined yet. We have found that in human BAC cells, suppression of RAD51C, one of the key effectors of the RAD51-independent HR pathway or SSA in yeast[45], inhibits SSA-mediated HR activities. Moreover, suppression of RAD51C in RS2 cells, in which RAD51 is strongly suppressed, also inhibits the elevated HR activity as evident by our HR assay using the substrate plasmid FG1. Our data suggest that, following a strong suppression of RAD51, balance of recombinational repair is shifted from RAD51-dependent classical HR towards RAD51C-mediated more erroneous SSA pathway, thus leading to increase in DNA damage and instability.
In summary, our data show that BAC cells depend on homologous recombination, probably because of ongoing damage to their DNA. In the absence of RAD51, a less precise and mutagenic HR pathway requiring RAD51C is activated which further disrupts genomic integrity and stability. In fact our unpublished data also show that BAC cells have increased level of spontaneous DNA damage including more abasic sites. Our previous studies also show that elevated HR in BAC cells contributes to genomic instability, telomere maintenance as well as tumor growth[38]. This study highlights an important point that targteing RAD51 alone may not inhibit HR and associated genomic instability but a combined inhibition of RAD51 and RAD51C may make BAC cells static. Role RAD51C in SSA in BAC cells is currently being evaluated in our laboratory.
Acknowledgments
FUNDING
This work was supported in part by grants from National Cancer Institute R01CA125711 to MAS and National Institute of Health Grant P01 CA155258 to NCM.
Footnotes
Conflict-of-interest statement: The author(s) declare(s) that there is no conflict of interest regarding the publication of this paper.
Contributor Information
Jagannath Pal, Department of Adult Oncology, Harvard (Dana Farber) Cancer Institute, Boston, MA, the United States. VA Health Care System, West Roxbury, MA, the United States. Multi-disciplinary Research Units (MRUs), Pt J.N.M. Medical College, Raipur, CG, India.
Purushothama Nanjappa, Department of Adult Oncology, Harvard (Dana Farber) Cancer Institute, Boston, MA, the United States. VA Health Care System, West Roxbury, MA, the United States.
Subodh Kumar, Department of Adult Oncology, Harvard (Dana Farber) Cancer Institute, Boston, MA, the United States. VA Health Care System, West Roxbury, MA, the United States.
Jialan Shi, Department of Adult Oncology, Harvard (Dana Farber) Cancer Institute, Boston, MA, the United States. VA Health Care System, West Roxbury, MA, the United States. Department of Medicine, Harvard Medical School, Boston, MA, the United States.
Leutz Buon, Department of Adult Oncology, Harvard (Dana Farber) Cancer Institute, Boston, MA, the United States.
Nikhil C. Munshi, Department of Adult Oncology, Harvard (Dana Farber) Cancer Institute, Boston, MA, the United States. VA Health Care System, West Roxbury, MA, the United States. Department of Medicine, Harvard Medical School, Boston, MA, the United States
Masood A. Shammas, Department of Adult Oncology, Harvard (Dana Farber) Cancer Institute, Boston, MA, the United States. VA Health Care System, West Roxbury, MA, the United States
References
- 1.Farber E. The multistep nature of cancer development. Cancer Res. 1984;44:4217–4223. [PubMed] [Google Scholar]
- 2.Shammas MA, Shmookler Reis RJ, Koley H, Batchu RB, Li C, Munshi NC. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma. Blood. 2009;113:2290–2297. doi: 10.1182/blood-2007-05-089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye CJ, Liu G, Bremer SW, Heng HH. The dynamics of cancer chromosomes and genomes. Cytogenet Genome Res. 2007;118:237–246. doi: 10.1159/000108306. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 6.Pathak S, Dave BJ, Gagos S. Chromosome alterations in cancer development and apoptosis. In Vivo. 1994;8:843–850. [PubMed] [Google Scholar]
- 7.Akagi T, Ito T, Kato M, Jin Z, Cheng Y, Kan T, Yamamoto G, Olaru A, Kawamata N, Boult J, et al. Chromosomal abnormalities and novel disease-related regions in progression from Barrett’s esophagus to esophageal adenocarcinoma. Int J Cancer. 2009;125:2349–2359. doi: 10.1002/ijc.24620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai JC, Liu D, Liu KH, Zhang HP, Zhong S, Xia NS. Microsatellite alterations in phenotypically normal esophageal squamous epithelium and metaplasia-dysplasia-adenocarcinoma sequence. World J Gastroenterol. 2008;14:4070–4076. doi: 10.3748/wjg.14.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finley JC, Reid BJ, Odze RD, Sanchez CA, Galipeau P, Li X, Self SG, Gollahon KA, Blount PL, Rabinovitch PS. Chromosomal instability in Barrett’s esophagus is related to telomere shortening. Cancer Epidemiol Biomarkers Prev. 2006;15:1451–1457. doi: 10.1158/1055-9965.EPI-05-0837. [DOI] [PubMed] [Google Scholar]
- 10.Paulson TG, Maley CC, Li X, Li H, Sanchez CA, Chao DL, Odze RD, Vaughan TL, Blount PL, Reid BJ. Chromosomal instability and copy number alterations in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:3305–3314. doi: 10.1158/1078-0432.CCR-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabinovitch PS, Reid BJ, Haggitt RC, Norwood TH, Rubin CE. Progression to cancer in Barrett’s esophagus is associated with genomic instability. Lab Invest. 1989;60:65–71. [PubMed] [Google Scholar]
