Abstract
Biomedical research involving nanoparticles has produced useful products with medical applications. However, the potential toxicity of nanoparticles in biofluids, cells, tissues, and organisms is a major challenge. The ‘-omics’ analyses provide molecular profiles of multifactorial biological systems instead of focusing on a single molecule. The ‘omics’ approaches are necessary to evaluate nanotoxicity because classical methods for the detection of nanotoxicity have limited ability in detecting miniscule variations within a cell and do not accurately reflect the actual levels of nanotoxicity. In addition, the ‘omics’ approaches allow analyses of in-depth changes and compensate for the differences associated with high-throughput technologies between actual nanotoxicity and results from traditional cytotoxic evaluations. However, compared with a single omics approach, integrated omics provides precise and sensitive information by integrating complex biological conditions. Thus, these technologies contribute to extended safety evaluations of nanotoxicity and allow the accurate diagnoses of diseases far earlier than was once possible in the nanotechnology era. Here, we review a novel approach for evaluating nanotoxicity by integrating metabolomics with metabolomic profiling and transcriptomics, which is termed “metabotranscriptomics”.
Keywords: Integrated omics, Metabolomics, Metabotranscriptomics, Nanotoxicity, Profile, Transcriptomics
INTRODUCTION
Several recent reports suggest breakthrough applications of nanoparticles (NPs) in biomedical and clinical fields (1–4). NPs have unique physiochemical properties because of their size and large surface area-to-volume ratio, rendering them more reactive and thermodynamically unstable than bulk materials (5, 6). Moreover, NPs are easily absorbed and readily interact with the human body when delivered through inhalation, penetration, and ingestion (7, 8). However, issues regarding their toxicity and safety due to adverse biological effects have led to widespread concerns about the possible negative effects of NPs (9, 10). Specifically, NP-induced oxidative stress is difficult to evaluate with classical methods (11, 12). The practical application of NPs requires studies of biological toxicity. However, there are limitations associated with the evaluation of nanotoxicity using traditional methods of analysis because of the complexities at the nanolevel, underscoring the need for advanced omics approaches.
Omics facilitates the collective characterization and quantification of many different molecules, such as DNA, RNA, proteins, lipids, and metabolites, in cells, tissues, organs, and organisms. The last few decades have produced developments in high-throughput technologies for omics, which enabled comprehensive understanding of processes and novel findings in biomedical studies (13–15). Thus, omics approaches address the complexity of biological systems via interpretations using bioinformatics analyses. Rapid developments in nanotechnology and the production of NPs, which are defined as engineered materials measuring less than 100 nm in one dimension, stress the importance of the potential toxicity of NPs (6, 7, 10, 16). The omics approaches including genomics, transcriptomics, proteomics, and metabolomics are used to evaluate nanotoxicity (17–20). However, a single omics approach provides limited insight into the intricate molecular pathways and the complex biological events in cells and organisms (21–23).
The concept of integrated omics was introduced by Dr. Hood, who suggested a systems biology approach based on the combination of different omics data to provide a comprehensive understanding of the multifactorial origins of biological research (24, 25). Palsson et al. suggested many approaches to generate multi-omic data sets and reduce the possibility of resource allocation for data generation versus data curation and integration (26). Integrated omics has been applied to a wide range of complex and intractable problems in biological studies. In particular, integrated omics was shown to facilitate the determination of cytotoxicity, especially nanotoxicity (27, 28) because cytotoxicity induced by NP treatment in cells could not be detected with traditional methods (29–31). Integrated omics provides a more comprehensive overview of the complexities associated with nanotoxicity compared with a single omics approach. Thus, these technologies contribute to expanding the safety evaluation of nanotoxicity and provide accurate diagnoses of diseases compared with the simple and fragmented interpretations using single omics approaches. Here, we divide this review into three sections as follows: (i) omics approaches for nanotoxicity; (ii) recent approaches for metabolomics and transcriptomics in nanotoxicity; and (iii) integration of omics for the analysis of nanotoxicity.
OMICS APPROACHES FOR NANOTOXICITY
Omics approaches provide a better understanding of cellular events by using large-scale data. In particular, high-throughput technologies in omics have enabled the use of large-scale data to generate novel findings related to NP toxicity and mechanisms of action (17–20). Thus, omics tools prevent the use of fragmented data that could lead to inappropriate conclusions about nanotoxicity. Although the molecular technologies for understanding nanotoxicity, stress responses, molecular damage, and varying responses to NPs have advanced in parallel with molecular cell biology and in vivo assessments, the traditional approaches for the safety evaluation of new NPs have limitations regarding their potential toxicity. Thus, omics techniques are well suited to evaluate nanotoxicity both in vitro and in vivo by providing a more comprehensive view than was previously possible.
