Abstract
Ion-mobility mass spectrometry (IM-MS) is an approach that can provide information on the stoichiometry, composition, protein contacts and topology of protein complexes. The power of this approach lies not only in its sensitivity and speed of analysis, but also in the fact that it is a technique that can capture the repertoire of conformational states adopted by protein assemblies. Here, we describe the array of available IM-MS based tools, and demonstrate their application to the structural characterization of various protein complexes, including challenging systems as amyloid aggregates and membrane proteins. We also discuss recent studies in which IM-MS was applied towards investigations of conformational transitions and stabilization effects induced by protein interactions.
Introduction
Proteins are inherently dynamic entities that sample multiple conformational states for their functional activities [1,2]. Therefore, a complete understanding of the structure–function relationships of proteins requires experimental methods that can capture the spread of the conformational states they adopt. However, this l complexity can present a significant challenge to many of the ‘classical’ high-resolution structural biology tools as X-ray crystallography [3], Nuclear Magnetic Resonance (NMR) [4] and Electron Microscopy (EM) [5]. Here, we will focus on a structural technique that can capture the conformational dynamics of a system, Ion-Mobility Mass Spectrometry (IM-MS). While IM-MS does not provide atomic resolution structures , it has the advantage that co-existing populations of a given assembly can be detected within a single spectrum.
IM-MS is a method that couples MS measurements with IM separation (recent reviews include [6–13]. By means of this method, the time it takes for a protein (or its various populated structural states) to transverse a weak electrical gradient in a gas-filled chamber is measured. The drift time depends not only on the mass and charge, but also on the shape of the analyzed protein complex. Larger ions collide more frequently with the neutral gas, hindering their progress and therefore increasing their drift time relative to more compact ions [14] (Figure 1a). This offers a way to distinguish between conformational states of the same protein. As the number of collisions is proportional to the surface area of the protein, the drift time measurement can be used to determine the rotationally averaged collision cross-section (CCS) value, indicating the three-dimensional shape of the protein. For intact proteins and protein complexes introduced from non-denaturing solutions (native IM-MS), this approach provides insights into the stoichiometry, composition, connectivity, topology and conformational heterogeneity of the analyzed biomolecules.
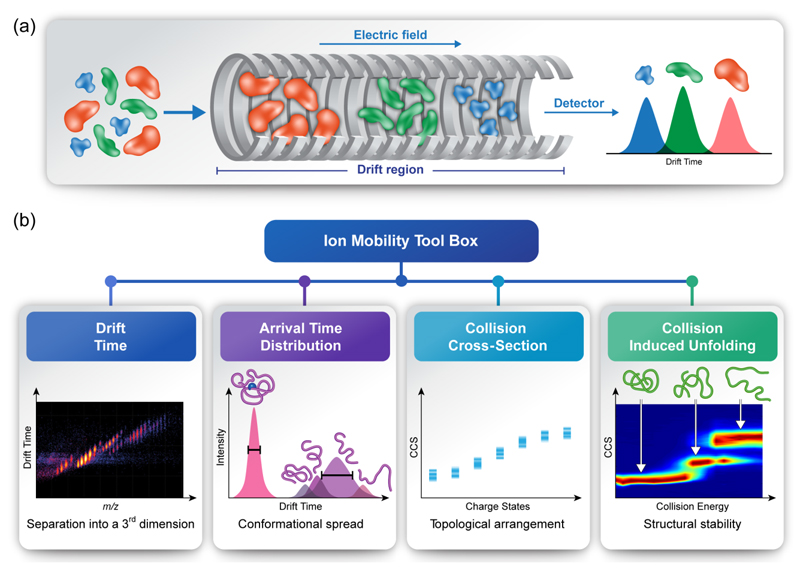
Figure 1. Structural characterization of protein complexes by IM-MS.
(a) Ion mobility is a method that measures the time it takes ions to travel through a gas-filled chamber, across which a weak electric field is applied. In the drift tube, ions experience two counter forces of electrostatic pulling through the cell and collisions with the buffer gas, which delay their movement. Ions with a larger surface area will experience more collisions with the buffer gas and, as a result, will take longer to traverse the drift tube, in comparison to smaller, more compact ions of the same molecular mass and charge, which will undergo fewer collisions with the buffer gas, and hence will display greater mobility and a shorter drift time. (b) IM-MS measurements enable the extraction of various structural properties. For example, mobility-based separation adds a third dimension to native MS analyses, and resolves heterogeneous samples that contain closely related species. Measurements of arrival time distributions reflect the conformational range of the examined species, and can be converted to CCS values. When measured over a range of collision energies, as is characteristic of CIU protocols, CCS can reflect the stabilities and intermediate folding states of the different protein species while they undergo in vacuo unfolding.
Different methods for applying the electric field and introducing the buffer gas gave rise to various IM-MS platforms, such as drift tube (DT) [11], traveling wave (TW) [10], differential mobility [15], transversal modulation [16], overtone [17], field asymmetric [18] and trapped IM-MS [19]. While each method has proven its worth, here we will focus on the two most common types of mobility techniques DT and TW. In DT, a homogeneous, linear electric field is used, and CCS values are determined directly from the measured drift time and the experimental conditions applied [11]. In TW, on the other hand, potential waves continually propagate through the drift tube [10]. Manipulation of the travelling wave frequency and height enables ion separation. Although this method enhances the duty cycle, absolute CCS values cannot be determined, and an indirect calibration procedure is required [20,21]. To date, both DT and TW IM-MS instruments utilizing a quadrupole time-of-flight platform (QTOF) are commercially available, and the application of the technique to structural studies is rapidly increasing [22,23].