- 12.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoler DL, Chen N, Basik M, Kahlenberg MS, Rodriguez-Bigas MA, Petrelli NJ, Anderson GR. The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc Natl Acad Sci U S A. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon E, Borrow J, Goddard AD. Chromosome aberrations and cancer. Science. 1991;254:1153–1160. doi: 10.1126/science.1957167. [DOI] [PubMed] [Google Scholar]
- 15.Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 16.Sweezy MA, Fishel R. Multiple pathways leading to genomic instability and tumorigenesis. Ann N Y Acad Sci. 1994;726:165–177. doi: 10.1111/j.1749-6632.1994.tb52810.x. [DOI] [PubMed] [Google Scholar]
- 17.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 18.West SC. The processing of recombination intermediates: mechanistic insights from studies of bacterial proteins. Cell. 1994;76:9–15. doi: 10.1016/0092-8674(94)90168-6. [DOI] [PubMed] [Google Scholar]
- 19.Poplawski T, Blasiak J. DNA homologous recombination repair in mammalian cells. Postepy Biochem. 2006;52:180–193. [PubMed] [Google Scholar]
- 20.Camenisch U, Naegeli H. Role of DNA repair in the protection against genotoxic stress. EXS. 2009;99:111–150. doi: 10.1007/978-3-7643-8336-7_5. [DOI] [PubMed] [Google Scholar]
- 21.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 22.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 23.Hey T, Lipps G, Sugasawa K, Iwai S, Hanaoka F, Krauss G. The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry. 2002;41:6583–6587. doi: 10.1021/bi012202t. [DOI] [PubMed] [Google Scholar]
- 24.Havre PA, Rice MC, Noe M, Kmiec EB. The human REC2/RAD51B gene acts as a DNA damage sensor by inducing G1 delay and hypersensitivity to ultraviolet irradiation. Cancer Res. 1998;58:4733–4739. [PubMed] [Google Scholar]
- 25.O’Regan P, Wilson C, Townsend S, Thacker J. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J Biol Chem. 2001;276:22148–22153. doi: 10.1074/jbc.M102396200. [DOI] [PubMed] [Google Scholar]
- 26.Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 27.Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 28.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 29.Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ. Evidence for simultaneous protein interactions between human Rad51 paralogs. Journal of Biological Chemistry. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- 30.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barlund M, Monni O, Kononen J, Cornelison R, Torhorst J, Sauter G, Kallioniemi O-P, Kallioniemi A. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Research. 2000;60:5340–5344. [PubMed] [Google Scholar]
- 32.Hansen LT, Lundin C, Spang-Thomsen M, Petersen LN, Helleday T. The role of RAD51 in etoposide (VP16) resistance in small cell lung cancer. International Journal of Cancer. 2003;105:472–479. doi: 10.1002/ijc.11106. [DOI] [PubMed] [Google Scholar]
- 33.Maacke H, Jost K, Opitz S, Miska S, Yuan Y, Hasselbach L, Luttges J, Kalthoff H, Sturzbecher HW. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000;19:2791–2795. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- 34.Sturgis EM, Clayman GL, Guan Y, Guo Z, Wei Q. DNA repair in lymphoblastoid cell lines from patients with head and neck cancer. Archives of Otolaryngology -- Head & Neck Surgery. 1999;125:185–190. doi: 10.1001/archotol.125.2.185. [DOI] [PubMed] [Google Scholar]
- 35.Pal J, Bertheau R, Buon L, Qazi A, Batchu RB, Bandyopadhyay S, Ali-Fehmi R, Beer DG, Weaver DW, Shmookler Reis RJ, et al. Genomic evolution in Barrett’s adenocarcinoma cells: critical roles of elevated hsRAD51, homologous recombination and Alu sequences in the genome. Oncogene. 2011;30:3585–3598. doi: 10.1038/onc.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biology. 2001;2:Research0032.1–0032.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Hong F. Cluster-Rasch models for microarray gene expression data. Genome Biology. 2001;2:Research0031.1–0031.13. doi: 10.1186/gb-2001-2-8-research0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu R, Pal J, Buon L, Nanjappa P, Shi J, Fulciniti M, Tai YT, Guo L, Yu M, Gryaznov S, et al. Targeting homologous recombination and telomerase in Barrett’s adenocarcinoma: impact on telomere maintenance, genomic instability and tumor growth. Oncogene. 2014;33:1495–1505. doi: 10.1038/onc.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noon AT, Goodarzi AA. 53BP1-mediated DNA double strand break repair: insert bad pun here. DNA Repair (Amst) 2011;10:1071–1076. doi: 10.1016/j.dnarep.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 42.Liao W, McNutt MA, Zhu WG. The comet assay: a sensitive method for detecting DNA damage in individual cells. Methods. 2009;48:46–53. doi: 10.1016/j.ymeth.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 43.Frankenberg-Schwager M, Gebauer A, Koppe C, Wolf H, Pralle E, Frankenberg D. Single-strand annealing, conservative homologous recombination, nonhomologous DNA end joining, and the cell cycle-dependent repair of DNA double-strand breaks induced by sparsely or densely ionizing radiation. Radiat Res. 2009;171:265–273. doi: 10.1667/RR0784.1. [DOI] [PubMed] [Google Scholar]
- 44.Bhargava R, Onyango DO, Stark JM. Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet. 2016;32:566–575. doi: 10.1016/j.tig.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]