A systematic understanding of molecular responses in biological systems has been emphasized following the growth in analytic technologies and bioinformatics. Developments in sequencing technologies have allowed researchers to gather genomic and transcriptomic data (genotypic features) with much higher coverage and cost-effectively. In proteomics and metabolomics, advances in NMR and mass spectroscopy enable the analysis of a broader range of the proteome or metabolome (phenotypic features) with high precision and sensitivity (32). However, despite these improvements, single omics approaches have a fundamental “blind spot” in unraveling complex biological responses. For example, even though transcriptomics allows detection of extensive genotypic changes, it may not facilitate the interpretation of nucleic acid modifications in the genome or address issues concerning coverage of repeat-rich regions and low abundance genes, and is thoroughly inadequate for the determination of the actual phenotype (33, 34). In metabolomics, for which one endpoint is the biological phenotype (35), amplification methods are unavailable for minor metabolites, and a quantitative analysis of a targeted process can only provide a partial representation of an entire metabolic pathway (22). In addition, it is frequently associated with errors and limitations involving the interpretation of causal mechanisms in biological processes (36). The integration of two or more omics methods is highly recommended for a more comprehensive and holistic understanding of biological systems than is possible with a single omics approach.
RECENT APPROACHES FOR METABOLOMICS AND TRANSCRIPTOMICS IN NANOTOXICITY
In this section, we introduce the main omics approaches: metabolomics and transcriptomics and their application to nanotoxicity studies.
Metabolomics
Metabolomics is the comprehensive analysis of chemical processes involving metabolites that drive cellular functions, such as cellular signaling cascades, homeostatic control, energy metabolism, and cell damage (37). Specifically, the metabolome represents the complete set of small-molecule chemicals found in biological fluids, cells, tissues, organisms, and biological samples; the metabolome directly links genotype with phenotype and is most related to the phenotype (35, 38). In contrast to other omics methods, metabolomics has great potential for the analysis and understanding of cellular biological mechanisms affected by NPs because metabolic changes accurately reflect the characteristic changes in biological fluids, cells, and tissues based on the quantitation of metabolome (27, 39–41).
Metabolomic profiling is necessary to evaluate potential toxicity using either nuclear magnetic resonance (NMR) or mass-spectrometry (MS). NMR is an effective tool for the determination of the structure of organic compounds ab initio and the quantitative analysis of a broad range of molecules (such as metabolic fingerprinting) in a crude extract without authentic standards (32, 42). In addition, NMR does not depend on hydrophobicity or metabolite dissociation value, and the results are comparatively more reproducible than those derived from MS (43). However, NMR has a relatively low sensitivity (> 1 nmol) and resolution and cannot detect NMR-inactive molecules (32). Thus, there are limitations for the comprehensive analysis of individual constituents within a sample (44). MS ionizes chemical species and sorts the ions based on their mass-to-charge ratio. It is one of the most widely used methods for the ultrasensitive and simultaneous detection of metabolites by coupling with gas or liquid chromatography (45, 46). Although different types of MS have a high sensitivity of detection, the sample preparation process is tedious, and the selectivity for different classes of metabolites has both advantages and complications (43). In particular, metabolomic profiling of the cellular components, and target tissue metabolic reactions with gas chromatography-mass chromatography (GC/MS), without targeting a single metabolite, provides a better understanding of the biofluids, cells, and clinical conditions (28, 47–51). Thus, metabolomics has been used in nanotoxicity investigations utilizing high-throughput quantitation methods (11, 52). However, a limitation of metabolomics is that it provides consequential data without identifying the pathways of cellular mechanism. However, by integrating it with transcriptomics, a better understanding of subtle effects such as nanotoxicity can be obtained (27, 28).
Transcriptomics
Transcriptomics refers to the set of all RNA molecules in a cell and involves techniques such as microarray analysis and next-generation sequencing (NGS), called RNA-Seq. This technique is widely used to screen the toxicity of related RNA molecules and to elucidate the toxicity mechanisms (53, 54). Biological analytes from environmental, industrial, and drug-induced toxic exposures have been analyzed using transcriptomics (54–61). Moreover, transcriptomics contributes to the comprehensive investigation of cellular responses induced by NPs using bioinformatics software (22). Even though transcriptomics can provide large data from NP-treated cells, they are qualitative and do not establish a direct relationship with the pathology. Nanotoxicologists are working to overcome these shortcomings of transcriptomics by dovetailing the molecular mechanisms using other omics methods such as proteomics and metabolomics (27, 62). Moreover, metabolomics facilitates the identification of a phenotype based on cellular response and provides quantitative data, to compensate for the limitations of transcriptomics (27).