Overall, when applying IM-MS, multiple features can be extracted, enabling in-depth analysis (Figure 1b), among them:
Drift Time – Separating ions according to their drift time adds an extra dimension to conventional MS measurements, yielding a three-dimensional spectrum containing information regarding the mass-to-charge (m/z) ratio, abundance of ions, and drift time. Consequently, the ion mobility capability not only provides structural information, but also enables to separate overlapping peaks by distributing the data into a third dimension, thereby allowing analysis of heterogeneous or polydisperse complexes of markedly similar composition.
Arrival Time Distribution – The structural heterogeneity of proteins and complexes is reflected in the full width at half maximum of the arrival time distributions (ATDs). Sharp peaks indicate a single conformation, while broader peaks are consistent with multiple states, indicating conformational flexibility of the protein assembly in solution. Thus, the impact of protein interactions and/or substrate binding on the conformational spread of the analyzed protein species may be determined.
Collision Cross-Section – By measuring the ATD of each charge state, it is possible to calculate, either directly (DT) or indirectly (TW), CCS values [11,20,21]. The derived CCS represents the effective area of the gas-phase ion that can interact with the drift gas, averaged over all orientations. Thus, it can be related to structural features of the ion. Considering that theoretical CCS values can be calculated for protein structure models [24,25], experimentally determined CCS can be utilized to help discriminate between different possible models. These in silico calculations are based on a variety of approximations as to what happens experimentally. The earliest developed [26] and most commonly used, is the projection approximation algorithm (PA), which essentially considers the ‘shadow’ that a given molecule would cast on a flat surface as representative of its collision cross section. In order to apply this approach to real molecules, and to the real effect of collisions on their rugged surfaces, empirical scaling factors derived from experiments have been introduced [27]. More accurate treatments calculate the momentum transferred as well as the effect of the interaction potential between the analyte ion and the buffer gas [24,25,28], but such more rigorous approaches require high computational power and are less well trained on large protein complexes.
Collision Induced Unfolding – The conformational stability and domain organization of intact proteins can be analyzed by collision induced unfolding (CIU) experiments. In this type of experiment, the collision energy (CE) is elevated in a stepwise manner, causing protein activation that may consequently induce conformational change or partial unfolding. As the activation step takes place before the drift tube, CCS values of compact and extended structures can be obtained for a single m/z value, enabling detection and quantification of conformational changes. Usually, the collision voltage at which the transitions between conformations occur, and the CCS values of the intermediate states, are evaluated and compared. Effects of ligand binding or association with protein partners may be visualized by changes in the CIU fingerprint. Similarly, protein isoforms that originate from various diversification mechanisms (i.e., alternative splicing, gene duplication, post-translational modifications) are expected to yield distinct CIU trajectories, providing insights into their different structural characteristics.
Recent studies demonstrating how the IM-MS toolbox might be harnessed for studying the structural properties of protein complexes are detailed below:
Characterization of protein structure
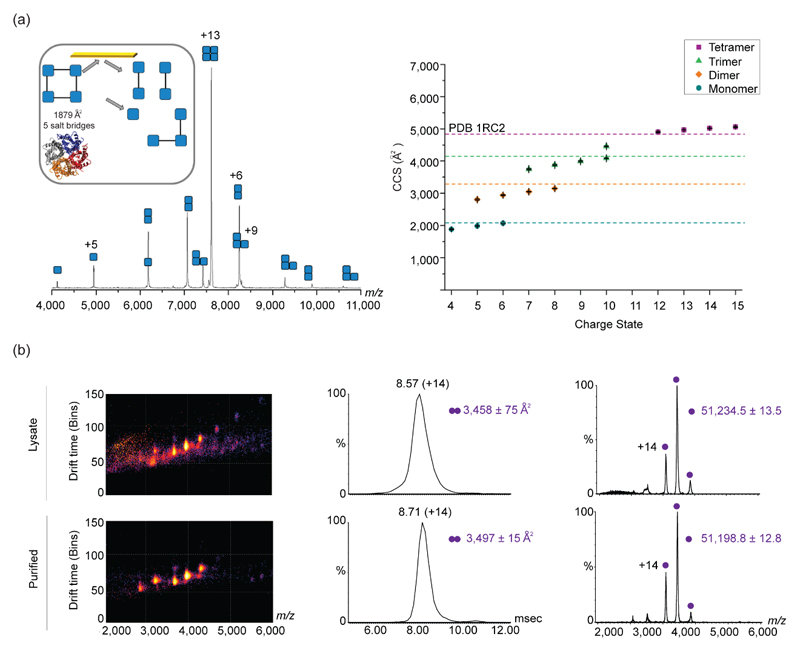
Understanding how protein structure is affected by a single amino acid substitution [29,30] or post-translational modification [31]•, are among the examples to which IM-MS has made valuable contributions. Such comparative structural studies are further enhanced by applying the CIU protocol, capable of probing the different structural characteristics of species with identical drift times. This was recently demonstrated for antibody isoforms possessing different numbers and/or patterns of disulfide bonding and glycosylation levels [32]. Moreover, by combining IM-MS with surface-induced dissociation (SID), which enables the selective disruption of the weaker interfaces of soluble protein complexes, it is possible to glean information on the native structure of both the intact assembly, and its subcomplexes [33]. The application of SID IM-MS has recently been extended to membrane protein complexes, demonstrating that the structure of such complexes and their generated subproducts, are retained within the mass spectrometer, even after having been liberated from detergent micelles [34]• (Figure 2a). A further advancement came with the realization that IM-MS analysis of recombinant protein complexes can be performed directly on crude cell lysates, saving the time, cost and labor of protein purification, and paving the way toward throughput IM-MS screening of engineered proteins (Figure 2b) [35]•.
