Abstract
Non-small-cell lung cancer (NSCLC) is commonly caused by a mutation in the epidermal growth factor receptor (EGFR) and subsequent aberrant EGFR signaling with uncontrolled kinase activity. A deletion mutation in EGFR exon 19 is frequently observed in EGFR gene mutations. We designed a DNAzyme to suppress the expression of mutant EGFR by cleaving the mutant EGFR mRNA. The DNAzyme (named Ex19del Dz) specifically cleaved target RNA and decreased cancer cell viability when transfected into gefitinib-resistant lung cancer cells harboring EGFR exon 19 deletions. The DNAzyme decreased EGFR expression and inhibited its downstream signaling pathway. In addition to EGFR downregulation, Ex19del Dz containing CpG sites activated Toll-like receptor 9 (TLR9) and its downstream signaling pathway via p38 kinase, causing an immunostimulatory effect on EGFR-mutated NSCLC cells. Thus, dual effects of this DNAzyme harboring the CpG site, such as TLR9 activation and EGFR downregulation, leads to apoptosis of EGFR-mutated NSCLC cells.
Keywords: CpG site, DNAzyme, EGFR mutation, Lung cancer, Toll-like receptor 9
INTRODUCTION
Non-small-cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancer incidents and is caused by various genetic defects. Mutations in epidermal growth factor receptor (EGFR) have been identified to be among the gene mutations that cause NSCLC (1). As a member of the Her/ErbB receptor family, EGFR is a tyrosine kinase receptor located on the cell surface. Normal activation of EGFR is achieved by binding with extracellular ligands such as epidermal growth factor (EGF) and tumor necrosis factor alpha (TNF-α). However, EGFR mutations contribute to the development of cancer through uncontrolled cell proliferation and metastasis without the binding of ligands (2).
EGFR genes in NSCLC patients are known to have mutations in exons 18–20, including deletions of amino acids from 746 to 750 in exon 19 or point mutations such as T790M in exon 20 and L858R in exon 21 (3). The exon 19 deletion and L858R point mutation increase the kinase activity of EGFR, resulting in the uncontrolled proliferation of cancer cells. Tyrosine kinase inhibitors (TKIs) such as erlotinib (TarcevaTM) and gefitinib (IressaTM) have been administered to patients over the last decade (4, 5). However, the conformational change of EGFR caused by the T790M point mutation leads to steric hindrance at adenosine triphosphate (ATP)-binding sites required for the binding of TKIs, inducing drug resistance in tumor cells (6).
For the efficient removal of pathogenic transcripts, RNA-cleaving deoxyribozyme (DNAzyme, Dz) has been utilized for oligonucleotide-based gene silencing (7, 8). The “10–23” type of DNAzyme includes a 15-nucleotide (nt) conserved catalytic core and flanking sequences of 6–12 bases as substrate-binding arms located on both sides of the core (9). They can bind to the target RNA through Watson-Crick base-pairing and cleave the RNA between purine-pyrimidine junctions through a phosphoester transfer reaction (9). DNAzyme is a more attractive material for gene silencing because DNAzyme does not require any accessory factors except divalent metal ions such as Mg2+, which are abundant in the cell cytosol (10). In addition, DNAzyme has several advantages over siRNA such as cost effectiveness, flexible chemical modifications, and relatively high stability in serum (11, 12).