INTEGRATION OF OMICS FOR ANAYLSIS OF NANOTOXICITY
Integration of omics
Integration of genomics, transcriptomics, proteomics, and metabolomics, facilitates a better understanding of the cellular biology because biological systems are dynamic and heterogeneous (21). Dr. Hood was a pioneer in terms of integrating the different data types and comparing them against a model with a focus on interdisciplinary and systems biology (25, 63). Compared with single omics approaches, integrated approaches provide a large volumet of accurate information related to pathophysiology (64–68). Here, we describe the integration of transcriptomics with metabolomics in a process called “metabotranscriptomics” for the analysis of nanotoxicity, to allow a comprehensive analysis of the treatment outcomes with NPs.
Metabotranscriptomics for nanotoxicity analysis
The use of NPs in biomedical research, such as in the diagnosis and treatment of diseases, has gained tremendous momentum (1, 7, 69). Recent research has shown that internalized NPs can cause cytotoxicity by inducing reactive oxygen species (ROS) and increasing endoplasmic reticulum (ER) stress (7, 27, 70, 71). Despite increasing research efforts, the underlying mechanisms of NP toxicity are not clearly understood because of limited studies and preliminary stages of research.
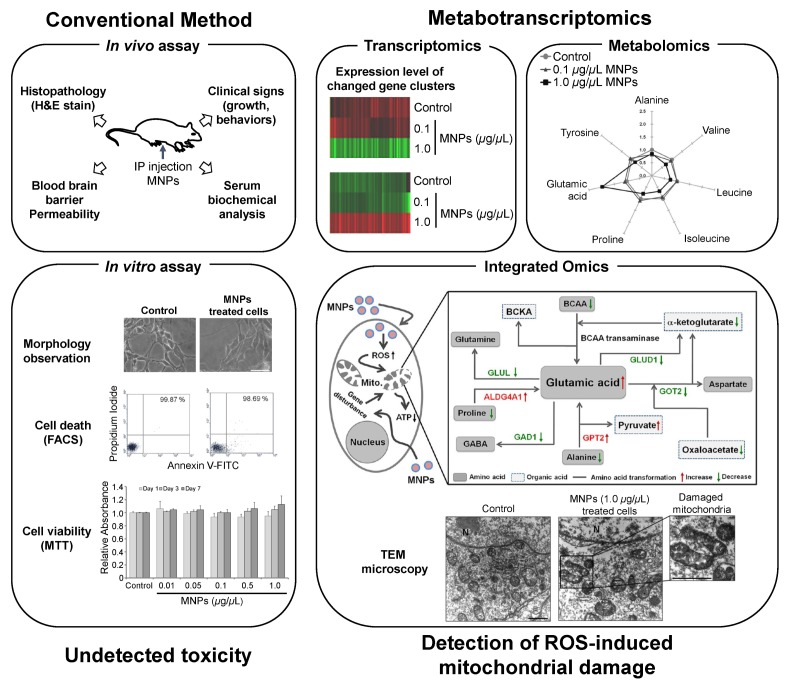
Magnetic NPs (MNPs) and MNPs coated with biocompatible compounds, which are defined as single-dimensional particles with magnetic properties, have been investigated in the context of novel applications in biochemistry, biology, medicine, antibody engineering, cell tracking, and imaging tools (72–75). MNPs@SiO2(RITC) are synthesized MNPs that consist of a cobalt ferrite core, CoFe2O4, and a silica shell containing chemically-bound Rhodamine B isothiocyanate (RITC) for cell staining, separation and MRI contrast (76). The cobalt ferrite core and RITC contained within the silica shell contribute to the stability of MNPs@SiO2(RITC) and prolonged red fluorescence at 540 nm without photobleaching. A study into the tissue distribution of MNPs@SiO2(RITC) in mice demonstrated their ability to cross the blood–brain barrier (BBB) without inducing functional deficits. Analyses using hematoxylin and eosin (H&E) staining found no abnormal histopathological lesions in organs after the intraperitoneal injection (IP) of MNPs@SiO2(RITC) into mice (29) (Fig. 1). Moreover, the injection did not induce any clinical changes (growth, body weight, behavior) or alterations in serum biochemical parameters (glucose, cholesterol, creatinine, and the ratio between the concentrations of the enzymes aspartate transaminase and alanine transaminase). In addition, a few in vitro studies such as FACS analysis, MTT assay, chromosome aberration assay, and cell cycle assay, failed to detecte any toxicity induced by NPs (27, 29–31) (Fig. 1).
Fig. 1.
Summary diagram comparing conventional methods and metabotranscriptomics approach for the assessment of MNPs@SiO2(RITC)-induced nanotoxicity (27, 29–31). MNPs: MNPs@SiO2(RITC), IP: Intraperitoneal, TEM: transmission electron microscopy.