Figure 2. Applying IM-MS for structural characterization of soluble and membrane proteins.
(a) Application of SID with IM-MS for the study of membrane proteins is shown for AqpZ. The SID spectrum for the 13+ charge state of AqpZ, shows the different subcomplexes that were dissociated after collision with the surface. The inset represents the structure of AqpZ and the interfacial analysis, as determined from the PISA software. The crystal structure (PDB 1RC2) is shown below. The right graph shows the CCS distribution of the different sub-complexes of AqpZ. Theoretical CCS values of the different structures, represented as dashed lines, were determined using MOBCAL and the scaled PA. Adapted with permission from [21]•. (b) IM-MS analysis of recombinant proteins directly from crude cell lysates produces comparable information to that obtained from purified proteins, as exemplified for Hsp31. The three-dimensional IM-MS spectra of the crude cell lysate and purified samples are shown on the left, while the arrival time distribution and two dimensional spectra of m/z over intensity are shown on the middle and right panels, respectively. Adapted with permission from [22]•.
On the whole, IM-MS has proven to be a reliable technique that is capable of detecting even subtle structural differences. Nevertheless, it is important to note that different factors may affect the structure of gas phase ions during analysis, and these must be taken into consideration when performing such measurements, and particularly when extracting CCS values. For example, in TW IM-MS experiments, care must be taken when choosing the appropriate calibrants, as the latter should fit the nature and character of the analyzed proteins [36,37]. Attention should also be given to the phenomenon of gas-phase compaction [38], particularly in instances of elongated, non-globular and intrinsically unstructured proteins, or proteins with flexible hinge regions in between structured domains [39,40]. Temperature constitutes an additional factor that may influence the conformational preferences of proteins [41] and protein complexes [42]•. Overall, when acquiring IM-MS data, especially when uncharacterized protein assemblies are studied, we recommend to define first the optimal experimental conditions, in order to ensure preservation of the native-like protein conformations, as previously described [38].
Analysis of protein interactions
The ability to capture multiple coexisting states within a single spectrum has turned IM-MS into a valuable tool for characterizing protein-protein and protein-ligand interactions. Recent examples include epitope mapping of antibody-antigen interactions [43]•, analysis of the changes in structure and stability imposed by protein-ligand binding [44–46], and real-time monitoring of the effect of small-molecule inhibitors on the conformational distribution and activity of enzymes [47]••.
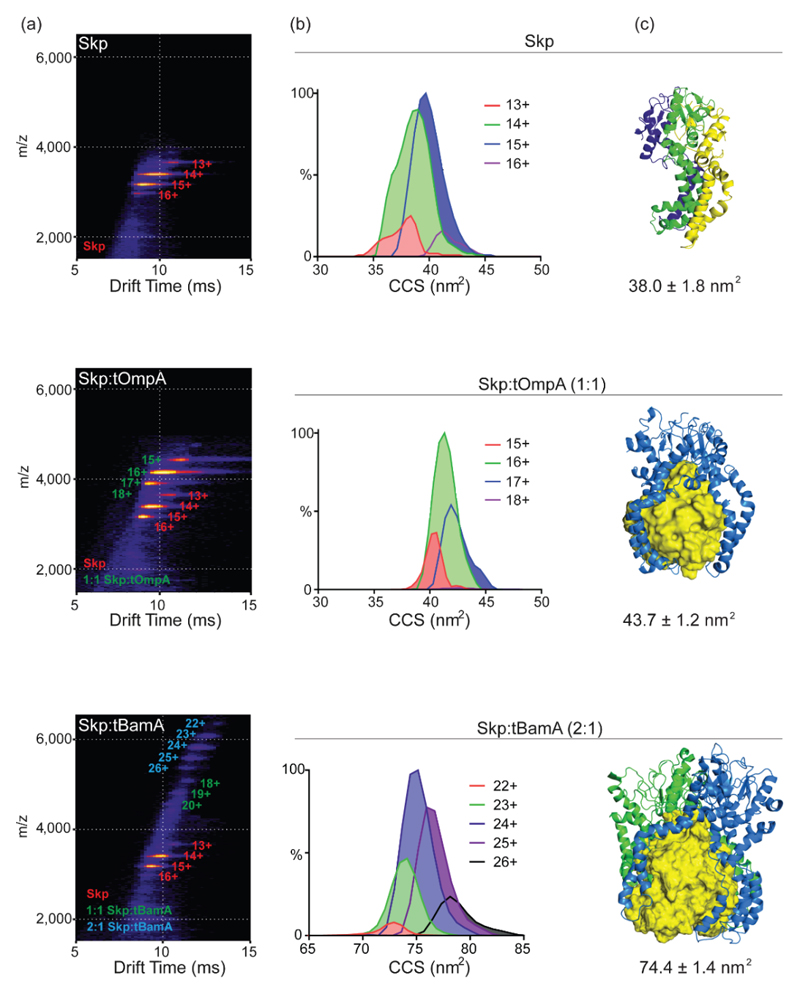
The investigation of the holdase chaperone Skp, and its interactions with a range of outer membrane proteins (OMPs) differing in size, not only serves as an additional, elegant example of the application of IM-MS, but also underscores the integration of the method with computer modeling and molecular dynamics simulations [48]•• (Figure 3). This study indicated that the CCS distribution of Skp in its free state is smaller than would be estimated by its crystal structure, demonstrating that the assembly collapses in the gas phase, possibly because the central cavity of the chaperone is empty. However, Skp binding to an 8-stranded OMP client resulted in CCS values comparable to that predicated by its crystal structure, suggesting that the client, which is sequestered within the central Skp cavity, prevents collapse of the chaperone in the gas phase. For larger OMPs containing 16-strands, which cannot be fully accommodated in the expanded cavity, it was demonstrated that sequestration is achieved by binding of an additional Skp complex to the substrate. Overall, the study provided a novel understanding of the mechanism by which Skp is able to bind, fold and release substrates that vary dramatically in size.