Oligonucleotide-based gene therapy has been known to induce an immunological challenge such as causing an innate immune response (13). The mammalian innate immune system responds to infectious agents by recognizing foreign organisms, termed pathogen-associated molecular patterns (PAMPs). A variety of pattern recognition receptors (PRRs) are involved in PAMP recognition (14). The most well understood of these receptors are the Toll-like receptors (TLRs), which were first discovered in Drosophila and have been assigned numbers 1 to 10 (TLR1-TLR10) in humans (15). Among these TLRs, TLR9, a member of the TLR family, recognizes unmethylated CpG sites in DNA molecules (16). The CpG site, which is commonly found in bacterial and viral DNA, indicates a consecutive arrangement of a cytosine (“C”) and a guanine (“G”) through a phosphodiester bond (“p”). The recognition of CpG-DNA by TLR9 activates p38 MAPK, which negatively regulates cell cycle progression by responding to cytokines and extracellular stress stimuli (17). Sustained activation of the p38 MAPK pathway leads to anti-proliferative signals for cancer cells (18). Several CpG-DNAs such as CpG-7909 have been demonstrated in clinical trials to act as TLR9 agonists to induce an immune response against cancer (19).
In this study, we designed a DNAzyme that can specifically cleave mutant EGFR (exon 19 deletions) mRNA to suppress the proliferation of lung cancer cells. We found that this DNAzyme that targets mutant EGFR mRNA (Ex19del Dz) could inhibit the expression of mutant EGFR, resulting in apoptosis in drug-resistant lung cancer cells. In addition, we found that the CpG sites in the DNAzyme activated the TLR9/P38 MAPK pathway via the innate immune response pathway.
RESULTS AND DISCUSSION
DNAzyme that targets and cleaves mutant EGFR mRNA
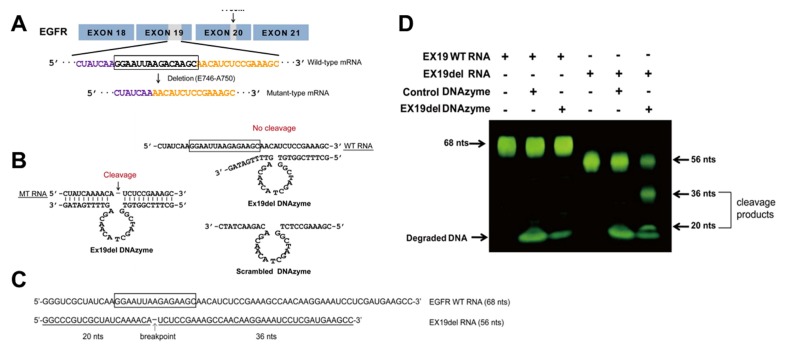
The DNAzyme targeting the exon 19 deletion sites in EGFR mRNA (Ex19del Dz) was designed using the 10–23 DNAzyme core format, which contained a 15-nt catalytic core and flanking sequences of side arms that are complementary to its RNA target. Mutant-type EGFR mRNA lacked 15 nt in exon 19 of the wild-type EGFR mRNA, which corresponded to a deletion of five amino acids (E746-A750) (Fig. 1A). Thus, Ex19del Dz was constructed to specifically bind to and cleave the 15 nt- deletion mutant-type mRNA (Fig. 1B). A scrambled DNAzyme as a negative control was constructed to contain the flanking sequences complementary to the Ex19del DNAzyme except for the catalytic core (Fig. 1B).
Fig. 1.
DNAzyme targeting mutant EGFR mRNA cleaves mutant RNA. (A) Sequence of mutant EGFR with a 15 nt-deletion in exon 19 (deletion from E746 to A750 in exon 19). (B) Sequences of Ex19del Dz (left) and scrambled DNAzyme (sc Dz, right). The DNAzyme-substrate mutant RNA (MT RNA) annealed complex is also shown. Ex19del DNAzyme was designed for cleavage of deletion type mRNA at a phosphodiester bond located between an unpaired adenine (A) and uracil (U). The site of cleavage by the DNAzyme is indicated by an arrow. (C) Sequences of each EGFR wild-type and deletion-type substrate RNA used for the cleavage reaction. (D) Exon 19 wild-type and mutant RNA substrates were incubated with Ex19del Dz and control DNAzyme (sc Dz) at 37°C for 3 h. The RNA products were analyzed by denaturing 10% PAGE (w/v) and visualized with SYBR Gold. DNAzyme was degraded with DNase to stop the reaction.