Previous studies were confined to the pathophysiological effects of MNPs@SiO2(RITC). Treatment with MNPs@SiO2(RITC) yielded 24 metabolites, nine of which were considered to be significantly altered. However, assessment using only metabolomics does not yield convincing data and has low reliability. Integrating the metabolomic profiling with transcriptomics will allow a more sensitive and detailed toxicological evaluation of cellular responses to NPs and identify novel nanotoxicological biomarkers (27). Metabolic profiling of MNPs@SiO2(RITC)-treated human embryonic kidney 293 (HEK293) cells revealed that changes in amino acids (AAs), organic acids (OAs) and a few metabolites were related to ROS generation (27), which triggered mitochondrial damage (Fig. 1).
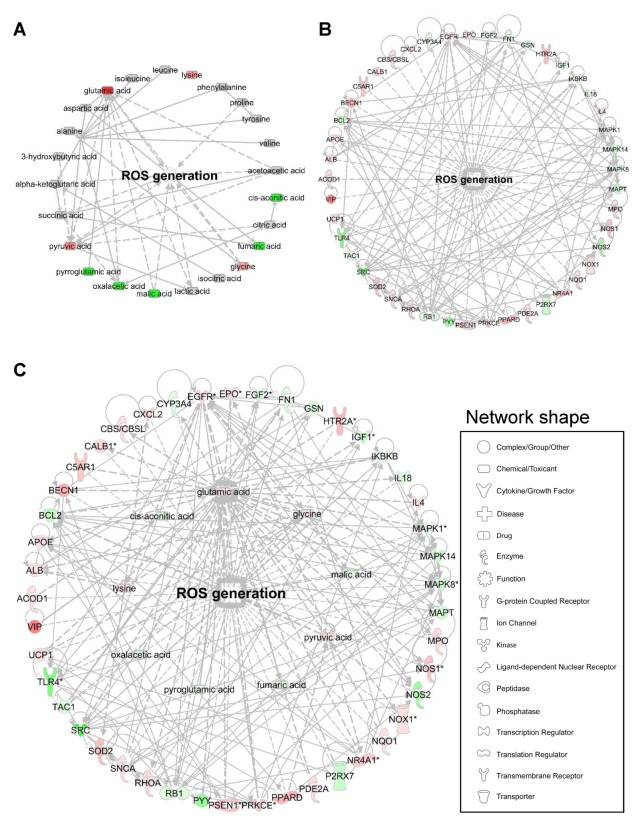
The percent compositions and normalized values of AAs and OAs clearly revealed a marked increase in glutamic acid and pyruvate levels, and decrease in other AAs, such as alanine, valine, leucine, isoleucine, proline, and tyrosine, and other OAs, such as α-ketoglutarate, oxaloacetate, fumarate, and malate, in the group treated with MNPs@SiO2(RITC) (27). However, there is a limitation associated with linking ROS generation with metabolic changes (Fig. 2A). For the transcriptome, the expression levels of 45 ROS-generation-related genes were altered. Specifically, 26 genes were upregulated and 19 genes were downregulated, and these genes were found to be connected with direct relationships (Fig. 2B). Thus, we combined transcriptomics and metabolomics for these data using Ingenuity Pathway Analysis (IPA Ver. 8.5, Ingenuity Systems, http://www.ingenuity.com), which is a web-based bioinformatics software for the identification of biological functions. The datasets of differentially expressed genes and metabolites were combined, to elucidate the interactions between differentially expressed genes and altered metabolites (Fig. 2C), and determine biological changes related to ROS generation. The integration of the metabolic profile and transcriptome revealed a direct correlation between the metabolites and genes related to ROS generation. These results demonstrate the importance of metabotranscriptomics for the detailed analyses of nanotoxicity. Although integrated omics approaches facilitate a comprehensive analysis of cellular pathways in biological systems, several new challenges need to be overcome before they can be used in nanotoxicity investigations.
Fig. 2.
Bioinformatics of ROS generation using ingenuity pathway analysis (IPA), (A) metabolomics, (B) transcriptomics, and (C) metabotranscriptomics based on a previous report (27). Red and green areas indicate up- and downregulated metabolites, respectively, in cells treated with MNPs@SiO2(RITC) compared with control cells. Differentially regulated metabolites obtained from the metabolic profile (more than a ± 20% change) and microarray data (genes with a > 3-fold change) are shown. In the representation of the genetic networks, the red and green colors indicate up- and down-regulated genes, respectively. Network shape indicates categorization of molecules and function. Information pertaining to the corresponding genes can be found in NCBI (https://www.ncbi.nlm.nih.gov/).
CONCLUSION
Here, we reviewed the cutting-edge metabotranscriptomic approaches for nanotoxicity evaluation. The traditional methods of detection are limited by their ability to measure nanotoxicity. The introduction of advanced tools has led to the integration of omics, especially metabolomic profiling and transcriptomics, to provide extensive information on biological conditions. The technological progress in the molecular diagnosticscan pave the way to the development of additional omics techniques. In addition to the integration of transcriptomics and metabolomics, the combination of transcriptomics, genomics, and proteomics in nanotoxicity studies can be used to facilitate the analyses of subtle changes in cellular physiology and molecular biology. Future studies in nanotoxicity will require the integration and multidisciplinary use of omics methods. This integration is expected to produce major advances in toxicity research and encourage the discovery of novel biomarkers for nanotoxicity for a more complete understanding of the effects of NPs in biomedical studies.