Figure 3. Investigation of the multivalent chaperone of outer membrane proteins.
(a) IM-MS plots of Skp alone, Skp bound with an 8-stranded (tOmpA) or with a 16-stranded (tBamA) OMP, the latter is larger than the Skp central cavity. Charge states corresponding to Skp, 1:1 Skp:OMP and 2:1 Skp:OMP are depicted on the plots in red, green and blue, respectively. (b) Representative CCS distributions of the major populations in each sample. Peak heights were normalized to MS peak intensities. (c) Representative structures from MD simulations obtained after 10 ns of in vacuo simulation. Skp is represented in cartoon format and OMPs are depicted as yellow surfaces. In the top panel, the different Skp subunits are colored green, blue and yellow. In the bottom panel, two Skp complexes, colored in blue and green are in complex with the OMP. Theoretical CCS values obtained by the molecular dynamics simulations are shown below. Adapted with permission from [48]••.
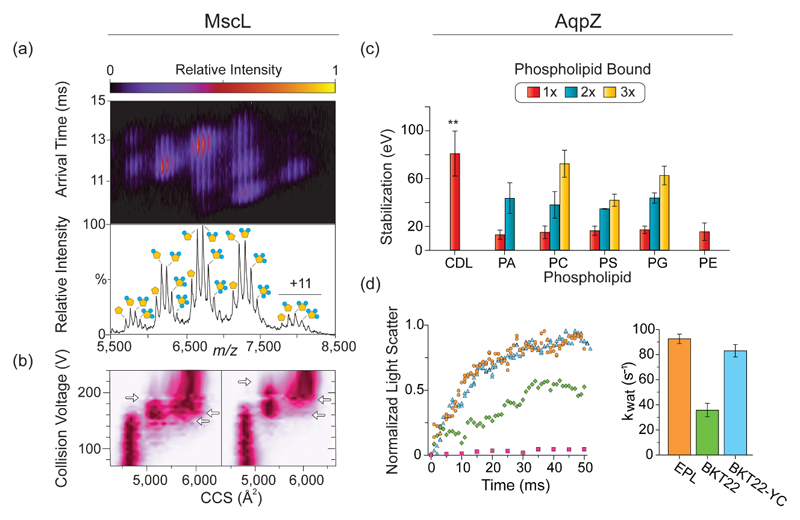
The ability to monitor the effects of ligand binding on protein stability is not limited to small molecules or globular proteins: indeed a method [49] complemented with software [50]•• for quantitative analysis of lipid binding to membrane proteins was recently reported. In this approach, membrane proteins solubilized in different types of lipids were sprayed into the mass spectrometer and changes in mass and CIU plots were quantified, to assess the effects of these interactions on protein stability. Results indicated that specific lipids stabilize particular membrane proteins in different ways (Figure 4). Moreover, comparison of the IM-MS approach with solution-phase techniques, as circular dichroism and differential scanning fluorimetry, demonstrated that this method is more sensitive, especially for lipid interactions with membrane proteins.
Figure 4. Quantification of the stabilizing effects of protein-lipid interactions by IM-MS.
(a) IM-MS reveals that several phosphatidylinositol phosphate (PI) lipids bind to MscL. The corresponding mass spectrum is shown below. (b) CIU diagrams of collision voltage, plotted against CCS, for the +12 ions of apo MscL (left), and MscL bound to 4 PI molecules (right), show the differences in gas-phase stability of the unfolding intermediates. Collision voltages at which transitions occur are shown by the horizontal arrows. (c) Quantification of AqpZ stabilization by various lipids, as calculated from CIU parameters. Results show that cardiolipin (CDP) has the strongest stabilizing effect on AqpZ. (d) The stabilization effect of CDP was validated by a functional assay that probed the water permeability of AqpZ reconstituted into liposomes containing various lipids: total polar lipid extracts from E. coli (EPL) (orange); cardiolipin-deficient lipid extracts (BKT22) (green); lipid extracts in which cardiolipin was restored (BKT22-YC) (cyan); and empty liposomes (EPL) (pink). The calculated rate constants of water transport (Kwat) ± SE are shown on the right. Adapted with permission from [49].
Amyloid structure and assembly
In the last decade, IM-MS has become an important tool in the investigation of amyloids, which are challenging systems due to their polydispersity, polymorphic, and transient nature. The advantage of IM-MS in studying these systems lies in its ability to distinguish and separate the different oligomeric species, without affecting the equilibrium of the ensemble [51,52]. Several recent examples include investigations into the effect of zinc binding on the aggregation of amylin [53], the mechanism of inhibition of amyloid β aggregation [54], and the regulatory mechanism that controls higher-order oligomerization of the microtubule-associated protein tau during its normal and pathological activities [55]. Another interesting study investigated the aggregation profiles of the homotetrameric protein transthyretin, purified directly from human blood serum. Transthyretin is known to misfold, aggregate, and cause different types of amyloidosis. IM-MS measurements pointed to differences in the assembly state and stability of this protein between healthy individuals and symptomatic patients suffering from this disease [56]•.