To examine the specific RNA cleavage activity of Ex19del Dz, an RNA cleavage assay was carried out using short RNA substrates, wild-type EGFR RNA (Ex19 WT RNA, 68 nt) and mutant-type RNA (Ex19del RNA, 56 nt) (Fig. 1C). RNA substrates were incubated with the DNAzyme at 37°C for 3 h, and the DNAzyme was then degraded with DNase I to stop the reaction. The degraded DNAzyme and cleaved RNA substrates were resolved by agarose gel electrophoresis (Fig. 1D). When the Ex19del Dz was incubated with wild-type EGFR RNA, RNA cleavage was not observed, while mutant RNA was significantly cleaved into two fragments by Ex19del Dz. The control scrambled DNAzyme did not have any effect on either RNA substrate. These results indicate that Ex19del Dz specifically recognizes and cleaves the target RNA (EGFR Ex19del RNA) without affecting the non-target EGFR WT RNA.
Cleavage of mutant EGFR transcripts reduced EGFR expression and cell proliferation
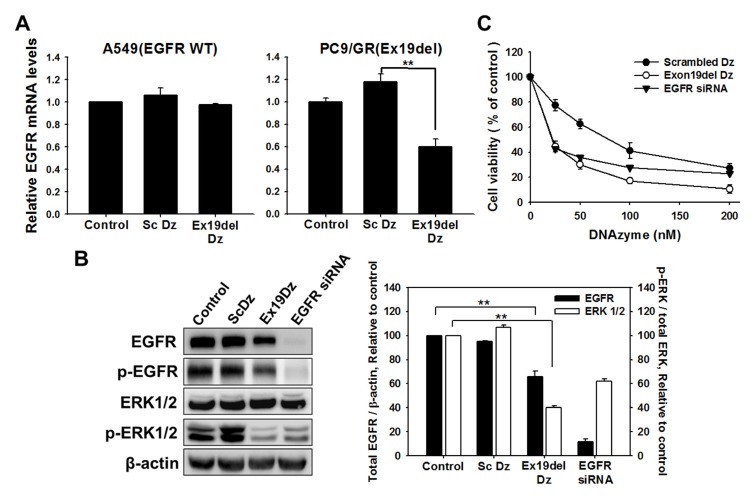
To determine whether Ex19del Dz can induce mRNA cleavage and further suppress the expression of EGFR in cells, the DNAzyme was transfected into lung cancer cells harboring the exon19 deletion mutant EGFR gene (PC9/GR) or wild-type EGFR gene (A549). The scrambled DNAzyme was also used as a control to assess nonspecific effects of the transfected oligonucleotides. When the Ex19del Dz was transfected into the mutant EGFR cells, the expression of EGFR transcripts was reduced by about 40% compared to the control or the group treated with the scrambled Dz (Fig. 2A). In A549 cells, however, transfection with the DNAzyme did not significantly decrease the amount of EGFR mRNA (Fig. 2A), indicating that Ex19del Dz selectively reduced the amount of mutant EGFR mRNA lacking 15 nt in exon 19.
Fig. 2.
EGFR expression was decreased after transfection of Ex19del DNAzyme into lung cancer cells. Oligo DNAs (100 nM) such as scrambled DNAzyme and Ex19del DNAzyme were transfected into PC9/GR lung cancer cells (5 × 105 cells). 48 h after transfection, total RNA and cellular proteins were obtained for analysis. (A) Real-time PCR for detection of EGFR cDNA was performed in DNAzyme-transfected cells, as described in the graph. cDNA amplified from EGFR transcripts was relatively quantified by normalization to control cells that were untreated. **P < 0.01. (B) Western blot analysis revealing the inactivation of EGFR protein in DNAzyme-transfected cells. Histograms show the levels of total EGFR including phospho-EGFR (black bar) expressed relative to the loading control (β-actin) and phospho-ERK1/2 (white bar) relative to total ERK1/2, which were normalized to the non-treated control. **P < 0.01. (C) The viability of cancer cells treated with different DNAzymes and EGFR siRNA was measured using methylene blue 48 h after treatment with DNAzyme or siRNA.