ACKNOWLEDGEMENTS
This work was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education, Science and Technology (2015R1D1A1A09060192); Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0093826, 2016R1C1B2016135); and the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2016M3C7A1904392).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Stark WJ. Nanoparticles in biological systems. Angew Chem Int Ed Engl. 2011;50:1242–1258. doi: 10.1002/anie.200906684. [DOI] [PubMed] [Google Scholar]
- 2.Havel H, Finch G, Strode P, et al. Nanomedicines: From Bench to Bedside and Beyond. AAPS J. 2016;18:1373–1378. doi: 10.1208/s12248-016-9961-7. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, Yu C. Advances in silica based nanoparticles for targeted cancer therapy. Nanomedicine. 2016;12:317–332. doi: 10.1016/j.nano.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Deng R, Lin D, Zhu L, et al. Nanoparticle interactions with co-existing contaminants: Joint toxicity, bio-accumulation and risk. Nanotoxicology. 2017:1–56. doi: 10.1080/17435390.2017.1343404. [DOI] [PubMed] [Google Scholar]
- 5.Vayssieres L, Chaneac C, Tronc E, Jolivet JP. Size Tailoring of Magnetite Particles Formed by Aqueous Precipitation: An Example of Thermodynamic Stability of Nanometric Oxide Particles. J Colloid Interface Sci. 1998;205:205–212. doi: 10.1006/jcis.1998.5614. [DOI] [PubMed] [Google Scholar]
- 6.Auffan M, Rose J, Bottero JY, Lowry GV, Jolivet JP, Wiesner MR. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat Nanotechnol. 2009;4:634–641. doi: 10.1038/nnano.2009.242. [DOI] [PubMed] [Google Scholar]
- 7.Krug HF, Wick P. Nanotoxicology: an interdisciplinary challenge. Angew Chem Int Ed Engl. 2011;50:1260–1278. doi: 10.1002/anie.201001037. [DOI] [PubMed] [Google Scholar]
- 8.Bouallegui Y, Ben Younes R, Bellamine H, Oueslati R. Histopathology and analyses of inflammation intensity in the gills of mussels exposed to silver nanoparticles: role of nanoparticle size, exposure time, and uptake pathways. Toxicol Mech Methods. 2017:1–10. doi: 10.1080/15376516.2017.1337258. [DOI] [PubMed] [Google Scholar]
- 9.Service RF. Is nanotechnology dangerous? Science. 2000;290:1526–1527. doi: 10.1126/science.290.5496.1526. [DOI] [PubMed] [Google Scholar]
- 10.Boyes WK, Thornton BLM, Al-Abed SR, et al. A comprehensive framework for evaluating the environmental health and safety implications of engineered nanomaterials. Crit Rev Toxicol. 2017:1–44. doi: 10.1080/10408444.2017.1328400. [DOI] [PubMed] [Google Scholar]
- 11.Schnackenberg LK, Sun J, Beger RD. Metabolomics techniques in nanotoxicology studies. Methods Mol Biol. 2012;926:141–156. doi: 10.1007/978-1-62703-002-1_10. [DOI] [PubMed] [Google Scholar]
- 12.Masoud R, Bizouarn T, Trepout S, et al. Titanium Dioxide Nanoparticles Increase Superoxide Anion Production by Acting on NADPH Oxidase. PLoS One. 2015;10:e0144829. doi: 10.1371/journal.pone.0144829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuncbag N, Gosline SJ, Kedaigle A, Soltis AR, Gitter A, Fraenkel E. Network-Based Interpretation of Diverse High-Throughput Datasets through the Omics Integrator Software Package. PLoS Comput Biol. 2016;12:e1004879. doi: 10.1371/journal.pcbi.1004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norris JL, Farrow MA, Gutierrez DB, et al. Integrated, High-Throughput, Multiomics Platform Enables Data-Driven Construction of Cellular Responses and Reveals Global Drug Mechanisms of Action. J Proteome Res. 2017;16:1364–1375. doi: 10.1021/acs.jproteome.6b01004. [DOI] [PubMed] [Google Scholar]
- 15.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu X, Li D, Gao Y, Mu L, Zhou Q. Knowledge gaps between nanotoxicological research and nanomaterial safety. Environ Int. 2016;94:8–23. doi: 10.1016/j.envint.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Reyes VC, Li M, Hoek EM, Mahendra S, Damoiseaux R. Genome-wide assessment in Escherichia coli reveals time-dependent nanotoxicity paradigms. ACS Nano. 2012;6:9402–9415. doi: 10.1021/nn302815w. [DOI] [PubMed] [Google Scholar]
- 18.Bo Y, Jin C, Liu Y, Yu W, Kang H. Metabolomic analysis on the toxicological effects of TiO(2) nano-particles in mouse fibroblast cells: from the perspective of perturbations in amino acid metabolism. Toxicol Mech Methods. 2014;24:461–469. doi: 10.3109/15376516.2014.939321. [DOI] [PubMed] [Google Scholar]