IM-MS analysis has also been also used to investigate the formation of heterogeneous pre-fibrillar, oligomeric species produced by the co-incubation of the amyloid-β peptide (Aβ-40) with the human islet amyloid polypeptide [57]. Comparison between the conformations, dynamics and relative gas-phase stabilities of homo- and hetero- assembled species showed that cross-polymerization resulted in the formation of unique structures, with distinct degrees of stability and oligomerization rates. Interestingly, the measured stabilities for the hetero-oligomeric species were found to lie between the values measured for each of the homo-oligomers, suggesting that mixed species may exert significant effects on the progress of fibril formation, and thus may have biological consequences in vivo.
Recently, IM-MS analysis has been taken a step further by combining IM-MS with gas-phase infrared (IR) spectroscopy [58]••. The benefit of this hybrid approach is that it enables acquisition not only of information concerning tertiary/quaternary, but also of secondary structures. This method, for example, has been used to resolve the secondary structures of oligomers formed by amyloidogenic peptides. The analysis showed that higher-order oligomers exist in different conformations, with CCS values ranging between compact spherical aggregates through extended oligomers. Conformer-specific IR analysis revealed a correlation between the size increase and β-sheet content, and pinpointed the onset of this structural transition from compact and unordered, to an extended β-sheet structure. Overall, this combined IM-MS technique offers novel means for examining the secondary structural transitions of self-assembled systems.
Conclusions
The variety of studies presented herein, covering diverse biological systems, demonstrate the strength of IM-MS. This method may be applied not only to investigations of soluble protein complexes, but also to analysis of challenging systems that are often resistant to crystallization, such as membrane proteins and amyloid aggregates. Considering that, by means of this technique, all co-existing populations can be probed in a single spectrum, its main advantages lie not only in examining complex samples containing mixtures of proteins, but also in its ability to probe binding assays and assembly pathways, while quantifying the conformational heterogeneity and relative stabilities of each species identified. Owing to the ease with which this method can be implemented, and the recent development of multiple software packages such as PULSAR [50], Amphitrite [59], ORIGAMI [60] or CIUsuite [61], all of which enable automated analysis of large and complex datasets, we anticipate that the applications of IM-MS will greatly increase.
Moreover, it is expected that in the future, the IM-MS method will be expanded to include high resolution instruments such as FTICR and Orbitrap platforms, paving the way toward defining mobility times of different post-transnationally modified species of a single protein assembly, alongside MSn analyses [62,63]. This task is not trivial, however, due to the slow acquisition rate of these instruments, in comparison to the IM separation time. Nevertheless, progress in this direction is already being made [64]. An additional, promising direction involves the integration of IM-MS with other gas-phase techniques that can provide complementary structural information, such as the use of IM-MS as a pre-selection tool to perform gas phase IR [58,65] or FRET [66] measurements, and we expect that additional applications will arise.
Highlights.
IM-MS provides valuable tools for structural characterization of protein complexes.
The spread of conformational states adopted by a protein can be captured by IM-MS
We discuss the application of IM-MS for investigating diverse protein systems
Acknowledgements
We thank P. Barran for helpful discussions. We are grateful for the support of a Starting Grant from the European Research Council (ERC) (Horizon 2020)/ERC Grant Agreement No. 636752, and from the Israel Science Foundation grant (ISF) No. 300/17.
References
- 1.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papaleo E, Saladino G, Lambrughi M, Lindorff-Larsen K, Gervasio FL, Nussinov R. The role of protein loops and linkers in conformational dynamics and allostery. Chem Rev. 2016;116:6391–6423. doi: 10.1021/acs.chemrev.5b00623. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y. A glimpse of structural biology through X-ray crystallography. Cell. 2014;159:995–1014. doi: 10.1016/j.cell.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 4.Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez-Leiro R, Scheres SH. Unravelling biological macromolecules with cryo-electron microscopy. Nature. 2016;537:339–346. doi: 10.1038/nature19948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewing MA, Glover MS, Clemmer DE. Hybrid ion mobility and mass spectrometry as a separation tool. J Chromatogr A. 2016;1439:3–25. doi: 10.1016/j.chroma.2015.10.080. [DOI] [PubMed] [Google Scholar]
- 7.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 8.May JC, McLean JA. Ion mobility-mass spectrometry: time-dispersive instrumentation. Anal Chem. 2015;87:1422–1436. doi: 10.1021/ac504720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyttenbach T, Pierson NA, Clemmer DE, Bowers MT. Ion mobility analysis of molecular dynamics. Annu Rev Phys Chem. 2014;65:175–196. doi: 10.1146/annurev-physchem-040513-103644. [DOI] [PubMed] [Google Scholar]
- 10.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 11.Mason EA, S HW., Jr Mobility of gaseous lons in weak electric fields. Ann Phys (NY) 1958;4:233–270. [Google Scholar]