We next monitored the expression of the EGFR and EGFR downstream signaling factor ERK1/2 by western blot analysis. As shown in Fig. 2B, expression of EGFR was significantly decreased by about 40% by Ex19del Dz. Furthermore, as the EGFR expression level was downregulated, the phosphorylation of ERK1/2 was also suppressed by about 60%. These effects induced by Ex19del Dz were also observed with EGFR-targeting siRNA. In contrast, the control DNAzyme (scrambled Dz) did not show any effect on EGFR expression and ERK1/2 phosphorylation. Taken together, these results indicate that the Ex19del DNAzyme efficiently suppresses transcription of EGFR by cleaving mRNA, resulting in a decrease in mutant EGFR expression and attenuation of downstream signaling in lung cancer cells.
We next tested the viability of lung cancer cells following EGFR knockdown with the DNAzyme. PC9/GR cells were transfected with Ex19del Dz for 6 h, and then, cell viability was measured using the methylene blue assay (Fig. 2C). EGFR siRNA, the effects of which were previously reported (20), was also used, and scrambled Dz (sc Dz) was used as a control to compare the cytotoxic effects of oligonucleotides transfected into the cells. Ex19del Dz significantly reduced cell viability in a dose-dependent manner. Contrary to our expectations, the control DNAzyme (sc Dz) lacking the complementary sequence to the target mutant RNA sequence also caused significant cell death, albeit lesser than that caused by Ex19del Dz and EGFR siRNA. Importantly, Ex19del Dz reduced cell viability more than siRNA. Considering the nonspecific sequence-independent effect caused by the scrambled DNAzyme, Ex19del Dz targeting the EGFR mutant sequence induced markedly more cancer cell death via a nonspecific oligonucleotide effect and sequence-specific EGFR mRNA cleavage effect. Thus, we suggest that Ex19del Dz targeting the EGFR mutant RNA sequence is superior to EGFR siRNA for suppressing lung cancer cell proliferation due to an additional nonspecific oligonucleotide effect on cell viability.
Oligo DNAs containing CpG sites are responsible for cellular apoptosis
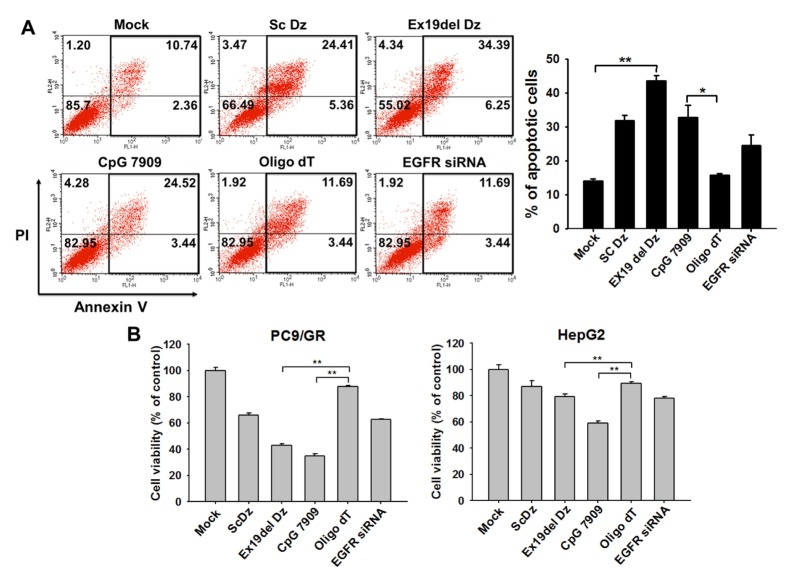
We hypothesized that the nonspecific cellular response incurred by oligo DNAs (sc Dz and Ex19del Dz), as shown in Fig. 2C, was an innate immune response caused by non-methylated CpG dinucleotides. DNA harboring nonmethylated CpG sites is recognized by TLR9 on the endoplasmic reticulum (ER) and endosomes, resulting in the induction of inflammatory innate immune responses and cancer cell death mediated via p38 MAPK (18, 21). Thus, we determined whether the CpG-DNA would affect the fate of lung cancer cells using flow cytometry analysis (Fig. 3A). DNA composed of only