- 19.Verano-Braga T, Miethling-Graff R, Wojdyla K, et al. Insights into the cellular response triggered by silver nanoparticles using quantitative proteomics. ACS Nano. 2014;8:2161–2175. doi: 10.1021/nn4050744. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Li L, Zhang PF, et al. Differential Regulation of Gene and Protein Expression by Zinc Oxide Nanoparticles in Hen’s Ovarian Granulosa Cells: Specific Roles of Nanoparticles. PLoS One. 2015;10:e0140499. doi: 10.1371/journal.pone.0140499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prohaska SJ, Stadler PF. The use and abuse of -omes. Methods Mol Biol. 2011;719:173–196. doi: 10.1007/978-1-61779-027-0_8. [DOI] [PubMed] [Google Scholar]
- 22.Van Assche R, Broeckx V, Boonen K, et al. Integrating -Omics: Systems Biology as Explored Through C. elegans Research. J Mol Biol. 2015;427:3441–3451. doi: 10.1016/j.jmb.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Sun YV, Hu YJ. Integrative Analysis of Multi-omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv Genet. 2016;93:147–190. doi: 10.1016/bs.adgen.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hood L. Leroy Hood expounds the principles, practice and future of systems biology. Drug Discov Today. 2003;8:436–438. doi: 10.1016/S1359-6446(03)02710-7. [DOI] [PubMed] [Google Scholar]
- 25.Hood L. Systems biology: integrating technology, biology, and computation. Mech Ageing Dev. 2003;124:9–16. doi: 10.1016/S0047-6374(02)00164-1. [DOI] [PubMed] [Google Scholar]
- 26.Palsson B, Zengler K. The challenges of integrating multi-omic data sets. Nat Chem Biol. 2010;6:787–789. doi: 10.1038/nchembio.462. [DOI] [PubMed] [Google Scholar]
- 27.Shim W, Paik MJ, Nguyen DT, et al. Analysis of changes in gene expression and metabolic profiles induced by silica-coated magnetic nanoparticles. ACS Nano. 2012;6:7665–7680. doi: 10.1021/nn301113f. [DOI] [PubMed] [Google Scholar]
- 28.Phukan G, Shin TH, Shim JS, et al. Silica-coated magnetic nanoparticles impair proteasome activity and increase the formation of cytoplasmic inclusion bodies in vitro. Sci Rep. 2016;6:29095. doi: 10.1038/srep29095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JS, Yoon TJ, Yu KN, et al. Toxicity and tissue distribution of magnetic nanoparticles in mice. Toxicol Sci. 2006;89:338–347. doi: 10.1093/toxsci/kfj027. [DOI] [PubMed] [Google Scholar]
- 30.Park KS, Tae J, Choi B, et al. Characterization, in vitro cytotoxicity assessment, and in vivo visualization of multimodal, RITC-labeled, silica-coated magnetic nanoparticles for labeling human cord blood-derived mesenchymal stem cells. Nanomedicine. 2010;6:263–276. doi: 10.1016/j.nano.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Beck GR, Jr, Ha SW, Camalier CE, et al. Bioactive silica-based nanoparticles stimulate bone-forming osteoblasts, suppress bone-resorbing osteoclasts, and enhance bone mineral density in vivo. Nanomedicine. 2012;8:793–803. doi: 10.1016/j.nano.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan TW, Higashi RM, Lane AN. Integrating metabolomics and transcriptomics for probing SE anticancer mechanisms. Drug Metab Rev. 2006;38:707–732. doi: 10.1080/03602530600959599. [DOI] [PubMed] [Google Scholar]
- 33.Rehrauer H, Opitz L, Tan G, Sieverling L, Schlapbach R. Blind spots of quantitative RNA-seq: the limits for assessing abundance, differential expression, and isoform switching. BMC Bioinformatics. 2013;14:370. doi: 10.1186/1471-2105-14-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans TG. Considerations for the use of transcriptomics in identifying the ‘genes that matter’ for environmental adaptation. J Exp Biol. 2015;218:1925–1935. doi: 10.1242/jeb.114306. [DOI] [PubMed] [Google Scholar]
- 35.Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B. Metabolomics in human nutrition: opportunities and challenges. Am J Clin Nutr. 2005;82:497–503. doi: 10.1093/ajcn.82.3.497. [DOI] [PubMed] [Google Scholar]