- 12.Zheng X, Wojcik R, Zhang X, Ibrahim YM, Burnum-Johnson KE, Orton DJ, Monroe ME, Moore RJ, Smith RD, Baker ES. coupling front-end separations, ion mobility spectrometry, and mass spectrometry for enhanced multidimensional biological and environmental analyses. Annu Rev Anal Chem (Palo Alto Calif) 2017;10:71–92. doi: 10.1146/annurev-anchem-061516-045212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konijnenberg A, Butterer A, Sobott F. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim Biophys Acta. 2017;1834:1239–1256. doi: 10.1016/j.bbapap.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to clusters and biomolecules. J Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 15.Rus J, Moro D, Sillero JA, Royuela J, Casado A, E-M F, de la Morab JF. IMS–MS studies based on coupling a differential mobility analyzer (DMA) to commercial API–MS systems. IJMS. 2010;298:30–40. [Google Scholar]
- 16.Vidal-de-Miguel G, Macia M, Barrios C, Cuevas J. Transversal modulation ion mobility spectrometry (IMS) coupled with mass spectrometry (MS): exploring the IMS-IMS-MS possibilities of the instrument. Anal Chem. 2012;87:1925–1932. doi: 10.1021/ac504178n. [DOI] [PubMed] [Google Scholar]
- 17.Zucker SM, Ewing MA, Clemmer DE. Gridless overtone mobility spectrometry. Anal Chem. 2013;85:10174–10179. doi: 10.1021/ac401568r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown LJ, Creaser CS. Field asymmetric waveform ion mobility spectrometry analysis of proteins and peptides: a review. Curr Anal Chem. 2013;9:192–198. [Google Scholar]
- 19.Michelmann K, Silveira JA, Ridgeway ME, Park MA. Fundamentals of trapped ion mobility spectrometry. J Am Soc Mass Spectrom. 2015;26:14–24. doi: 10.1007/s13361-014-0999-4. [DOI] [PubMed] [Google Scholar]
- 20.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 21.Smith DP, Knapman TW, Campuzano I, Malham RW, Berryman JT, Radford SE, Ashcroft AE. Deciphering drift time measurements from travelling wave ion mobility spectrometry-mass spectrometry studies. Eur J Mass Spectrom (Chichester) 2009;15:113–130. doi: 10.1255/ejms.947. [DOI] [PubMed] [Google Scholar]
- 22.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, et al. Conformational ordering of biomolecules in the gas phase: nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem. 2014;86:2107–2116. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pringle SD, Giles KW, Jason L, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int J Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 24.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural information from ion mobility measurements: effects of the long-range potential. J Phys Chem. 1996;100:16082–16086. [Google Scholar]
- 25.Shvartsburg AA, Jarrold MF. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem Phys Lett. 1996;261:86–91. [Google Scholar]
- 26.Mack E. Average cross-sectional areas of molecules by gaseous diffusion methods. J Am Chem Soc. 1925;47:2468–2482. [Google Scholar]
- 27.Bleiholder C, Johnson NR, Contreras S, Wyttenbach T, Bowers MT. Molecular structures and ion mobility cross sections: analysis of the effects of He and N2 buffer gas. Anal Chem. 2015;87:7196–7203. doi: 10.1021/acs.analchem.5b01429. [DOI] [PubMed] [Google Scholar]
- 28.Larriba-Andaluz C, Hogan CJ., Jr Collision cross section calculations for polyatomic ions considering rotating diatomic/linear gas molecules. J Chem Phys. 2014;141 doi: 10.1063/1.4901890. 194107. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Nissan G, Chotiner A, Tarnavsky M, Sharon M. Structural Characterization of missense mutations using high resolution mass spectrometry: a case study of the Parkinson's-related protein, DJ-1. J Am Soc Mass Spectrom. 2016;27:1062–1070. doi: 10.1007/s13361-016-1379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurneczko E, Cruickshank F, Porrini M, Clarke DJ, Campuzano ID, Morris M, Nikolova PV, Barran PE. Probing the conformational diversity of cancer-associated mutations in p53 with ion-mobility mass spectrometry. Angew Chem Int Ed Engl. 2013;52:4370–4374. doi: 10.1002/anie.201210015. [DOI] [PubMed] [Google Scholar]
- 31.Lossl P, Brunner AM, Liu F, Leney AC, Yamashita M, Scheltema RA, Heck AJ. Deciphering the Interplay among multisite phosphorylation, interaction dynamics, and conformational transitions in a tripartite protein system. ACS Cent Sci. 2016;2:445–455. doi: 10.1021/acscentsci.6b00053. [•This study examined the impact of phosphorylation on the conformational dynamics and interactions between a kinase and its activator. IM-MS analysis showed that the activator undergoes clear, rapid compaction in response to multisite phosphorylation, critical to the hetero-dimerization between the two proteins.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y, Han L, Buckner AC, Ruotolo BT. Collision induced unfolding of intact antibodies: rapid characterization of disulfide bonding patterns, glycosylation, and structures. Anal Chem. 2015;87:11509–11515. doi: 10.1021/acs.analchem.5b03291. [DOI] [PubMed] [Google Scholar]
- 33.Quintyn RS, Zhou M, Yan J, Wysocki VH. Surface-induced dissociation mass spectra as a tool for distinguishing different structural forms of gas-phase multimeric protein complexes. Anal Chem. 2015;87:11879–11886. doi: 10.1021/acs.analchem.5b03441. [DOI] [PubMed] [Google Scholar]