thymine (i.e., oligo dT) was used as a negative control for the CpG-mediated immune response. The oligo dT did not affect lung cancer cell fate, similar to the effect of the transfection vehicle control (mock control). In contrast, two oligo DNAs (i.e., Sc Dz and CpG7909) that do not recognize the mutant EGFR mRNA induced apoptosis in about 30% of PC9/GR cells, which was a two-fold enhancement in apoptosis compared with the negative controls. When PC9/GR cells were transfected with Ex19del Dz, the population of apoptotic cells significantly increased up to ~40%, which was the highest level of apoptosis induction. This result was consistent with the preceding data showing a significant reduction in cell viability by Ex19del Dz (Fig. 2C).
Fig. 3.
DNA containing the CpG site causes apoptotic cell death in lung cancer cells. (A) 100 nM individual oligo DNA was transfected into lung cancer cells (PC9/GR). After 48 h of incubation, cell fate was analyzed by flow cytometry (FACS analysis). The graph represents the estimated percentage of apoptotic cells determined in the FACS analysis. *P < 0.05 and **P < 0.01 (B) Lung cancer cells (PC9/GR) and liver cancer cells (HepG2) were seeded onto 24-well plates, and each oligo DNA (100 nM final concentration) and EGFR siRNA (50 nM) were transfected into the cells followed by incubation for 48 h. Cell viability was assessed using the methylene blue assay. Data are the mean ± S.D. of three independent experiments; *P < 0.05 and **P < 0.01 vs. the oligo dT group.
Next, we determined whether the oligodeoxynucleotide effect on cancer cell viability occurred in other cancer cells (i.e., hepatoma cells). As shown in Fig. 3B, cell viability was measured in lung cancer cells and hepatoma cells transfected with the oligonucleotides used in the flow cytometry experiment. The cell viability results were similar to those observed in the flow cytometry analysis of lung cancer epithelial cells (PC9/GR); both DNAzymes (scrambled Dz and Ex19del Dz) and CpG-DNA caused significant cell death, while oligo dT did not. In HepG2 hepatoma cells, CpG-dependent apoptosis was less obvious than that observed in the lung cancer cells. This result indicates that the cell death via innate immunity exerted by the CpG-DNA was more pronounced in lung cancer epithelial cells. This explanation is further supported by the fact that human lung cancer cells actively express the CpG-DNA receptor TLR9, which can recognize CpG-DNA on the ER membrane (22). More importantly, the viability of PC9/GR cells was reduced more by Ex19del Dz than by EGFR siRNA. Based on these results, we suggest that Ex19del Dz possesses a secondary action in addition to the suppression of EGFR expression compared with EGFR siRNA.