- 36.Moseley HN. Error Analysis and Propagation in Metabolomics Data Analysis. Comput Struct Biotechnol J. 2013;4 doi: 10.5936/csbj.201301006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukusaki E. Application of Metabolomics for High Resolution Phenotype Analysis. Mass Spectrom (Tokyo) 2014;3:S0045. doi: 10.5702/massspectrometry.S0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zlatkis A, Brazell RS, Poole CF. The role of organic volatile profiles in clinical diagnosis. Clin Chem. 1981;27:789–797. [PubMed] [Google Scholar]
- 40.Karlic H, Thaler R, Gerner C, et al. Inhibition of the mevalonate pathway affects epigenetic regulation in cancer cells. Cancer Genet. 2015;208:241–252. doi: 10.1016/j.cancergen.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zabala-Letona A, Arruabarrena-Aristorena A, Martin-Martin N, et al. mTORC1-dependent AMD1 regulation sustains polyamine metabolism in prostate cancer. Nature. 2017;547:109–113. doi: 10.1038/nature22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson DG, Watkins PB, Reily MD. Metabolomics in toxicology: preclinical and clinical applications. Toxicol Sci. 2011;120(Suppl 1):S146–170. doi: 10.1093/toxsci/kfq358. [DOI] [PubMed] [Google Scholar]
- 43.Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 44.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 46.Dudley E, Yousef M, Wang Y, Griffiths WJ. Targeted metabolomics and mass spectrometry. Adv Protein Chem Struct Biol. 2010;80:45–83. doi: 10.1016/B978-0-12-381264-3.00002-3. [DOI] [PubMed] [Google Scholar]
- 47.Paik MJ, Lee KA, Park CS, et al. Pattern recognition analysis of polyamines in the plasma of rat models with adenovirus infection. Clin Chim Acta. 2007;380:228–231. doi: 10.1016/j.cca.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Paik MJ, Li WY, Ahn YH, et al. The free fatty acid metabolome in cerebral ischemia following human mesenchymal stem cell transplantation in rats. Clin Chim Acta. 2009;402:25–30. doi: 10.1016/j.cca.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 49.Paik MJ, Ahn YH, Lee PH, et al. Polyamine patterns in the cerebrospinal fluid of patients with Parkinson’s disease and multiple system atrophy. Clin Chim Acta. 2010;411:1532–1535. doi: 10.1016/j.cca.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 50.Shin TH, Phukan G, Shim JS, et al. Restoration of Polyamine Metabolic Patterns in In Vivo and In Vitro Model of Ischemic Stroke following Human Mesenchymal Stem Cell Treatment. Stem Cells Int. 20162016:4612531. doi: 10.1155/2016/4612531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin TH, Lee S, Choi KR, et al. Quality and freshness of human bone marrow-derived mesenchymal stem cells decrease over time after trypsinization and storage in phosphate-buffered saline. Sci Rep. 2017;7:1106. doi: 10.1038/s41598-017-01315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lv M, Huang W, Chen Z, et al. Metabolomics techniques for nanotoxicity investigations. Bioanalysis. 2015;7:1527–1544. doi: 10.4155/bio.15.83. [DOI] [PubMed] [Google Scholar]
- 53.Vatakuti S, Pennings JL, Gore E, Olinga P, Groothuis GM. Classification of Cholestatic and Necrotic Hepatotoxicants Using Transcriptomics on Human Precision-Cut Liver Slices. Chem Res Toxicol. 2016;29:342–351. doi: 10.1021/acs.chemrestox.5b00491. [DOI] [PubMed] [Google Scholar]
- 54.Kohonen P, Parkkinen JA, Willighagen EL, et al. A transcriptomics data-driven gene space accurately predicts liver cytopathology and drug-induced liver injury. Nat Commun. 2017;8:15932. doi: 10.1038/ncomms15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dix DJ, Gallagher K, Benson WH, et al. A framework for the use of genomics data at the EPA. Nat Biotechnol. 2006;24:1108–1111. doi: 10.1038/nbt0906-1108. [DOI] [PubMed] [Google Scholar]
- 56.Williams TD, Mirbahai L, Chipman JK. The toxicological application of transcriptomics and epigenomics in zebrafish and other teleosts. Brief Funct Genomics. 2014;13:157–171. doi: 10.1093/bfgp/elt053. [DOI] [PubMed] [Google Scholar]
- 57.Cheng F, Theodorescu D, Schulman IG, Lee JK. In vitro transcriptomic prediction of hepatotoxicity for early drug discovery. J Theor Biol. 2011;290:27–36. doi: 10.1016/j.jtbi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang M, Chen M, Tong W. Is toxicogenomics a more reliable and sensitive biomarker than conventional indicators from rats to predict drug-induced liver injury in humans? Chem Res Toxicol. 2012;25:122–129. doi: 10.1021/tx200320e. [DOI] [PubMed] [Google Scholar]