- 34.Harvey SR, Liu Y, Liu W, Wysocki VH, Laganowsky A. Surface induced dissociation as a tool to study membrane protein complexes. Chem Commun (Camb) 2017;53:3106–3109. doi: 10.1039/c6cc09606a. [•Native IM-MS and surface-induced dissociation (SID) were applied to study integral membrane protein complexes. The results indicated that fragments produced from SID are consistent with the solved structures of these complexes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gan J, Ben-Nissan G, Arkind G, Tarnavsky M, Trudeau D, Noda Garcia L, Tawfik DS, Sharon M. Native mass spectrometry of recombinant proteins from crude cell lysates. Anal Chem. 2017;89:4398–4404. doi: 10.1021/acs.analchem.7b00398. [•This study demonstrates that native IM-MS measurements can be performed directly in crude cell lysates, providing immediate information on the identity, solubility, oligomeric state, overall structure, and stability, as well as ligand binding of protein complexes, without the need for purification.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allison TM, Landreh M, Benesch JLP, Robinson CV. Low charge and reduced mobility of membrane protein complexes has implications for calibration of collision cross section measurements. Anal Chem. 2016;88:5879–5884. doi: 10.1021/acs.analchem.6b00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen SJ, Giles K, Gilbert T, Bush MF. Ion mobility mass spectrometry of peptide, protein, and protein complex ions using a radio-frequency confining drift cell. Analyst. 2016;141:884–891. doi: 10.1039/c5an02107c. [DOI] [PubMed] [Google Scholar]
- 38.Michaelevski I, Eisenstein M, Sharon M. Gas-phase compaction and unfolding of protein structures. Anal Chem. 2010;82:9484–9491. doi: 10.1021/ac1021419. [DOI] [PubMed] [Google Scholar]
- 39.Devine PWA, Fisher HC, Calabrese AN, Whelan F, Higazi DR, Potts JR, Lowe DC, Radford SE, Ashcroft AE. Investigating the structural compaction of biomolecules upon transition to the gas-phase using ESI-TWIMS-MS. J Am Soc Mass Spectrom. 2017 doi: 10.1007/s13361-017-1689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jhingree JR, Bellina B, Pacholarz KJ, Barran PE. Charge mediated compaction and rearrangement of gas-phase proteins: a case study considering two proteins at opposing ends of the structure-disorder continuum. J Am Soc Mass Spectrom. 2017;28:1450–1461. doi: 10.1007/s13361-017-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Baba TJ, Woodall DW, Raab SA, Fuller DR, Laganowsky A, Russell DH, Clemmer DE. Melting proteins: evidence for multiple stable structures upon thermal denaturation of native ubiquitin from ion mobility spectrometry-mass spectrometry measurements. J Am Chem Soc. 2017;139:6306–6309. doi: 10.1021/jacs.7b02774. [DOI] [PubMed] [Google Scholar]
- 42.Ujma J, Giles K, Morris M, Barran PE. New high resolution ion mobility mass spectrometer capable of measurements of collision cross sections from 150 to 520 K. Anal Chem. 2016;88:9469–9478. doi: 10.1021/acs.analchem.6b01812. [•The development of a new variable temperature high resolution IM-MS instrument is described. Novel features of the instrument and results from variable temperature IM-MS measurements on a range of model systems are discussed.] [DOI] [PubMed] [Google Scholar]
- 43.Yefremova Y, Opuni KFM, Danquah BD, Thiesen HJ, Glocker MO. Intact transition epitope mapping (ITEM) J Am Soc Mass Spectrom. 2017;28:1612–1622. doi: 10.1007/s13361-017-1654-7. [•This study describes a method for rapid and accurate determination of protein antigen-derived epitopes, which is based on IM separation and subsequent dissociation of the immune complex by collision induced dissociation.] [DOI] [PubMed] [Google Scholar]
- 44.Byrne DP, Vonderach M, Ferries S, Brownridge PJ, Eyers CE, Eyers PA. cAMP-dependent protein kinase (PKA) complexes probed by complementary differential scanning fluorimetry and ion mobility-mass spectrometry. Biochem J. 2016;473:3159–3175. doi: 10.1042/BCJ20160648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai AL, Clerico EM, Blackburn ME, Patel NA, Robinson CV, Borbat PP, Freed JH, Gierasch LM. Key features of an Hsp70 chaperone allosteric landscape revealed by ion-mobility native mass spectrometry and double electron-electron resonance. J Biol Chem. 2017;292:8773–8785. doi: 10.1074/jbc.M116.770404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehmood S, Marcoux J, Gault J, Quigley A, Michaelis S, Young SG, Carpenter EP, Robinson CV. Mass spectrometry captures off-target drug binding and provides mechanistic insights into the human metalloprotease ZMPSTE24. Nat Chem. 2016;8:1152–1158. doi: 10.1038/nchem.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mironov GG, Clouthier CM, Akbar A, Keillor JW, Berezovski MV. Simultaneous analysis of enzyme structure and activity by kinetic capillary electrophoresis-MS. Nat Chem Biol. 2016;12:918–922. doi: 10.1038/nchembio.2170. [••A method based on kinetic capillary electrophoresis coupled on-line with UV detection and IM-MS is described for simultaneous monitoring in real-time of both enzyme structure and activity.] [DOI] [PubMed] [Google Scholar]