CpG-DNAzyme upregulates TLR9 expression in lung cancer cells
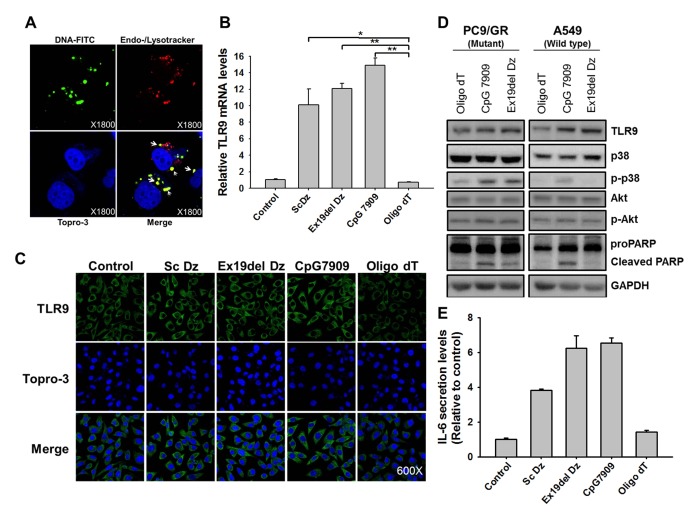
The CpG site in oligonucleotides is known to be recognized by TLR9, which is located on the ER membrane, and induces an innate immune response through cytokine production (21, 23). To observe the co-localization of transfected Dz and endosomes expressing TLR9, Ex19del Dz was labeled with FITC, and lysosomes were stained with a specific fluorescent dye. When FITC-labeled Ex19del Dz was transfected into cells, it mostly co-localized with lysosomes, visualized as yellow-colored fluorescence in the merged image (Fig. 4A). Thus, we assumed that Dz surrounded by a liposome was imported into the cell cytosol via endocytosis and moved to the lysosome, suggesting that Dz containing the CpG sites may recognize and activate TLR9.
Fig. 4.
CpG-DNAzyme induces an increase in TLR9 expression and subsequent p38 MAPK activation. (A) FITC-labeled Ex19del Dz (100 nM, green fluorescence) was transfected into lung cancer cells for 6 h, and the cells were fixed in 4% paraformaldehyde and imaged under a fluorescence confocal microscope. After fixation, endosomes and lysosomes were stained with Endo-/Lyso tracker dye (red fluorescence). Nuclei were stained with TOPRO-3 (blue fluorescence). Merged images indicate the co-localization of FITC-labeled DNAzyme and endosomes/lysosomes, as shown in yellow-colored fluorescence (indicated with arrows). (B) Oligo DNAs (100 nM) were transfected into PC9/GR lung cancer cells (5 × 105 cells). At 48 h after transfection, total RNAs were obtained for analysis. Real-time PCR for the detection of TLR9 cDNA was performed in oligo DNA-transfected cells, as described in the graph. cDNA amplified from TLR9 transcripts was relatively quantified by normalization to control cells that were untreated. *P < 0.01 and **P < 0.005 vs. the oligo dT group. (C) Confocal microscopic images of PC9/GR and HepG2 cells that were transfected with 100 nM individual oligo DNA as indicated. At 48 h after transfection, cells were fixed in 4% paraformaldehyde and incubated with primary antibody against TLR9. After 12 h of incubation, goat anti-rabbit IgG with fluorophore was used as the secondary antibody (green fluorescence). Nuclei were counterstained using TOPRO-3. (D) Oligo DNAs (100 nM) were transfected into lung cancer cells (PC9/GR and A549) for 6 h. After 24 h of incubation, total cellular proteins (30 μg) were obtained and resolved by 10% SDS-PAGE. In addition to probing PARP cleavage, TLR9, Akt, p38, and their phosphorylated forms were probed by western blot analysis. (E) Oligo DNAs (100 nM) were transfected into lung cancer cells (PC9/GR) for 24 h. Cell culture supernatants were harvested and 100 μl of supernatants was used for ELISA to detect IL-6. Bar graphs show the mean ± S.D. of three independent experiments, and the values were normalized to those of the non-treated control.
To compare the effects exerted by each oligo on TLR9 mRNA levels, we transfected each oligo DNA and siRNA into PC9/GR cells and quantitated TLR9 mRNA (Fig. 4B). The amount of TLR9 mRNA was decreased up to 50% by TLR9 siRNA. However, in the cells transfected with Ex19del Dz, the amount of TLR9 mRNA increased by about 12 times compared with that in the non-treated control cells 48 h after transfection. Sc Dz and CpG 7909 upregulated TLR9 transcripts by about 10 and 15 times, respectively. As expected, transfection of oligo dT into the cells did not increase TLR9 mRNA levels.