- 59.Chen M, Bisgin H, Tong L, et al. Toward predictive models for drug-induced liver injury in humans: are we there yet? Biomark Med. 2014;8:201–213. doi: 10.2217/bmm.13.146. [DOI] [PubMed] [Google Scholar]
- 60.Chen S, Xuan J, Couch L, et al. Sertraline induces endoplasmic reticulum stress in hepatic cells. Toxicology. 2014;322:78–88. doi: 10.1016/j.tox.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otava M, Shkedy Z, Talloen W, Verheyen GR, Kasim A. Identification of in vitro and in vivo disconnects using transcriptomic data. BMC Genomics. 2015;16:615. doi: 10.1186/s12864-015-1726-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lorscheidt S, Lamprecht A. Safety assessment of nanoparticles for drug delivery by means of classic in vitro assays and beyond. Expert Opin Drug Deliv. 2016;13:1545–1558. doi: 10.1080/17425247.2016.1198773. [DOI] [PubMed] [Google Scholar]
- 63.Hood L, Rowen L, Galas DJ, Aitchison JD. Systems biology at the Institute for Systems Biology. Brief Funct Genomic Proteomic. 2008;7:239–248. doi: 10.1093/bfgp/eln027. [DOI] [PubMed] [Google Scholar]
- 64.Shah SH, Newgard CB. Integrated metabolomics and genomics: systems approaches to biomarkers and mechanisms of cardiovascular disease. Circ Cardiovasc Genet. 2015;8:410–419. doi: 10.1161/CIRCGENETICS.114.000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rebollar EA, Antwis RE, Becker MH, et al. Using “Omics” and Integrated Multi-Omics Approaches to Guide Probiotic Selection to Mitigate Chytridiomycosis and Other Emerging Infectious Diseases. Front Microbiol. 2016;7:68. doi: 10.3389/fmicb.2016.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hartiala JA, Tang WH, Wang Z, et al. Genome-wide association study and targeted metabolomics identifies sex-specific association of CPS1 with coronary artery disease. Nat Commun. 2016;7:10558. doi: 10.1038/ncomms10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, Woo HR, Nam HG. Toward Systems Understanding of Leaf Senescence: An Integrated Multi-Omics Perspective on Leaf Senescence Research. Mol Plant. 2016;9:813–825. doi: 10.1016/j.molp.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 68.Berg KCG, Eide PW, Eilertsen IA, et al. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017;16:116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thakor AS, Gambhir SS. Nanooncology: the future of cancer diagnosis and therapy. CA Cancer J Clin. 2013;63:395–418. doi: 10.3322/caac.21199. [DOI] [PubMed] [Google Scholar]
- 70.Chen R, Huo L, Shi X, et al. Endoplasmic reticulum stress induced by zinc oxide nanoparticles is an earlier biomarker for nanotoxicological evaluation. ACS Nano. 2014;8:2562–2574. doi: 10.1021/nn406184r. [DOI] [PubMed] [Google Scholar]
- 71.Matysiak M, Kapka-Skrzypczak L, Brzoska K, Gutleb AC, Kruszewski M. Proteomic approach to nanotoxicity. J Proteomics. 2016;137:35–44. doi: 10.1016/j.jprot.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 72.Amemiya Y, Tanaka T, Yoza B, Matsunaga T. Novel detection system for biomolecules using nano-sized bacterial magnetic particles and magnetic force microscopy. J Biotechnol. 2005;120:308–314. doi: 10.1016/j.jbiotec.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 73.Jun YW, Seo JW, Cheon J. Nanoscaling laws of magnetic nanoparticles and their applicabilities in biomedical sciences. Acc Chem Res. 2008;41:179–189. doi: 10.1021/ar700121f. [DOI] [PubMed] [Google Scholar]
- 74.Larsen BA, Haag MA, Serkova NJ, Shroyer KR, Stoldt CR. Controlled aggregation of superparamagnetic iron oxide nanoparticles for the development of molecular magnetic resonance imaging probes. Nanotechnology. 2008;19:265102. doi: 10.1088/0957-4484/19/26/265102. [DOI] [PubMed] [Google Scholar]
- 75.Kang T, Li F, Baik S, Shao W, Ling D, Hyeon T. Surface design of magnetic nanoparticles for stimuli-responsive cancer imaging and therapy. Biomaterials. 2017;136:98–114. doi: 10.1016/j.biomaterials.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 76.Yoon TJ, Kim JS, Kim BG, Yu KN, Cho MH, Lee JK. Multifunctional nanoparticles possessing a “magnetic motor effect” for drug or gene delivery. Angew Chem Int Ed Engl. 2005;44:1068–1071. doi: 10.1002/anie.200461910. [DOI] [PubMed] [Google Scholar]