- 48.Schiffrin B, Calabrese AN, Devine PWA, Harris SA, Ashcroft AE, Brockwell DJ, Radford SE. Skp is a multivalent chaperone of outer-membrane proteins. Nat Struct Mol Biol. 2016;23:786–793. doi: 10.1038/nsmb.3266. [••This study examined the interactions of a chaperone with different clients, including those that are too large to fit into its hydrophobic cavity. IM–MS, combined with computer modeling and molecular dynamics simulations, showed that large clients are held within an expanded chamber. Clients that are too big to fit into the extended cavity are sequestered by binding an additional chaperone.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A, Robinson CV. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat Commun. 2015;6:8551. doi: 10.1038/ncomms9551. [••This work describes the development of an IM-MS based method and software for quantifying the stabilizing effects of protein-ligand interactions in the gas phase. The method is based on assessing the effects of these interactions on protein stability through measuring resistance to unfolding, using the CIU approach.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bleiholder C, Bowers MT. The solution assembly of biological molecules using ion mobility methods: from amino acids to amyloid beta-protein. Annu Rev Anal Chem (Palo Alto Calif) 2017;10:365–386. doi: 10.1146/annurev-anchem-071114-040304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann W, von Helden G, Pagel K. Ion mobility-mass spectrometry and orthogonal gas-phase techniques to study amyloid formation and inhibition. Curr Opin Struct Biol. 2017;46:7–15. doi: 10.1016/j.sbi.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Erthal LC, Marques AF, Almeida FC, Melo GL, Carvalho CM, Palmieri LC, Cabral KM, Fontes GN, Lima LM. Regulation of the assembly and amyloid aggregation of murine amylin by zinc. Biophys Chem. 2016;218:58–70. doi: 10.1016/j.bpc.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Kocis P, Tolar M, Yu J, Sinko W, Ray S, Blennow K, Fillit H, Hey JA. Elucidating the Aβ42 anti-aggregation mechanism of action of tramiprosate in Alzheimer's disease: integrating molecular analytical methods, pharmacokinetic and clinical data. CNS Drugs. 2017;31:495–509. doi: 10.1007/s40263-017-0434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feinstein HE, Benbow SJ, LaPointe NE, Patel N, Ramachandran S, Do TD, Gaylord MR, Huskey NE, Dressler N, Korff M, et al. Oligomerization of the microtubule-associated protein tau is mediated by its N-terminal sequences: implications for normal and pathological tau action. J Neurochem. 2016;137:939–954. doi: 10.1111/jnc.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pont L, Benavente F, Vilaseca M, Gimenez E, Sanz-Nebot V. Characterisation of serum transthyretin by electrospray ionisation-ion mobility mass spectrometry: Application to familial amyloidotic polyneuropathy type I (FAP-I) Talanta. 2015;144:1216–1224. doi: 10.1016/j.talanta.2015.07.079. [•This study examined the aggregation profiles of the amyloidosis-forming protein, transthyretin. IM-MS measurements pointed to differences in the assembly state and stability of the protein between healthy individuals and symptomatic patients suffering from the disease.] [DOI] [PubMed] [Google Scholar]
- 57.Young LM, Mahood RA, Saunders JC, Tu LH, Raleigh DP, Radford SE, Ashcroft AE. Insights into the consequences of co-polymerisation in the early stages of IAPP and Abeta peptide assembly from mass spectrometry. Analyst. 2015;140:6990–6999. doi: 10.1039/c5an00865d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seo J, Hoffmann W, Warnke S, Huang X, Gewinner S, Schollkopf W, Bowers MT, von Helden G, Pagel K. An infrared spectroscopy approach to follow beta-sheet formation in peptide amyloid assemblies. Nat Chem. 2017;9:39–44. doi: 10.1038/nchem.2615. [••The first direct secondary-structure analysis of individual amyloid intermediates, using a combination of IM-MS and gas-phase infrared spectroscopy. The results showed that oligomers exist in different conformations, and as their sizes increase, they adopt a higher content of β-sheets. The method could also pinpoint the onset of the structural rearrangement from a compact and unordered, to an extended β-sheet structure.] [DOI] [PubMed] [Google Scholar]
- 59.Sivalingam GN, Yan J, Sahota H, Thalassinos K. Amphitrite: a program for processing travelling wave ion mobility mass spectrometry data. Int J Mass Spectrom. 2013;345–347:54–62. doi: 10.1016/j.ijms.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Migas LG, France A, Bellina B, Barran P. ORIGAMI: a software suite for activated ion mobility mass spectrometry (aIM-MS) applied to multimeric protein assemblies. bioRxiv. 2017 [Google Scholar]
- 61.Eschweiler JD, Rabuck-Gibbons JN, Tian Y, Ruotolo BT. CIUSuite: a quantitative analysis package for collision induced unfolding measurements of gas-phase protein ions. Anal Chem. 2015;87:11516–11522. doi: 10.1021/acs.analchem.5b03292. [DOI] [PubMed] [Google Scholar]
- 62.Ben-Nissan G, Belov ME, Morgenstern D, Levin Y, Dym O, Arkind G, Lipson C, Makarov AA, Sharon M. triple-stage mass spectrometry unravels the heterogeneity of an endogenous protein complex. Anal Chem. 2017;89:4708–4715. doi: 10.1021/acs.analchem.7b00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Loo RRO, Loo JA. Structural characterization of a thrombin-aptamer complex by high resolution native top-down mass spectrometry. J Am Soc Mass Spectrom. 2017 doi: 10.1007/s13361-017-1751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ibrahim YM, Garimella SV, Prost SA, Wojcik R, Norheim RV, Baker ES, Rusyn I, Smith RD. Development of an ion mobility spectrometry-orbitrap mass spectrometer platform. Anal Chem. 2016;88:12152–12160. doi: 10.1021/acs.analchem.6b03027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khanal N, Masellis C, Kamrath MZ, Clemmer DE, Rizzo TR. Glycosaminoglycan analysis by cryogenic messenger-tagging ir spectroscopy combined with IMS-MS. Anal Chem. 2017;89:7601–7606. doi: 10.1021/acs.analchem.7b01467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daly S, Kulesza A, Poussigue F, Simon AL, Min Choi C, Knight G, Chirot F, MacAleese L, Antoineab R, Dugourd P. Conformational changes in amyloid-beta (12–28) alloforms studied using action-FRET, IMS and molecular dynamics simulations. Chem Sci. 2015;6:5040–5047. doi: 10.1039/c5sc01463h. [DOI] [PMC free article] [PubMed] [Google Scholar]