The results showing enhanced TLR9 expression were further supported by TLR9 protein levels as examined by immunocytochemistry (Fig. 4C). After PC9/GR cells were transfected with each oligonucleotide for 6 h, the expression of TLR9 was significantly elevated by CpG-containing oligo DNAs, such as Sc Dz, Ex19del Dz, and CpG 7909. Based on these results, we suggest that Ex19del Dz upregulated TLR9 expression in the lung cancer epithelial cells because of the presence of CpG sites in the DNAzyme. The elevated level of TLR9 was responsible for the activation of the innate immune response, which caused apoptotic cell death. Thus, Ex19del Dz, which cleaves the mutant EGFR mRNA, has an auxiliary immunostimulatory effect due to the CpG site, and it was named CpG-DNAzyme.
CpG-DNAzyme activates p38 MAP kinase and IL-6 secretion through TLR9 signaling
To determine whether the CpG DNAzyme can activate the p38 MAPK and/or AKT pathway in lung cancer cells, subsequently leading to apoptosis, the activation (i.e., phosphorylation) of p38 and Akt was examined after transfection of each DNA into lung cancer cells (Fig. 4D). In PC9/GR cells, the levels of phosphorylated p38 were 1.8-times higher in cells transfected with the CpG-containing DNAs (Ex19del Dz and CpG 7909) than in cells transfected with oligo dT. CpG oligonucleotide induced a significant enhancement in p38 phosphorylation and subsequent proPARP cleavage in both cell groups. However, there was no detectable increase in Akt phosphorylation with CpG-containing DNAs. Interestingly, the CpG DNAzyme targeting the mutant EGFR gene resulted in p38 activation and proPARP cleavage only in PC9/GR cells containing the mutant EGFR gene, not in the other lung cancer cells (A549 cells) harboring the wild-type EGFR gene. This result indicates that CpG-containing DNAs induced apoptosis in lung cancer cells by activating the p38 MAPK pathway but not the AKT pathway. More importantly, Ex19del Dz induced an apoptotic response by activating p38 and PARP cleavage only in the EGFR mutant lung cancer cells, suggesting that CpG-DNAzyme targeting EGFR induces apoptotic cell death by EGFR knockdown in addition to the CpG-induced TLR9 activation.
It has been reported that the secretion of IL-6 is mediated by NF-κB signaling through TLR9 activation (24). Thus, we next examined CpG-dependent IL-6 production using an ELISA. As shown in Fig. 4E, after treatment with CpG motifs containing DNA (i.e., Sc Dz, Ex19del Dz, and CpG 7909) IL-6 was released at significantly higher levels than those in the non-treated control or oligo dT groups. Importantly, Ex19del Dz activated IL-6 secretion to a similar extent as CpG 7909, which was used as a TLR9 agonist. Taken together, we demonstrated that CpG sites in Ex19del Dz induced the activation of the TLR9-p38 MAPK pathway and cytokine secretion in PC9/GR cells.
In conclusion, Ex19del Dz targeting the exon 19 deletion in mutant EGFR RNA has dual effects on apoptosis in lung cancer cells by downregulating EGFR through a specific RNA-cleaving activity and nonspecifically activating TLR9-p38 MAPK through CpG sequences. This RNA-cleaving DNAzyme with dual effects caused a stronger suppression of NSCLC proliferation than siRNA. We thus suggest that the Ex19del DNAzyme can be used as a therapeutic agent that acts by inducing apoptosis in lung cancer cells through CpG sequences in DNA and knocking down EGFR through cleavage of transcripts.
MATERIALS AND METHODS
Cell culture and oligonucleotide transfection, RNA cleavage with DNAzyme, Cell proliferation assay, Western blot analyses, Flow cytometry, RT-PCR, Immunocytochemistry, and ELISA methods are described in “Supplementary information”.
Supplementary Information
ACKNOWLEDGEMENTS
This work was supported by a grant from the Ministry of Health and Welfare, Republic of Korea (HI15C2917).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 2.Ciardiello F, Tortora G. A novel approach in the treatment of cancer: targeting the epidermal growth factor receptor. Clin Cancer Res. 2001;7:2958–2970. [PubMed] [Google Scholar]
- 3.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 4.Riely GJ, Politi KA, Miller VA, Pao W. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12:7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- 5.Paez J, Jänne P, Lee J, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dass CR, Choong PF, Khachigian LM. DNAzyme technology and cancer therapy: cleave and let die. Mol Cancer Ther. 2008;7:243–251. doi: 10.1158/1535-7163.MCT-07-0510. [DOI] [PubMed] [Google Scholar]
- 8.Kim JE, Yoon S, Choi BR, et al. Cleavage of BCR-ABL transcripts at the T315I point mutation by DNAzyme promotes apoptotic cell death in imatinib-resistant BCR-ABL leukemic cells. Leukemia. 2013;27:1650–1658. doi: 10.1038/leu.2013.60. [DOI] [PubMed] [Google Scholar]
- 9.Santoro S, Joyce G. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci U S A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khachigian L. Catalytic DNAs as potential therapeutic agents and sequence-specific molecular tools to dissect biological function. J Clin Invest. 2000;106:1189–1195. doi: 10.1172/JCI11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman SK. Catalytic DNA (deoxyribozymes) for synthetic applications-current abilities and future prospects. Chem Commun (Camb) 2008:3467–3485. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]
- 12.Appaiahgari MB, Vrati S. DNAzyme-mediated inhibition of Japanese encephalitis virus replication in mouse brain. Mol Ther. 2007;15:1593–1599. doi: 10.1038/sj.mt.6300231. [DOI] [PubMed] [Google Scholar]
- 13.Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11(Suppl 1):S10–17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 14.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Aderem A, Ulevitch R. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 16.Ahmad-Nejad P, Häcker H, Rutz M, Bauer S, Vabulas R, Wagner H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur J Immunol. 2002;32:1958–1968. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 18.Lee KJ, Kim YK, Krupa M, et al. Crotamine stimulates phagocytic activity by inducing nitric oxide and TNF-α via p38 and NFκ-B signaling in RAW 264.7 macrophages. BMB Rep. 2016;49:185–190. doi: 10.5483/BMBRep.2016.49.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YH, Girardi M, Duvic M, et al. Phase I trial of a Toll-like receptor 9 agonist, PF-3512676 (CPG 7909), in patients with treatment-refractory, cutaneous T-cell lymphoma. J Am Acad Dermatol. 2010;63:975–983. doi: 10.1016/j.jaad.2009.12.052. [DOI] [PubMed] [Google Scholar]
- 20.da Rosa MR, Falcao AS, Fuzii HT, et al. EGFR signaling downstream of EGF regulates migration, invasion, and MMP secretion of immortalized cells derived from human ameloblastoma. Tumour Biol. 2014;35:11107–11120. doi: 10.1007/s13277-014-2401-3. [DOI] [PubMed] [Google Scholar]
- 21.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 22.Droemann D, Albrecht D, Gerdes J, et al. Human lung cancer cells express functionally active Toll-like receptor 9. Respir Res. 2005;6:1. doi: 10.1186/1465-9921-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latz E, Verma A, Visintin A, et al. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 24.Tsujimura H, Tamura T, Kong HJ, et al. Toll-Like Receptor 9 Signaling Activates NF-κB through IFN Regulatory Factor-8/IFN Consensus Sequence Binding Protein in Dendritic Cells. J Immunol. 2004;172:6820–6827. doi: 10.4049/jimmunol.172.11.6820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.